Abstract
Members of the Snail family of transcription factors have been shown to induce epithelial-mesenchymal transition (EMT), a fundamental mechanism of embryogenesis and progressive disease. Here, we show that Snail and Slug promote formation of β-catenin–T-cell factor (TCF)-4 transcription complexes that bind to the promoter of the TGF-β3 gene to increase its transcription. Subsequent transforming growth factor (TGF)-β3 signaling increases LEF-1 gene expression causing formation of β-catenin–lymphoid enhancer factor (LEF)-1 complexes that initiate EMT. TGF-β1 or TGF-β2 stimulates this signaling mechanism by up-regulating synthesis of Snail and Slug. TGF-β1- and TGF-β2-induced EMT were found to be TGF-β3 dependent, establishing essential roles for multiple TGF-β isoforms. Finally, we determined that β-catenin–LEF-1 complexes can promote EMT without upstream signaling pathways. These findings provide evidence for a unified signaling mechanism driven by convergence of multiple TGF-β and TCF signaling molecules that confers loss of cell–cell adhesion and acquisition of the mesenchymal phenotype.
INTRODUCTION
Epithelial-mesenchymal transition (EMT) is an essential mechanism that guides proper development during several phases of embryogenesis (Hay, 1995). If this mechanism is stimulated within the adult organism, it promotes pathological conditions such as organ fibrosis (Kalluri and Neilson, 2003) or tumor metastasis (Thiery, 2002). This transition to an invasive phenotype is characterized by loss of cell adhesion and apical-basal polarity, followed by a shift in cytoskeletal dynamics toward front end-back end polarity and cell migration (Nawshad et al., 2005).
The most common biochemical change associated with EMT is the loss of E-cadherin expression. E-cadherin transcriptional repressors such as Snail (SNAI1), Slug (SNAI2), ZEB-1, SIP-1, E12/E47 (Peinado et al., 2004), and Twist (Yang et al., 2004) have traditionally been implicated in promoting EMT in various systems of embryonic development and tumor progression. Other common initiators of EMT include members of the transforming growth factor (TGF)-β family. In particular, TGF-β1 has been found to be a potent initiator of EMT in cancer cells stimulating cell invasion and metastasis (Akhurst and Derynck, 2001) and also in inducing EMT associated with renal fibrosis (Zeisberg and Kalluri, 2004). TGF-β2 has been shown to induce endothelial-mesenchymal transition essential for cardiac development (Camenisch et al., 2002). TGF-β3 is necessary to stimulate EMT during craniofacial development, particularly in the medial-edge epithelial cells of the palate (Nawshad and Hay, 2003; Nawshad et al., 2007). These TGF-β ligands bind to a heterodimeric complex of receptors composed of TβRII (ligand binding receptor) and TβRI (signaling receptor), which promotes phosphorylation of R-Smad proteins (Smad2 or 3) and their subsequent binding with the co-Smad, Smad4. This Smad complex then shuttles to the nucleus where it aids in transcription of target genes (Shi and Massague, 2003; Massague, 2004; Ten Dijke and Hill, 2004).
Another molecule that has been implicated in E-cadherin repression is lymphoid enhancer factor (LEF)-1, a transcription factor typically associated with Wnt signaling (Jamora et al., 2003). Wnt, which has been found to be necessary for stimulating EMT during neural crest formation (Garcia-Castro and Bronner-Fraser, 1999), promotes stabilization of cytoplasmic β-catenin. This occurs through the actions of the protein Dishevelled by its phosphorylation of glycogen synthase kinase (GSK)-3β, which dissociates a ubiquitin complex (that degrades cytoplasmic β-catenin) composed of GSK-3β, Axin, and adenomatosis polyposis coli (APC). This inhibition causes β-catenin to bind with members of the T-cell factor (TCF) family of transcription factors (including LEF-1), which can enter the nucleus to help promote or suppress transcription of target genes (Jamora et al., 2003; Waterman, 2004).
Cross-talk between TGF-β and Wnt signaling pathways has recently been confirmed, demonstrating that LEF-1 can be functionally activated by binding with either β-catenin or Smad proteins (Labbe et al., 2000; Nishita et al., 2000). During palatogenesis, TGF-β3 stimulates EMT in the medial-edge epithelium by promoting increased gene expression of LEF-1 (Nawshad and Hay, 2003; LaGamba et al., 2005; Nawshad et al., 2007). Furthermore, TGF-β1 has recently been found to promote β-catenin–LEF-1 signaling and EMT in Madin-Darby canine kidney (MDCK) cells (Medici et al., 2006).
Snail family transcription factors are well known to stimulate EMT, but it has been difficult to establish where to place Snail within the grander scheme of classical EMT signaling pathways. Previous studies have been unable to demonstrate a β-catenin–LEF-1-induced up-regulation of Snail or Slug (Kim et al., 2002; Peinado et al., 2003). We hypothesized that the inverse is more likely: that E-cadherin repressors such as Snail or Slug (by reducing levels of E-cadherin, the membrane substrate for β-catenin) may induce formation of β-catenin–LEF-1 complexes that initiate EMT. In the study described here, we provide novel evidence that expression of Snail or Slug in DLD1 colon carcinoma, A375 melanoma, and MDCKII cells promotes formation of β-catenin–LEF-1 transcription complexes through a β-catenin–TCF-4–dependent up-regulation of TGF-β3 expression and signaling. We also demonstrate that TGF-β3 is an essential mediator of TGF-β1- or TGF-β2-induced EMT through its unique ability to promote synthesis of LEF-1. Because TGF-β1 and TGF-β2 can promote transcription of Snail or Slug, we establish that these transcriptional repressors are functional downstream of TGF-β1 or TGF-β2 and upstream of TGF-β3 and β-catenin–LEF-1 complexes in the EMT signaling cascade. Finally, we show that activated LEF-1 is the essential molecule for EMT, because it can induce this transition independently of its upstream signaling pathways.
MATERIALS AND METHODS
Cell Culture
DLD1 colon carcinoma, MDCKII, and A375 melanoma cells were acquired from the American Tissue Culture Collection (Manassas, VA). Cells were grown in culture with RPMI 1640 medium for DLD1 and A375 cells or DMEM (Invitrogen, Carlsbad, CA) for MDCKII cells +10% fetal bovine serum +1% penicillin/streptomycin. Fetal bovine serum was removed for all experimental conditions. Recombinant TGF-β1, TGF-β2, or TGF-β3 (R&D Systems, Minneapolis, MN) was added to the culture medium at a concentration of 10 ng/ml for all relative experiments. Snail (provided by Dr. Angela Nieto, Cajal Institute, Madrid, Spain), Slug (provided by Dr. Tom Jessell, Columbia University, New York, NY), and pcDNA3-E-cadherin (provided by Dr. Barry Gumbiner, University of Virginia, Charlottesville, VA) expression plasmids (500 ng) were transfected into cells using Lipofectamine and Plus reagents (Invitrogen) according to the manufacturer's guidelines. Dominant-negative (DN) Smad4 (provided by Dr. Diane Simeone, University of Michigan, Ann Arbor, MI) and DN LEF-1 (provided by Dr. Marian Waterman, University of California, Irvine, CA) adenoviral constructs, which produce proteins that lack the ability to bind DNA, were added at dilutions of 1:100 as described previously (Nawshad and Hay, 2003). Phosphoinositide-3-kinase (PI3K) inhibitor LY294002 (at a dilution of 1:50) and mitogen-activated protein kinase kinase (MEK)1/2 inhibitor U0126 (at a dilution of 1:20) (Cell Signaling Technology, Danvers, MA) were added for 1 h before treatment with TGF-β1, TGF-β2, or TGF-β3. TGF-β1/-β2/-β3 neutralizing antibody and TGF-β3 neutralizing antibody (R&D Systems), which does not cross-react with TGF-β1 or TGF-β2, were used according to the manufacturer's guidelines. Antisense oligodeoxynucleotides (Integrated DNA Technologies, Coralville, IA) were used at a concentration of 4.0 μg/ml and were synthesized using the following sequences: TCF-1, 5′-GAGTAGACGGTCTCTTTGTA-3′; LEF-1, 5′-CCTCCTCCGGAGAGTTGGGG-3′; TCF-3, 5′-CCCCCGGCGGCGAGCTGGGG-3′; TCF-4, 5′-CCACCGCCGTTCAGCTGCGG-3′; β-catenin, 5′-GTGGTCCACAGAACTTCTC-3′; and negative control, 5′-TTCCTCTCTTTTCTCTCCCT-3′.
