Abstract
The tricellular tight junction (tTJ) forms at the convergence of bicellular tight junctions (bTJs) where three epithelial cells meet in polarized epithelia, and it is required for the maintenance of the transepithelial barrier. Tricellulin is a four transmembrane domain protein recently identified as the first marker of tTJ, but little is known about how tricellulin is localized at tTJs. As for the molecular mechanism of association of tricellulin with tight junctions (TJs), we found that tricellulin was incorporated into claudin-based TJs independently of binding to zona occludens-1. Unexpectedly, exogenous expression of tricellulin increased cross-links of TJ strands in the plasma membrane. As for the molecular mechanisms for localization of tricellulin at tricellular junctions, we found that knockdown of occludin caused mislocalization of tricellulin to bTJs, implying that occludin supports tricellular localization of tricellulin by excluding tricellulin from bTJs.
INTRODUCTION
Among junctional complexes, tight junctions (TJs) are specialized junctions for the prevention of the leakage of solutes through the paracellular pathway of epithelial cells (Tsukita et al., 2001). Ultrathin section electron microscopy has demonstrated that TJs consist of discrete sites of apparent fusion involving the outer leaflet of the plasma membranes of adjacent cells (Farquhar and Palade, 1963). In freeze-fracture electron microscopy, TJs occur as a set of continuous, anastomosing intramembranous particle strands (TJ strands) (Staehelin et al., 1969). Over the past 10 years, a variety of molecular components of TJs have been identified (Matter and Balda, 2003; Furuse and Tsukita, 2006). Among them, occludin was the first integral membrane protein of TJs to be identified. Occludin contains four transmembrane domains (Furuse et al., 1993). Using occludin as a probe, another integral membrane component, claudin, was identified (Furuse et al., 1998a). Claudins also bear four transmembrane domains, but they do not show any sequence similarity to occludin. Claudins comprise a family of up to 24 members, and the combination of claudin subtypes determines the tightness and functional specificity of TJs (Turksen and Troy, 2004; Furuse and Tsukita, 2006).
More precisely, TJs can be divided into two groups: bicellular TJs (bTJs), which are formed between two adjacent cells; and tricellular TJs (tTJs), which are formed where three cells meet. The structure of tTJs has been examined extensively by freeze-fracture replica electron microscopy (Friend and Gilula, 1972; Staehelin, 1973; Wade and Karnovsky, 1974; Walker et al., 1985). As TJ strands approach a tricellular contact region, their network extends basolaterally and TJ strands orient vertically. These vertically oriented TJs form central tubes between the cells, and these tubes could be a weak point for the epithelial barrier (Staehelin, 1973; Ikenouchi et al., 2005). tTJs are thus specialized structures; however, the importance of tTJs has been largely underestimated.
TJs completely disappear during the epithelial-mesenchymal transition (Hay, 1995). Recently, Snail, a zinc-finger type transcription factor, was shown to play a central role in the epithelial-mesenchymal transition (Nieto, 2002). In our previous study, we showed that the transcription of claudins and occludin was abolished by direct binding of Snail to their promoter regions (Ikenouchi et al., 2003). Therefore, we hypothesized that investigation of proteins whose expression is down-regulated by Snail might be a good tool for identification of further, as yet unidentified, integral membrane proteins of bTJs/tTJs. We compared the expression profiles of parental epithelial cells and epithelial cells transformed by exogenous expression of Snail by using Gene chip analysis, and we thereby identified several tetraspan proteins as putative targets of Snail in addition to claudins and occludin. Interestingly, one of them was uniquely concentrated at tricellular contacts of epithelial cells, and we named this protein tricellulin (Ikenouchi et al., 2005).
In our previous report, we showed that tricellulin mainly localized at tTJs in epithelial cells and that the loss of tricellulin affected the organization of tTJ and the barrier function of epithelial cells. Moreover, the staining of occludin at bTJs seemed to be thinner in tricellulin-deficient cells compared with wild-type cells. In the present study, we examined molecular mechanisms, which enable tricellulin to be incorporated into TJ strands and to be concentrated at tricellular junctions.