RNA Interference (RNAi)
Small interfering RNA (siRNA) gene expression knockdown studies were performed using the TriFECTa RNAi kit (Integrated DNA Technologies) and corresponding protocol. Each 27mer RNAi duplex was transfected into cells using X-tremeGene siRNA transfection reagent (Roche Diagnostics, Indianapolis, IN) following the manufacturer's guidelines. siRNA was synthesized (Integrated DNA Technologies) using the following sequences: Snail, 5′-CCACAGAAAUGGCCAUGGGAAGGCCUC-3′; Slug, 5′-UCCGAAUAUGCAUCUUCAGGGCGCCCA-3′; LEF-1, 5′-CCGGGAUUUGCGCGCGGAGAACGCCGG-3′; and negative control, 5′-UCACAAGGGAGAGAAAGAGAGGAAGGA-3′.
Immunocytochemistry, Immunoprecipitation, and Immunoblotting
Immunofluorescence, immunoprecipitation, and Western blotting were performed using the following antibodies at concentrations (and using protocols) recommended by the respective manufacturers: TGF-β1, TGF-β2, TGF-β3 (R&D Systems), Smad4, TCF-1, LEF-1, TCF-3, TCF-4, Snail (Santa Cruz Biotechnology, Santa Cruz, CA), Slug (provided by Dr. Tom Jessell, Columbia University), β-catenin, vimentin, APC (Sigma-Aldrich, St. Louis, MO), P-Smad2/3, P-GSK-3β (Cell Signaling Technology), GSK-3β, fibronectin (BD Biosciences Transduction Laboratories, Lexington, KY), α-tubulin (Calbiochem, San Diego, CA). Horseradish peroxidase-conjugated secondary antibodies (Millipore Bioscience Research Reagents, Temecula, CA) were used at a dilution of 1:5000. Fluorescein- and rhodamine-conjugated secondary antibodies (Pierce Chemical, Rockford, IL) were used at a dilution of 1:250. Images were acquired using a Nikon 80i fluorescence microscope. Adjustments of image size, brightness, and contrast were made using Adobe Photoshop CS (Adobe Systems, Mountain View, CA).
Real-Time Quantitative Polymerase Chain Reaction (PCR)
RNA extractions were performed using the RNeasy Mini kit (QIAGEN, Valencia, CA) and protocol. RNA samples were submitted to a core facility (Biopolymers Facility, Department of Genetics, Harvard Medical School, Boston, MA) where real-time PCR experiments were conducted using the SYBER Green PCR system (Applied Biosystems, Foster City, CA) on an ABI 7500 cycler, with 40 cycles per sample. Cycling temperatures were as follows: denaturing, 95°C; annealing, 60°C; and extension, 70°C. The following primers were used: Snail, forward 5′-ACCACTATGCCGCGCTCTT-3′ and reverse, 5′-GGTCGTAGGGCTGCTGGAA-3′; Slug, forward 5′-TGTTGCAGTGAGGGCAAGAA-3′ and reverse 5′-GACCCTGGTTGCTTCAAGGA-3′; TGF-β3, forward 5′-AAGTGGGTCCATGAACCTAA-3′ and reverse 5′-GCTACATTTACAAGACTTCAC-3′; LEF-1, forward 5′-CCGAAGAGGAAGGCGATTTAGC-3′ and reverse 5′-GGTCCCTTGTTGTAGAGGCC-3′; Vimentin, forward 5′-TCTACGAGGAGGAGATGCGG-3′ and reverse 5′-GGTCAAGACGTGCCAGAGAC-3′; fibronectin, forward 5′-CCCACCGTCTCAACATGCTTAG-3′; reverse 5′-CTCGGCTTCCTCCATAACAAGTAC-3′; α-smooth muscle actin (SMA), forward 5′-CAATGGCTCTGGGCTCTGTAAG-3′ and reverse 5′-TGTTCTATCGGGTACTTCAGGGTC-3′; cyclin D1, forward 5′-ATGCCAACCTCCTCAACGAC-3′ and reverse, 5′-GGCTCTTTTTCACGGGCTCC-3′; α-actinin 1, forward 5′-GAAGAAATCCAGACCCTAGCACG-3′ and reverse 5′-GAGATGACCTCCAGCAGCAG-3′; E-cadherin, forward 5′-GTCAGTTCAGACTCCAGCCC-3′ and reverse 5′-AAATTCACTCTGCCCAGGACG-3′; Keratin 7, forward 5′-TCACCATTAACCAGAGCCTGC-3′ and reverse 5′-GGGCCTCAAAGATGTCTGGG-3′; and glyceraldehyde-3-phosphate dehydrogenase, forward 5′-ACCACAGTCCATGCCATCAC-3′ and reverse: 5′-TCCACCCTGTTGCTGTA-3′.
Chromatin Immunoprecipitation (ChIP)
The ChIP assay was performed using the ChIP-IT kit (Active Motif, Carlsbad, CA) and protocol. PCR analysis was performed on DNA isolated by ChIP using a PTC-100 thermal cycler (MJ Research, South San Francisco, CA), with 35 cycles per sample. Cycling temperatures were as follows: denaturing 94°C; annealing, 58°C; and extension, 70°C. The following primers were used to isolate the TCF binding region within the TGF-β3 gene promoter: forward, 5′-CCGAGGTGCTGGTGACCCTG-3′ and reverse, 5′-CCAGTGAGTAGGTGGGGAGA-3′. Samples were run on 1.5% agarose gels containing ethidium bromide (1.5 μl) and visualized with a ChemiDoc XRS Imager (Bio-Rad, Hercules, CA).
Plasmid Construction and Site-directed Mutagenesis
A recombinant plasmid was made by inserting the −1-kb fragment of the human TGF-β3 gene promoter into the pGL3 luciferase plasmid vector (Promega, Madison, Wi). The insert was amplified by PCR using a human genomic DNA template with addition of KpnI and HindIII restriction sites to primers matching those found in the pGL3 vector. The following primers (Integrated DNA Technologies) were used: forward, 5′-TTGGTACCCCAAGGGAATGAGCGAGAGA3′ and reverse, 5′-CCCAAGCTTGTGTGAGCTGGGAAGAGAGG-3′. Restriction digest with KpnI and HindIII enzymes (New England Biolabs, Ispwich, MA) was performed for both the insert and vector, followed by ligation with T4 DNA ligase (New England Biolabs). Two base-pair mutagenesis PCR within the TCF binding region was performed using the following primer sequences: forward, 5′-GAGGCAGCATGAACGACGTCATTTAGAAAG-3′ and reverse, 5′-CCTTCTAAATGACGTCGTTCACGCTGCCTC-3′. Mutagenesis caused formation of a unique AatII restriction site within the recombinant plasmid. The sequence changes for the −704 bp to −688 bp TCF binding site were as follows: wild type, 5′-GCGTGAACAAAGTCATT-3′; and mutant, 5′-GCGTGAACGACGTCATT-3′. Mutagenesis was confirmed by restriction digest with AatII and HindIII enzymes (New England Biolabs).
Luciferase Reporter Gene Assays
Luciferase reporter gene assays were conducted using the Luciferase Assay System (Promega) and its corresponding protocol. All plasmids (500 ng) were transfected into cells using Lipofectamine and Plus reagents (Invitrogen) according to the manufacturer's guidelines. Light units were measured with a Luminometer TD-20/20 (Turner Designs, Sunnyvale, CA). Assays were normalized for transfection efficiency by cotransfecting cells with a β-gal control plasmid and were detected with the Luminescent β-gal control assay kit (Clontech, Mountain View, CA). Experimental (Luciferase) results were divided by the β-gal results to provide normalized data. The pTOPFLASH-Lux reporter construct was provided by Dr. Hans Clevers (Netherlands Institute for Developmental Biology, Utrecht, The Netherlands). The p3TP-Lux reporter plasmid was provided by Dr. Joan Massague (Memorial Sloan-Kettering Cancer Center, New York, NY).
Invasion/Migration Assays
Invasion and migration were assessed using the Innocyte Cell Migration Assay kit (EMD Biosciences, San Diego, CA). Transwell plates (96-well) containing 8-μm pores were coated with 250 ng/ml type I collagen (BD Biosciences, San Jose, CA) and 100 ng/ml Laminin (Invitrogen). Cells migrated toward 10% serum into the lower chambers. Migrated cells in the lower chamber were stained with a Calcein-acetoxymethyl ester fluorescent dye. Excitation max (485 nm)/emission max (520 nm) was assessed using a standard fluorescent plate reader (BD FACSArray bioanalyzer; BD Biosciences).
RESULTS
Snail and Slug Promote Activated β-Catenin–LEF-1 Transcription Complexes
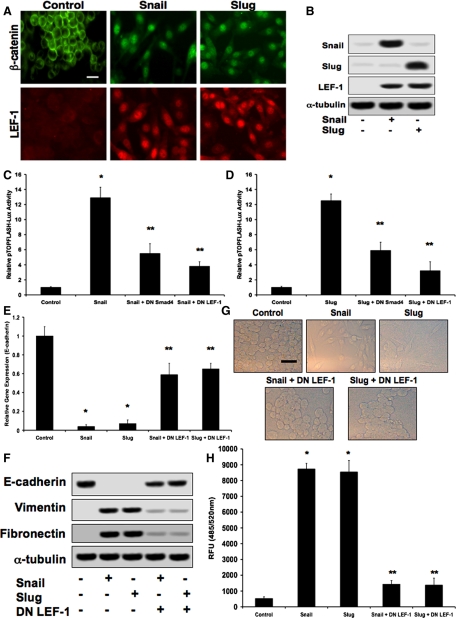
EMT typically occurs 36–72 h after initial exposure to EMT-inducing stimuli in vitro (Peinado et al., 2003; Medici et al., 2006) and in vivo (Boyer et al., 1999; Camenisch et al., 2002; Nawshad and Hay, 2003), so we made a time-dependent assessment of the intracellular signaling mechanisms that occur during this transitional period. Because E-cadherin repression by Snail or Slug should reduce the membrane substrate for β-catenin, we examined the protein localization of β-catenin and LEF-1 by immunocytochemistry. On transfection of DLD1 cells with plasmids expressing either Snail or Slug, we observed a shift of β-catenin from the cell membrane and cytoplasm to the nucleus within 48 h. Nuclear expression of LEF-1 was also observed at this time point (Figure 1A). Immunoblotting confirmed increased expression of LEF-1 after transfection with Snail or Slug plasmids (Figure 1B). Using a pTOPFLASH-Lux reporter plasmid (containing LEF-1 binding sites), we observed significant up-regulation of LEF-1 transcriptional activity in cells treated with Snail or Slug. Furthermore, treatment of cells with DN Smad4 or DN LEF-1 adenoviral constructs inhibited TOPFLASH luciferase activity (Figure 1, C and D). Real-time PCR analysis showed that levels of E-cadherin transcripts were decreased upon treatment of cells with Snail or Slug constructs. These decreases were prevented by addition of DN LEF-1 (Figure 1E). EMT was confirmed by immunoblotting for the epithelial marker E-cadherin and the mesenchymal markers vimentin and fibronectin. E-cadherin was lost upon exposure to Snail or Slug plasmids 48 h post transfection, whereas vimentin and fibronectin were greatly increased. These changes were prevented in the presence of a DN LEF-1 construct (Figure 1F). Phase-contrast imaging confirmed the change in cell morphology characteristic of EMT. These images are representative of the phenotype change throughout all relative experiments. Transwell migration assays showed that DN LEF-1 also prevented Snail- or Slug-induced increases in cell invasion/migration (Figure 1H). Similar results were observed with MDCKII and A375 cells (data not shown).
Figure 1.
Snail and Slug promote β-catenin–LEF-1 activity. (A) Immunocytochemistry demonstrating translocation of β-catenin to the nuclei of DLD1 cells transfected with either Snail or Slug expression plasmids, as well as increased expression and nuclear localization of LEF-1. Bar, 10 μm. (B) Immunoblotting confirming increased expression of Snail, Slug, and LEF-1 in response to the Snail and Slug constructs. (C and D) pTOPFLASH-Lux reporter assay assessing increased LEF-1 transcriptional activity upon cotransfection with Snail or Slug expression plasmids. Addition of DN Smad4 and DN LEF-1 adenoviral constructs significantly inhibited luciferase activity. Data represent mean±SD; n=3; *p<0.01 compared with control; **p<0.05 compared with Snail or Slug. (E) Real-time quantitative PCR analysis showing reduction of E-cadherin expression in cells exposed to Snail or Slug plasmids. These inhibitions were mostly prevented by DN LEF-1. (F) Biochemical confirmation of EMT by immunoblotting, showing decreased E-cadherin expression, as well as increased vimentin and fibronectin expression in the presence of the Snail or Slug constructs. Addition of a DN LEF-1 plasmid prevented these changes. (G) Phase contrast imaging showing the EMT phenotype in Snail- or Slug-treated cells, which is prevented by DN LEF-1. (H) Cell invasion/migration assays observing increased levels in cells transfected with Snail or Slug plasmids. Addition of DN LEF-1 prevented these increases. Data represent mean±SD; n=3; *p<0.0001 compared with control.
Snail and Slug Increase TGF-β3 Expression and Signaling
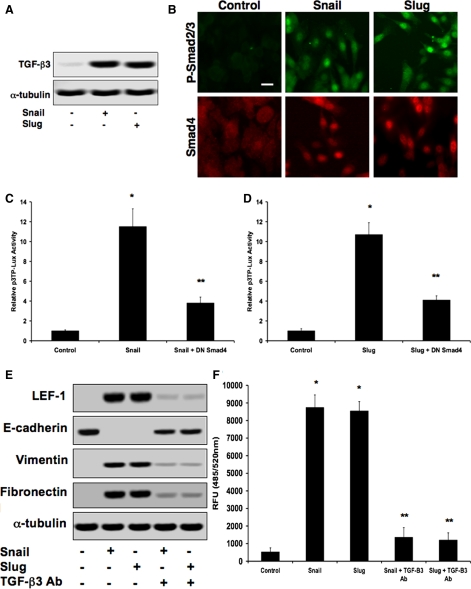
Because treatment with DN Smad4 inhibited LEF-1 activity, we examined protein levels of TGF-β3 (which has recently been linked to LEF-1 gene expression [Nawshad and Hay, 2003]) via immunoblotting. We found that protein expression of TGF-β3 dramatically increased 48 h after transfection in DLD1 cells with either Snail or Slug expression plasmids (Figure 2A). TGF-β3 signaling was confirmed in these cells by demonstrating nuclear localization of phosphorylated Smad2/3 (P-Smad2/3) and Smad4 via immunocytochemistry (Figure 2B). Smad transcriptional activity was confirmed in these cells by using a p3TP-Lux reporter plasmid (containing TGF-β response elements). The addition of a DN Smad4 construct greatly reduced luciferase activity (Figure 2, C and D). Immunoblotting showed decreased expression of E-cadherin and increased expression of LEF-1, vimentin, and fibronectin 48 h after transfection with Snail or Slug constructs. Presence of a TGF-β3 neutralizing antibody prevented these changes (Figure 2E). TGF-β3 blocking antibody also inhibited invasion and migration caused by Snail or Slug activity (Figure 2F). Similar results were found using MDCKII and A375 cells (data not shown).