MATERIALS AND METHODS
Antibodies and Other Reagents
Mouse anti-zona occludens (ZO)-1 monoclonal antibody (mAb), rat anti-occludin mAb, rat anti-tricellulin mAb, and rabbit anti-tricellulin polyclonal antibody (pAb) were raised and characterized previously (Saitou et al., 1997; Ikenouchi et al., 2005). Rabbit anti-claudin-3 pAb and mouse anti-claudin-1 mAb were purchased from Zymed Laboratories (South San Francisco, CA). Mouse anti-green fluorescent protein (GFP) mAb and rat anti-hemagglutinin (HA) mAb were purchased from Roche Applied Science (Indianapolis, IN). Mouse anti-Myc mAb (9E10) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-FLAG mAb (M2) was purchased from Sigma-Aldrich (St. Louis, MO).
Expression Vectors
To construct a tricellulin expression vector with an HA epitope tag or a GFP tag at the COOH terminus, cDNA fragments encoding full-length mouse tricellulin (amino acids 1–555) were produced by polymerase chain reaction (PCR) and subcloned into the vector pCAGGS-cHA or pCAGGS-cGFP.
To generate deletion mutant constructs of tricellulin with a HA epitope tag at the COOH terminus, cDNA was amplified by PCR using tricellulin cDNA as a template with the following primers: sense primer from position 1 containing a SmaI site and antisense primer from position 1185 containing a SmaI site for HA-delC; and sense primer from position 541 containing a SmaI site and antisense primer from position 1667 containing a SmaI site for HA-delN. Each fragment amplified by PCR was directly digested with SmaI, added with a HA epitope tag at the COOH terminus and ligated into an EcoRI-digested and blunted pCAGGSneodelEcoRI vector (Niwa et al., 1991).
For production of FLAG-tagged claudin-3 lacking its COOH-terminal YV, the cDNA fragment was amplified by PCR using the sense primer from position 1 and antisense primer from 651 containing a stop codon and a FLAG epitope tag was added at the COOH terminus.
Cell Culture and Transfection
Madin-Darby canine kidney (MDCK) II cells, L cells, and MTD1A cells were grown in DMEM supplemented with 10% fetal calf serum. Stable L cell transfectants expressing claudin-1/claudin-3 were characterized previously (Itoh et al., 1999). Occludin knockdown MDCK cells were established based on the previous report by Yu et al. (2005). The DNA oligonucleotides encoding short hairpin DNA sequence targeting dog occludin were annealed, phosphorylated, and ligated to the H1 promoter vector as described previously (Ikenouchi et al., 2005). The oligonucleotides were as follows: 5′-GATCCCCGTGAAGAGTACATGGCTGCTTCAAGAGAGCAGCCATGTACTCTTCACTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGTGAAGAGTACATGGCTGCTCTCTTGAAGCAGCCATGTACTCTTCACGGG-3′.
Selection with 4 μg/ml puromycin was initiated 24 h after transfection, and three independent clones were isolated. Transfection was performed using Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as described previously (Ikenouchi et al., 2007). Briefly, cells cultured on coverslips were fixed with 3% Formalin in phosphate-buffered saline (PBS) for 10 min at room temperature, treated with 0.2% Triton X-100 in PBS for 5 min, and washed with PBS. Blocking was done by incubating the fixed cells with 5% bovine serum albumin in PBS for 10 min at room temperature (RT). After the antibodies had been diluted with the blocking solution, the cells were incubated at RT for 1 h with the primary antibody and then for 30 min with the secondary antibody.
Freeze-Fracture Analysis
Cells were fixed in 2% glutaraldehyde in 0.1 M phosphate buffer (PB), pH 7.3, at 4°C, washed three times with 0.1 M PB, immersed in 30% glycerol in 0.1 M PB for 2 h, and then frozen in liquid nitrogen. Frozen samples were fractured at −100°C and platinum-shadowed unidirectionally at an angle of 45° in a freeze etching system (BAF060; BalTec, Hudson, NH). The samples were then immersed in household bleach, and replicas that floated off the samples were washed with distilled water. The replicas were picked up on Formvar-coated grids and examined with an H-7500 electron microscope (Hitachi, Tokyo, Japan) at 100 kV. The total length of TJ strands was measured based on three representative freeze fracture images by using ImageJ 1.40 picture analyzing software (http://rsbweb.nih.gov/ij/). The number of cross-points of TJ strands was counted using printed photographs. The mean length of TJ strands between cross-points was calculated based on the total length of TJ strands and the number of cross-points.