Figure 2.
Snail and Slug increase TGF-β3 expression and signaling. (A) Immunoblotting showing increased levels of TGF-β3 after transfection of DLD1 cells with Snail or Slug expression plasmids. (B) Immunocytochemistry confirming nuclear localization of P-Smad2/3 and Smad4 post transfection. Bar, 10 μm. (C and D) Increased Smad transcriptional activity determined using a p3TP-Lux reporter plasmid. Addition of a DN Smad4 adenoviral construct significantly reduced luciferase activity. Data represent mean±SD; n=3; *p<0.001 compared with control; **p<0.01 compared with Snail or Slug. (E) Immunoblotting showing that a TGF-β3 neutralizing antibody (TGF-β3 Ab) prevented Snail- or Slug-induced changes in LEF-1, E-cadherin, vimentin, and fibronectin expression. (F) TGF-β3 neutralizing antibody prevented Snail- or Slug-induced invasion/migration. Data represent mean±SD; n=3; *p<0.001 compared with control.
Snail and Slug have been described as transcriptional repressors (Peinado et al., 2004), so it is not likely that they directly increase TGF-β3 expression, rather TGF-β3 expression is more likely the result of a secondary effect of Snail or Slug activity. Consistent with such an indirect effect of Snail and Slug on TGF-β3 expression are the results of promoter analyses of the human, murine and canine TGF-β3 genes (data not shown). Analyses of available genomic DNA sequences from GenBank (National Center for Biotechnology Information) were performed upstream of the TGF-β3 gene start site (−4 kb), as well as for Intron 1 using MatInspector software (Genomatix, Munich, Germany). However, there was no evidence of Snail or Slug binding regions on each promoter.
Snail and Slug Up-Regulate TGF-β3 Gene Expression through β-Catenin–TCF-4
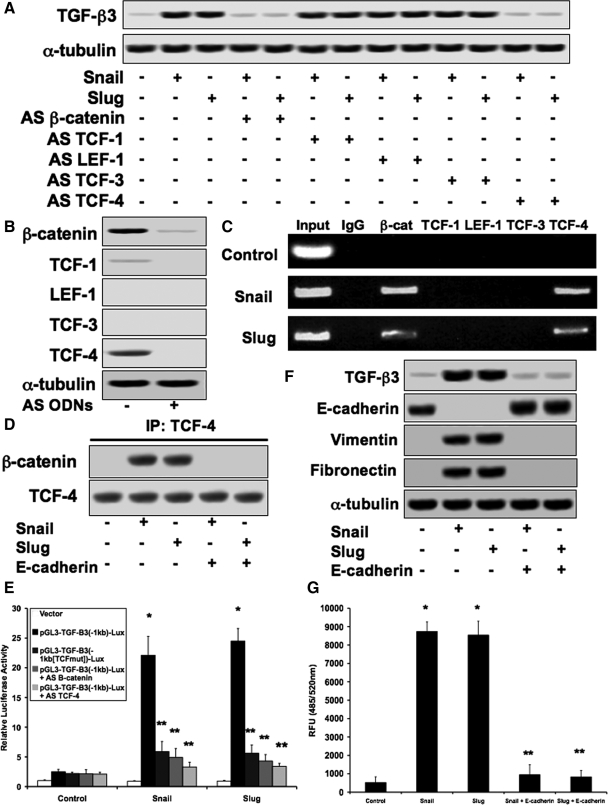
Because Snail and Slug reduce expression of E-cadherin, the membrane substrate for β-catenin (Peinado et al., 2004), we assessed the potential involvement of β-catenin in regulating expression of TGF-β3. Immunoblotting was performed to detect TGF-β3 from lysates transfected with Snail or Slug constructs in the presence of antisense (AS) oligodeoxynucleotides (ODNs) against β-catenin and all four TCF transcription factors. We found that AS β-catenin and AS TCF-4 inhibited Snail or Slug induced up-regulation of TGF-β3, whereas AS TCF-1, AS LEF-1, and AS TCF-3 had no effect (Figure 3A). Control immunoblotting confirmed expression knockdown with the AS ODNs (Figure 3B). Promoter analysis revealed a TCF binding site 700 bp upstream of the start site of the human, murine, and canine TGF-β3 gene promoters (data not shown). Chromatin immunoprecipitation was performed using antibodies against β-catenin and the TCF proteins. Immunoglobulin G was used as a negative control. PCR was conducted using primers coding for the TCF binding site within the TGF-β3 gene promoter and showed binding of β-catenin–TCF-4 to this site, whereas the other TCF proteins did not (Figure 3C). Formation of the β-catenin–TCF-4 complex by Snail or Slug transfection was observed by immunoprecipitation. Addition of a pcDNA3-E-cadherin expression plasmid prevented formation of this complex (Figure 3D). A recombinant reporter plasmid was constructed by inserting the −1-kb region of the human TGF-β3 gene promoter into a pGL3 luciferase vector. Cotransfection of this reporter with Snail of Slug expression plasmids greatly increased luciferase activity. However, luciferase activity was inhibited upon site directed mutagenesis of the TCF binding site within the reporter plasmid, or treatment with AS β-catenin or AS TCF-4 (Figure 3E). Immunoblotting showed that an E-cadherin expression plasmid was sufficient to prevent changes in expression of TGF-β3, E-cadherin, vimentin, and fibronectin caused by Snail or Slug activity (Figure 3F). The E-cadherin plasmid also prevented Snail- or Slug-induced invasion and migration (Figure 3G). Similar data were found with MDCKII and A375 cells (data not shown).
Figure 3.
Snail and Slug increase TGF-β3 expression through β-catenin–TCF-4. (A) Immunoblotting demonstrating that Snail or Slug-induced increases in TGF-β3 expression were inhibited with AS ODNs against β-catenin or TCF-4, but not TCF-1, LEF-1, or TCF-3 in DLD1 cells. (B) Control immunoblotting showing significant knockdown of expression of each target in the presence of the AS ODNs. (C) Chromatin immunoprecipitation establishing that β-catenin and TCF-4 interact with the TCF binding site on the promoter region of the TGF-β3 gene in the presence of Snail or Slug expression plasmids. (D) Immunoprecipitation confirming formation of a β-catenin–TCF-4 complex when cells were transfected with Snail or Slug constructs. Cotransfection with an E-cadherin expression plasmid prevented formation of this complex. (E) Cotransfection of cells with a recombinant reporter plasmid containing the −1-kb region of the TGF-β3 gene promoter showed increased luciferase activity in the presence of Snail or Slug expression plasmids. Site-directed mutagenesis of the TCF binding site within the promoter insert, as well as AS β-catenin or AS TCF-4 prevented these increases. Data represent mean±SD; n=3; *p<0.001 compared with control; **p<0.05 compared with pGL3-TGF-β3(−1kb)-Lux. (F) Immunoblotting showing that the E-cadherin expression plasmid prevented Snail- or Slug-induced changes in protein expression of TGF-β3, E-cadherin, vimentin, and fibronectin. (G) The E-cadherin expression construct also inhibited Snail- or Slug-induced invasion/migration. Data represent mean±SD; n=3; *p<0.0001 compared with control.
TGF-β3 Induces Formation of β-Catenin–LEF-1 Transcription Complexes
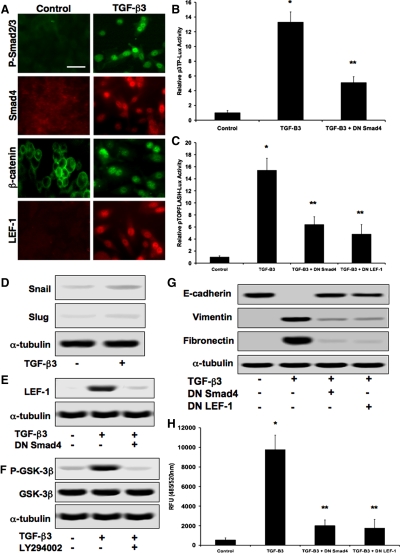
We next addressed the question of whether TGF-β3 directly stimulates synthesis of LEF-1 as well as the formation of β-catenin–LEF-1 transcription complexes. Immunocytochemistry confirmed nuclear localization of P-Smad2/3, Smad4, β-catenin, and LEF-1 in cells exposed to TGF-β3 (Figure 4A). Smad transcriptional activity in these cells was confirmed with the p3TP-Lux reporter plasmid, with decreased luciferase activity in the presence of DN Smad4 (Figure 4B). LEF-1 transcriptional activity was confirmed using the pTOPFLASH-Lux reporter construct, with low levels of luciferase activity using DN Smad4 or DN LEF-1 (Figure 4C). Immunoblotting showed little difference in expression of Snail or Slug in cells treated with exogenous TGF-β3 (Figure 4D). Control DLD1 cells seemed to lack significant levels of LEF-1 as observed by immunoblotting, but addition of exogenous TGF-β3 resulted in increased expression levels. The addition of DN Smad4 to cells treated with TGF-β3 prevented this up-regulation, suggesting that it is caused by a Smad-dependent mechanism (Figure 4E). Because the formation of β-catenin–LEF-1 complexes would normally require stabilization of cytoplasmic β-catenin, we assessed levels of GSK-3β phosphorylation that might be induced by TGF-β3. Immunoblotting with a phospho-specific GSK-3β antibody revealed that TGF-β3 did increase levels of P-GSK-3β. The same experiments were performed in the presence of a PI3K inhibitor (LY294002), which revealed an inhibition of phosphorylation, suggesting that TGF-β3 causes GSK-3β phosphorylation through PI3K signaling (Figure 4F). Immunoblotting showed that E-cadherin levels were decreased, whereas vimentin and fibronectin were increased 48 h after treatment with TGF-β3. Exposure to DN Smad4 or DN LEF-1 inhibited these changes (Figure 4G). DN Smad4 and DN LEF-1 also prevented TGF-β3–induced invasion/migration (Figure 4H). Similar results for the effects of TGF-β3 were observed using MDCKII and A375 cells (data not shown).
Figure 4.
TGF-β3 stimulates β-catenin–LEF-1 activity. (A) Addition of recombinant TGF-β3 to DLD1 cells conferred nuclear localization of p-Smad2/3, Smad4, β-catenin, and LEF-1 as observed via immunocytochemistry. Bar, 10 μm. (B) Smad transcriptional activity detected with a p3TP-Lux reporter plasmid, showing that transcription is increased by TGF-β3 and inhibited by DN Smad4. Data represent mean±SD; n=3; *p<0.0001 compared with control; **p<0.01 compared with TGF-B3. (C) pTOPFLASH-Lux confirmed LEF-1 transcriptional activity caused by TGF-β3, whereas DN Smad4 and DN LEF-1 greatly reduced luciferase activity. Data represent mean±SD; n=3; *p<0.001 compared with control; p < 0.05 compared with TGF-B3. (D) TGF-β3 had minimal effect on Snail or Slug protein expression. (E) Immunoblotting confirmed TGF-β3-induced LEF-1 synthesis, which was hindered by the addition of DN Smad4, suggesting that LEF-1 expression is Smad-dependent. (F) Immunoblotting showing that upon treatment of cells with TGF-β3, phosphorylation of GSK-3β (P-GSK-3β) was significantly increased but prevented with a PI3K inhibitor (LY294002). (G) Immunoblotting also confirmed decreases in E-cadherin and increases in vimentin and fibronectin in cells exposed to TGF-β3. These changes were inhibited upon exposure to DN Smad4 or DN LEF-1 constructs. (H) DN Smad4 and DN LEF-1 inhibited TGF-β3-induced increases in cell invasion/migration. Data represent mean±SD; n=3; *p<0.01 compared with control.
Double immunostaining and immunoprecipitation was performed to confirm one-to-one correspondence of DLD1 cells treated with Snail, Slug, or TGF-β3 and those with nuclear localization of P-Smad2/3–Smad4 or β-catenin–LEF-1 (Supplemental Figure S1). Real-time quantitative PCR was used to show time-dependent up-regulation of TGF-β3, LEF-1, vimentin, and fibronectin, as well as down-regulation of E-cadherin expression, in DLD1 cells treated with Snail, Slug, or TGF-β3 (Supplemental Figure S2).
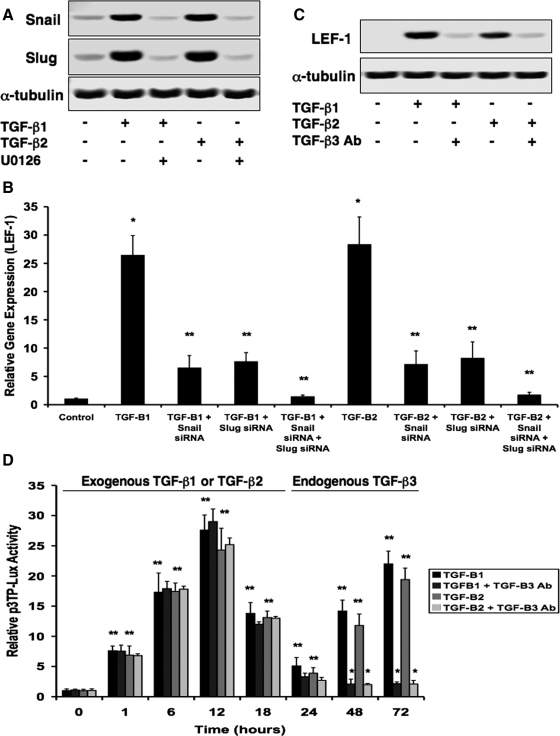
TGF-β1 or TGF-β2 Stimulates the Snail (or Slug)-TGF-β3-β-catenin–LEF-1 Signaling Cascade
The recent finding that TGF-β1 can promote up-regulation of Snail gene expression (Peinado et al., 2003) led us to question whether it could be a stimulator of the Snail (or Slug)-TGF-β3-β-catenin-LEF-1 signaling cascade. Using real-time quantitative PCR with RNA extracted from DLD1 cells treated with TGF-β1 or TGF-β2 at time points of 0 (control), 24, 48, and 72 h, we found that TGF-β1 and TGF-β2 promote increased expression of Snail and Slug, followed by TGF-β3, then LEF-1, vimentin, and fibronectin. This cascade correlated with decreased expression levels of E-cadherin (Supplemental Figure S3A). Increases in Snail and Slug protein expression were confirmed by immunoblotting. Addition of an MEK1/2 inhibitor (U0126) prevented these increases, suggesting that TGF-β1 or TGF-β2 promotes these up-regulations through mitogen-activated protein kinase (MAPK) signaling (Figure 5A). TGF-β1 or TGF-β2 increased levels of LEF-1 mRNA as assessed by real-time PCR. Snail siRNA and/or Slug siRNA prevented these up-regulations (Figure 5B). We then questioned why both TGF-β1 (or TGF-β2) and TGF-β3 isoforms were necessary for this signaling pathway, particularly because both signal through Smad proteins. Immunoblotting assays showed increased levels of LEF-1 expression induced by TGF-β1 or TGF-β2. However, when the experiments were repeated in the presence of a TGF-β3 neutralizing antibody, LEF-1 expression was significantly reduced. These results suggested that up-regulation of LEF-1 is TGF-β3-dependent (Figure 5C). We also showed via Western blotting that (much like TGF-β3) TGF-β1 and TGF-β2 caused up-regulation of P-GSK-3β, which was prevented in the presence of a PI3K inhibitor (LY294002) (Supplemental Figure S3B). Snail or Slug plasmid transfection showed no effects on TGF-β1 or TGF-β2 protein levels (Supplemental Figure S3C). Snail- and Slug-dependent TGF-β1 or TGF-β2-induced formation of β-catenin–TCF-4 complexes that bind to the TGF-β3 gene promoter were confirmed by immunoprecipitation and chromatin immunoprecipitation, respectively (Supplemental Figure S3, D and E). Finally, we established that E-cadherin protein was heavily reduced upon treatment of cells with either TGF-β1 or TGF-β2 for 48 h, whereas vimentin and fibronectin were greatly up-regulated. Addition of Snail siRNA, Slug siRNA, AS TCF-4, TGF-β3 neutralizing antibody, or DN LEF-1 prevented these changes (Supplemental Figure S3, F and G). These inhibitors also prevented TGF-β1- or TGF-β2-induced invasion/migration (Supplemental Figure S3, H and I). Similar results were found with MDCKII and A375 cells (data not shown).