RESULTS
Tricellulin Is Incorporated into Claudin-based TJs and Affects Secondary Organization of TJ Strands
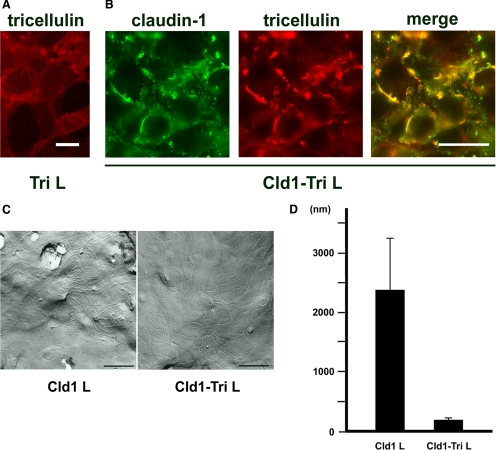
Cultured L fibroblasts are a useful system to examine the association between membrane proteins of TJs because this mouse fibroblast cell line lacks both TJs and the expression of claudins and occludin (Furuse et al., 1998b). In L cell transfectants expressing tricellulin (Tri L cells), tricellulin was not concentrated at cell–cell contacts, but it was found all over the plasma membrane, indicating that tricellulin alone did not show a homophilic interaction (Figure 1A and Supplemental Figure S1).
Figure 1.
Tricellulin is incorporated into claudin-based TJs and affects secondary organization of TJ strands. (A) Immunofluorescence staining of stable L cell transfectants expressing tricellulin (Tri L) with anti-tricellulin mAb. Bar, 10 μm. (B) Immunofluorescence staining of stable L cell transfectants expressing claudin-1 and HA-tagged tricellulin (Cld1-Tri L) with anti-claudin-1 mAb (green) and anti-HA mAb (red). Bar, 20 μm. (C) Freeze-fracture images of cell contact planes of Cld1 L cells (left) and Cld1-Tri L cells (right). Bars, 495 nm. (D) The mean length of TJ strands between cross-points were shorter in Cld1-Tri L cells (205 ± 18 nm) than in Cld1 L cells (2391 ± 865 nm), indicating that the number of cross-links of TJ strands increased in Cld1-Tri L cells.
In our previous study, we showed in situ by immunoreplica electron microscopy that tricellulin localized at TJ strands (Ikenouchi et al., 2005). Therefore, we transfected claudin-1–expressing L cells with an expression vector encoding HA-tagged tricellulin (Cld1-Tri L cells). Immunofluorescence microscopy revealed that claudin-1 was concentrated in Cld1-Tri L cells as planes at regions of cell contact, as reported previously (Furuse et al., 1998b). Interestingly, tricellulin was concentrated at cell–cell borders as planes together with claudin-1 in Cld1-Tri L cells (Figure 1B). Next, we performed freeze-fracture analyses of Cld1 L cells and Cld1-Tri L cells. Unexpectedly, TJ strands of Cld1-Tri L cells showed an increased number of cross-links compared with parental Cld1 L cells (Figure 1C and Supplemental Figure S2). The mean length of TJ strands between cross-points was shorter in Cld1-Tri L cells (205 ± 18 nm) than in Cld1 L cells (2391 ± 865 nm) (Figure 1D). This suggests that tricellulin affects two-dimensional organization of TJ strands.
Tricellulin Resides in TJ Strands Independently of Interaction with ZO-1
Next, we wondered whether binding to ZO-1 is required for the incorporation of tricellulin into claudin-based TJ strands because ZO-1 is known to link the transmembrane proteins occludin, claudins, and junction adhesion molecules to many cytoplasmic proteins and the actin cytoskeleton (Tsukita et al., 2001).