Figure 5.
TGF-β1 or TGF-β2 initiates this Snail- or Slug-induced signaling cascade to promote EMT. (A) TGF-β1 and TGF-β2 caused up-regulations of Snail and Slug as observed by immunoblotting. Addition of a MEK1/2 inhibitor (U0126) prevented these up-regulations. (B) Real-time quantitative PCR showing that LEF-1 expression was increased upon treatment of cells with TGF-β1 or TGF-β2 but was prevented by the addition of Snail siRNA and/or Slug siRNA. Data represent mean±SD; n=3; *p<0.001 compared with control; **p<0.001 compared with TGF-B1 or TGF-B2. (C) Immunoblotting for LEF-1 protein expression showing that TGF-β1 or TGF-β2 causes increased levels, but in the presence of a TGF-β3 neutralizing antibody (TGF-β3 Ab), LEF-1 expression was greatly reduced. (D) Time course-dependent p3TP-Lux luciferase assay showing that TGF-β/Smad activity increased through 12 h posttreatment with either TGF-β1 or TGF-β2. Reporter activity was then reduced by 24 h followed by continuous increases through 72 h as a result of endogenous expression of TGF-β3, because a TGF-β3 neutralizing antibody (TGF-β3 Ab) prevented the second wave of transcriptional activity. Data represent mean±SD; n=3; *p<0.05 compared with 0 h; **p<0.0001 compared with TGF-B1 or TGF-B2.
We next assessed the effects of the three TGF-β isoforms on each other's expression by immunoblotting with lysates from cells treated with each isoform for 48 h. We found that TGF-β1 increased expression of TGF-β3 but not TGF-β2. TGF-β2 also increased levels of TGF-β3 but not TGF-β1. Cells treated with TGF-β3 had no effect on expression of either TGF-β1 or TGF-β2 (Supplemental Figure S3J). Using the p3TP-Lux reporter plasmid, we measured activity of TGF-β signaling in a time course-dependent manner. We found that treatment of DLD1 cells with exogenous TGF-β1 or TGF-β2 caused luciferase activity to peak at 12 h after exposure. Signaling was then lost by 24 h, but it was restored through 72 h by the observed endogenous expression of TGF-β3, as confirmed by the presence of a TGF-β3 neutralizing antibody (Figure 5D). These results were confirmed in MDCKII and A375 cells (data not shown).
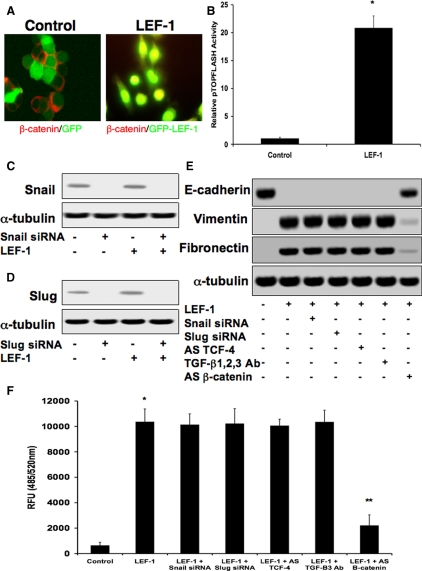
β-Catenin–LEF-1 Is Sufficient to Promote EMT without Upstream TGF-β or Snail/Slug Signaling
Because DLD1 cells already have excessive amounts of cytoplasmic β-catenin due to an APC mutation, we were able to induce EMT within 48 h after the addition of an adenoviral LEF-1 expression construct used as described previously (Kim et al., 2002) (Figure 6A). Increased LEF-1 transcriptional activity was confirmed using the pTOPFLASH-Lux reporter plasmid (Figure 6B). We found that LEF-1 had a 67.8-fold increase in gene expression upon exposure to the adenovirus by real-time PCR. Expression of genes associated with β-catenin–LEF-1-induced EMT were increased including vimentin, fibronectin, α-SMA, α-actinin 1, and cyclin D1 (positive control; Waterman, 2004), whereas expression of the epithelial markers E-cadherin and keratin 7 decreased (Supplemental Figure S4). To find out whether LEF-1 can promote EMT without its upstream signaling pathways, we assessed LEF-1 function in DLD1 cells in the presence of Snail, Slug, TCF-4, and TGF-β inhibitors. Control immunoblotting showed that Snail siRNA fully inhibited Snail expression in the presence or absence of LEF-1. LEF-1 expression did not alter Snail protein levels (Figure 6C). Similar results were found for Slug expression using Slug siRNA (Figure 6D). Expression of E-cadherin and vimentin using immunoblotting showed that LEF-1 caused full inhibition of E-cadherin, as well as massive increases in vimentin and fibronectin. LEF-1 expression in the presence of Snail siRNA, Slug siRNA, AS TCF-4, or a blocking antibody against TGF-β1, TGF-β2, and TGF-β3 did not affect LEF-1-induced EMT, whereas AS β-catenin did (Figure 6E). These inhibitors showed similar effects for LEF-1-induced invasion/migration (Figure 6F). siRNA-mediated knockdown of LEF-1 was observed by real-time quantitative PCR, showing >90% decreased expression in DLD1 cells treated with TGF-β1, TGF-β2, Snail, Slug, or TGF-β3. These decreases in LEF-1 were sufficient to prevent expression changes of E-cadherin and vimentin induced by these factors (Supplemental Figure S5).
Figure 6.
β-catenin–LEF-1 is sufficient to promote EMT without upstream TGF-β or Snail/Slug signaling. (A) Due to the APC mutation in DLD1 cells, exposure to an adenovirus containing an LEF-1 expression plasmid was sufficient to induce EMT via β-catenin–LEF-1 complexes alone, as show by immunocytochemistry. (B) LEF-1 transcriptional activity was confirmed with the pTOPFLASH-Lux reporter plasmid. Data represent mean±SD; n=3; *p<0.01 compared with control. (C) Immunoblotting showing that Snail protein expression was greatly reduced in the presence of Snail siRNA. Exogenous LEF-1 had no effect on Snail expression with or without the presence of Snail siRNA. (D) Similar results were found for Slug expression in the presence of Slug siRNA. (E) EMT was observed by assessing protein expression of E-cadherin and vimentin. Exposure to adenoviral LEF-1 caused complete inhibition E-cadherin expression, while increasing levels of vimentin and fibronectin. Presence of Snail siRNA, Slug siRNA, AS TCF-4, or a neutralizing antibody against TGF-β1, TGF-β2, and TGF-β3 (TGF-β1, 2, 3 Ab) had no effect, but AS β-catenin prevented LEF-1-induced EMT. (F) Similar results were observed for LEF-1-induced cell invasion/migration.
DISCUSSION
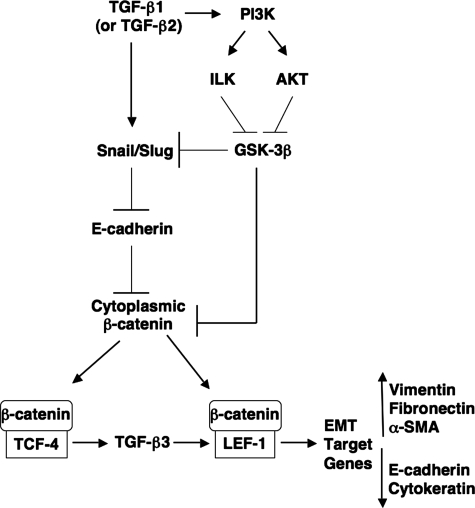
Our results provide evidence for an intricate signaling mechanism by which TGF-β1 (the most common TGF-β family member involved in EMT; Akhurst and Derynck, 2001) promotes epithelial cells to the invasive, mesenchymal phenotype. We found that TGF-β1 causes up-regulation in expression levels of the E-cadherin repressors Snail and Slug, which peak 24 h after treatment. TGF-β2, a common initiator of embryonic EMT (Behnan et al., 2005; Boyer et al., 1999), was found to do the same. Both Snail and Slug cause a β-catenin–TCF-4-induced up-regulation of TGF-β3, which in turn is responsible for increases in LEF-1 expression. Initial E-cadherin repression via Snail and Slug reduce the level of substrate for β-catenin in the cell membrane. Cytoplasmic β-catenin (as well as Snail) is stabilized by phosphorylation and functional inactivation of GSK-3β (Zhou et al., 2004). TGF-β1, TGF-β2, and TGF-β3 all signal through PI3K, which causes phosphorylation of GSK-3β through its downstream signaling molecules ILK and AKT (Hannigan et al., 2005). The surplus of β-catenin within the cytoplasm is then free to associate with the LEF-1 that was up-regulated by TGF-β3/Smad signaling. These β-catenin–LEF-1 transcription complexes then cause increased expression of target genes (vimentin, fibronectin, α-SMA) that drive formation of the mesenchymal phenotype (Figure 7).
Figure 7.
Schematic diagram of the proposed signaling mechanism that stimulates EMT. TGF-β1 (or TGF-β2) promotes the up-regulation of E-cadherin repressors Snail and Slug via MAPK signaling. Snail and Slug inhibit E-cadherin gene expression, thus limiting β-catenin substrate binding at the cell membrane. Through this mechanism, β-catenin–TCF-4 complexes up-regulate synthesis of TGF-β3. TGF-β3, in turn, through Smad signaling can up-regulate levels of LEF-1, a new substrate for β-catenin. β-catenin is further stabilized in the cytoplasm by the actions of TGF-β1, TGF-β2, and TGF-β3 to signal through PI3K. PI3K can then signal molecules such as ILK and AKT, which can phosphorylate and inactivate GSK-3β, a protein that targets both Snail and β-catenin for degradation through the ubiquitin proteosome pathway. This high amount of cytoplasmic β-catenin achieved through GSK-3β phosphorylation and E-cadherin repression will promote association with LEF-1, upon which these β-catenin–LEF-1 complexes will translocate to the nucleus to promote transcription of EMT target genes (vimentin, fibronectin).
The most significant finding in this study is that Snail and Slug activity promotes TGF-β3 signaling. Because Snail and Slug are known to be transcriptional repressors (Peinado et al., 2004), and because we found no evidence of Snail or Slug binding sites on the TGF-β3 gene promoter, this increase in TGF-β3 expression occurs as a secondary effect of Snail or Slug activity. The initial reduction of E-cadherin expression reduces the membrane substrate for β-catenin, which then associates with TCF-4, thus forming a complex that interacts with the TCF binding site on the TGF-β3 gene promoter to enhance its transcription. This allows TGF-β3 to up-regulate LEF-1, another TCF factor activated by β-catenin, that promotes EMT. This establishes an essential role for multiple TCF proteins in this EMT signaling mechanism. TCF-1, TCF-3, and TCF-4 were all significantly expressed in two of the three cell lines tested, and AS knockdown only had effects on TGF-β3 expression for TCF-4. Although they are not expressed at the same levels, we would expect to see some effect if indeed TCF-1 or TCF-3 were important in regulation of TGF-β3 expression. LEF-1 was not expressed in any of the cell types under control conditions, but it was expressed after TGF-β3 was functional. Furthermore, when we added adenoviral LEF-1 to DLD1 cells, we detected no significant changes in TGF-β3 expression (data not shown). Therefore, the binding site seems to be specific for TCF-4.
Another interesting aspect of the mechanism we describe here is that two forms of TGF-β (TGF-β1 [or TGF-β2] and TGF-β3) seem to be necessary for the full cascade to function. TGF-β1 and TGF-β2 cause increased expression of the E-cadherin repressors Snail and Slug, which will decrease levels of β-catenin localized at the cell membrane. All three TGF-β isoforms can signal through PI3K to promote stabilization of cytoplasmic β-catenin by ILK- or AKT-dependent phosphorylation of GSK-3β (Hannigan et al., 2005). The unique function of TGF-β3 seems to be the promotion of increased LEF-1 expression, which can subsequently bind with β-catenin to form transcription complexes that induce EMT. How exactly these three ligands differ in downstream signaling potential remains to be defined, as they all signal through Smad proteins. Whereas they all have the potential to use both Smad2 and Smad3, recent evidence has shown that TGF-β3 primarily uses Smad2 as its R-Smad protein during EMT (Nawshad and Hay, 2003; Shiomi et al., 2006). Therefore, it is tempting to hypothesize that TGF-β1 and TGF-β2 primarily use Smad3 as their R-Smad during EMT. What is known is that LEF-1 up-regulation seems to be Smad dependent, whereas increases in Snail seems to be driven by Smad-independent (MAPK) signaling.
These findings complement our previous studies of TGF-β1-induced EMT. We recently reported that TGF-β1 signaling promotes EMT through cooperative activities of Smad-dependent and Smad-independent signaling pathways (Medici et al., 2006). We found that signaling was active 2–3 d after treatment with TGF-β1, whereas these signals would typically be lost over a period of several hours rather than days. We now find that the continuous TGF-β signaling observed at these later time points is a result of endogenous TGF-β3 expression, rather than the initial treatment with TGF-β1. Furthermore, the ability of TGF-β1-induced Snail expression to promote TGF-β3 signaling (which is essential for LEF-1 expression) provides significantly new insights into the mechanisms of EMT signaling.
These data regarding sequential expression of individual TGF-β isoforms are supported by in vivo studies of embryonic EMT. Both TGF-β2 and TGF-β3 have been implicated in EMT during endocardial cushion development (Romano and Runyan, 2000; Camenisch et al., 2002). Studies have shown that TGF-β2 is highly expressed in the endocardium 9.5–12.5 d post conception (dpc) and is eventually marked by overlap with TGF-β3, which is heavily expressed from 14.5 dpc (Molin et al., 2003). Also, regarding EMT in palate development, TGF-β2 is expressed in palatal shelves before fusion and TGF-β3 has been proved to be the key mediator of EMT in palate medial-edge epithelial cells (Nawshad et al., 2007). Furthermore, both TGF-β2 and TGF-β3 knockout mice demonstrate a cleft palate phenotype (Kaartinen et al., 1995; Sanford et al., 1997). Elevated levels of Snail have been found in palates of TGF-β3 knockout mice (Martinez-Alvarez et al., 2004). Because we found that Snail up-regulates TGF-β3, these elevated levels of Snail by palate cells may represent an attempt to compensate for the lack of TGF-β3 expression. Although TGF-β2 seems to promote most embryonic EMT, pathological forms of EMT such as tumor metastasis and organ fibrosis are more commonly driven by TGF-β1 (Thiery, 2003). Our results suggest that cooperation between TGF-β1 and TGF-β3 may stimulate EMT in these pathological conditions.
The signal transduction pathways that govern EMT are extensive, with cross-talk mechanisms forming a complex web of signaling. Here, we provide the foundation for a unified mechanism that promotes EMT. We show that the transcriptional repressor proteins Snail and Slug can promote TGF-β signal transduction by up-regulating synthesis of TGF-β3 through the transcription factor TCF-4. We also provided evidence of how TGF-β3 can be integrated with other pathways (including β-catenin–LEF-1) that induce EMT, establishing essential and unique roles for multiple TGF-β isoforms in stimulating this change in cell morphology. Furthermore, by using an APC-null cancer cell line (DLD1), we were able to demonstrate that active LEF-1 can promote EMT without Snail, Slug, TCF-4, or any of the TGF-β isoforms. This role of LEF-1 may provide a basis for developing new anti-metastasis therapeutics for carcinomas and may help the understanding of mechanisms of EMT in other systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Rector (Harvard Medical School) for assistance with real-time quantitative PCR. We also thank N. Fukai (Harvard School of Dental Medicine) for technical assistance. This work was supported by grants R01-DE11142 (to E.D.H.) and P01-AR048564 (to B.R.O.) from the National Institutes of Health.
Abbreviations used:
- APC
adenomatosis polyposis coli
- AS
antisense
- DN
dominant negative
- EMT
epithelial-mesenchymal transition
- GSK
glycogen synthase kinase
- LEF
lymphoid enhancer factor
- MAPK
mitogen-activated protein kinase
- MDCK
Madin-Darby canine kidney
- ODN
oligodeoxynucleotide
- PI3K
phosphoinositide 3-kinase
- TCF
T-cell factor
- TGF
transforming growth factor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0506) on September 17, 2008.
REFERENCES
- Akhurst R. J., Derynck R. TGF-beta signaling in cancer–a double-edged sword. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- Behnan S. M., Guo C., Gong T. W., Shum L., Gong S. G. Gene and protein expression of transforming growth factor beta 2 gene during murine primary palatogenesis. Differentiation. 2005;73:233–239. doi: 10.1111/j.1432-0436.2005.00022.x. [DOI] [PubMed] [Google Scholar]
- Boyer A. S., Ayerinskas I. I., Vincent E. B., McKinney L. A., Weeks D. L., Runyan R. B. TGFbeta2 and TGFbeta3 have separate and sequential activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev. Biol. 1999;208:530–545. doi: 10.1006/dbio.1999.9211. [DOI] [PubMed] [Google Scholar]
- Camenisch T. D., Molin D. G., Person A., Runyan R. B., Gittenberger-de Groot A. C., McDonald J. A., Klewer S. E. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev. Biol. 2002;248:170–181. doi: 10.1006/dbio.2002.0731. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro M, Bronner-Fraser M. Induction and differentiation of the neural crest. Curr. Opin. Cell Biol. 1999;11:695–698. doi: 10.1016/s0955-0674(99)00038-1. [DOI] [PubMed] [Google Scholar]
- Hay E. D. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- Hannigan G., Troussard A. A., Dedhar S. Integrin-linked kinase: a cancer therapeutic target unique among its ILK. Nat. Rev. Cancer. 2005;5:51–63. doi: 10.1038/nrc1524. [DOI] [PubMed] [Google Scholar]
- Jamora C., DasGupta R., Kocieniewski P., Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen V., Voncken J. W., Shuler C., Warburton D., Bu D., Heisterkamp N., Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- Kalluri R., Neilson E. G. Epithelial-mesenchymal transition and its implication for fibrosis. J. Clin. Invest. 2003;11:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Lu Z., Hay E. D. Direct evidence for a role of beta-catenin/LEF-1 signalling pathway in the induction of EMT. Cell Biol. Int. 2002;26:463–476. doi: 10.1006/cbir.2002.0901. [DOI] [PubMed] [Google Scholar]
- Labbe E., Letamendia A., Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc. Natl. Acad. Sci. USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGamba D., Nawshad A., Hay E. D. Microarray analysis of gene expression during epithelial-mesenchymal transformation. Dev. Dyn. 2005;234:132–142. doi: 10.1002/dvdy.20489. [DOI] [PubMed] [Google Scholar]
- Martinez-Alvarez C., Blanco M. J., Perez R., Rabadan M. A., Aparicio M., Resel E., Martinez T., Nieto M. A. Snail family members and cell survival in physiological and pathological cleft palates. Dev. Biol. 2004;265:207–218. doi: 10.1016/j.ydbio.2003.09.022. [DOI] [PubMed] [Google Scholar]
- Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- Medici D., Hay E. D., Goodenough D. A. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol. Biol. Cell. 2006;17:1871–1879. doi: 10.1091/mbc.E05-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin D. G., Bartram U., Van der Heiden K., Van Iperen L., Speer C. P., Hierck B. P., Poelmann R. E., Gittenberger-de-Groot A. C. Expression patterns of Tgfbeta1-3 associate with myocardialisation of the outflow tract and the development of the epicardium and the fibrous heart skeleton. Dev. Dyn. 2003;227:431–444. doi: 10.1002/dvdy.10314. [DOI] [PubMed] [Google Scholar]
- Nawshad A., Hay E. D. TGFβ3 signaling activates transcription of the LEF1 gene to induce epithelial-mesenchymal transformation during mouse palate development. J. Cell Biol. 2003;163:1291–1301. doi: 10.1083/jcb.200306024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawshad A., LaGamba D., Polad A., Hay E. D. Transforming growth factor beta signaling during epithelial-mesenchymal transformation: implications for embryogenesis and tumor metastasis. Cells Tiss. Org. 2005;179:11–23. doi: 10.1159/000084505. [DOI] [PubMed] [Google Scholar]
- Nawshad A., Medici D., Liu C. C., Hay E. D. TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF-1 transcription complex. J. Cell Sci. 2007;120:1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita M., Hashimoto M. K., Ogata S., Laurent M. N., Ueno N., Shibuya H., Cho K. W. Interaction between Wnt and TGF-beta signaling pathways during formation of Spemann's organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- Peinado H., Quintanilla M., Cano A. Transforming growth factor beta-1 induces Snail transcription factor in epithelial cell lines: implications for epithelial mesenchymal transitions. J. Biol. Chem. 2003;23:21113–21123. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- Peinado H., Portillo F., Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 2004;48:365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- Romano L. A., Runyan R. B. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev. Biol. 2000;223:91–102. doi: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- Sanford L. P., Ormsby I., Gittenberger-de Groot A. C., Sariola H., Friedman R., Boivin G. P., Cardell E. L., Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shiomi N., Cui X. M., Yamamoto T., Saito T., Shuler C. F. Inhibition of SMAD2 expression prevents murine palatal fusion. Dev. Dyn. 2006;235:1785–1793. doi: 10.1002/dvdy.20819. [DOI] [PubMed] [Google Scholar]
- Ten Dijke P., Hill C. S. New insights into TGF-beta-Smad signaling. Trends Biochem. Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Thiery J. P. Epithelial-mesenchymal transitions in tumor progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thiery J. P. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Waterman M. L. Lymphoid enhancer factor/T cell factor expression in colorectal cancer. Cancer Metastasis Rev. 2004;23:41–52. doi: 10.1023/a:1025858928620. [DOI] [PubMed] [Google Scholar]
- Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R. A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Zeisberg M., Kalluri R. The role of epithelial-to-mesenchymal transition in renal fibrosis. J. Mol. Med. 2004;82:175–181. doi: 10.1007/s00109-003-0517-9. [DOI] [PubMed] [Google Scholar]
- Zhou B. P., Deng J., Xia W., Xu J., Li Y. M., Gunduz M., Hung M. C. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.