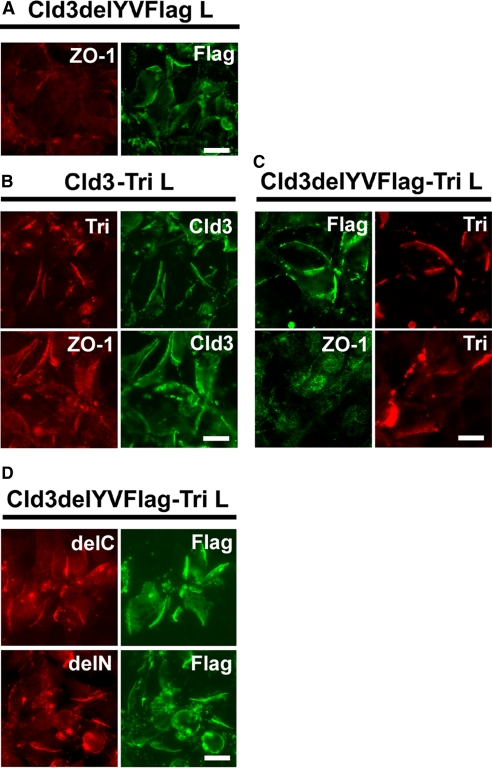
To elucidate this question, we established L cell transfectants coexpressing a FLAG-tagged claudin-3 mutant lacking the COOH-terminal domain (Cld3delYVFlag L) (Figure 2A). As reported previously, in claudin-3 expressing L (Cld3 L) cells, claudin-3 recruited ZO-1 to cell–cell contacts (Itoh et al., 1999). In L cell transfectants coexpressing claudin-3 and tricellulin (Cld3-Tri L), claudin-3 was concentrated with tricellulin and ZO-1 as planes at cell–cell contacts (Figure 2B). In Cld3delYVFlag L cells, Cld3delYVFlag was also concentrated at cell–cell contacts, indicating that it formed homophilic interactions. However, because it lacked the postsynaptic density 95/disc-large/zona occludens (PDZ) binding motif of claudin-3, it did not recruit endogenous ZO-1 to cell–cell contacts (Figure 2A).
Figure 2.
Tricellulin resides in TJ strands independently of binding to ZO-1. (A) Immunofluorescence staining of stable L cell transfectants expressing FLAG-tagged claudin-3 lacking the PDZ binding motif (Cld3delYVFlag L) with anti-FLAG mAb (green) and anti-ZO-1 mAb (red). In Cld3delYVFlag L cells, FLAG-tagged claudin-3 lacking the PDZ binding motif is concentrated at cell–cell contacts, whereas ZO-1 is no longer recruited to cell–cell contacts. Bar, 10 μm. (B) Immunofluorescence staining of stable L cell transfectants expressing claudin-3 and tricellulin (Cld3-Tri L) with anti-claudin-3 pAb (green) and anti-tricellulin mAb (red) or anti-claudin-3 pAb (green) and anti-ZO-1 mAb (red). In Cld3-Tri L cells, claudin-3 is concentrated at cell–cell contacts together with tricellulin and ZO-1. Bar, 10 μm. (C) Immunofluorescence staining of Cld3delYVFlag L cells expressing tricellulin (Cld3delYVFlag-Tri L) with anti-FLAG mAb (green) and anti-tricellulin mAb (red) or anti-ZO-1 pAb (green) and anti-tricellulin mAb (red). Tricellulin and FLAG-tagged mutant claudin-3 colocalize at cell–cell contacts, whereas ZO-1 is not recruited to cell–cell contacts, suggesting that tricellulin does not bind to ZO-1. Bar, 10 μm. (D) Cld3delYVFlag L cells expressing HA-tagged truncated tricellulin lacking the COOH-terminal domain (HA-delC) or the NH2-terminal domain (HA-delN) were stained with anti-FLAG mAb (green) and anti-HA mAb (red). Both truncated mutants colocalize with FLAG-tagged claudin-3 lacking PDZ binding motif at cell contact planes. Bar, 10 μm.
Nonetheless, tricellulin colocalized with Cld3delYVFlag as planes at cell–cell contacts in L cell transfectants coexpressing Cld3delYVFlag and tricellulin (Cld3delYVFlag-Tri L), indicating that the association between claudin and tricellulin does not require ZO-1 and that also tricellulin does not recruit ZO-1 to cell–cell contacts (Figure 2C). Subsequently, to examine whether cytoplamic regions of tricellulin are required for the interaction with claudins, HA-tagged COOH or NH2 deletion mutants of tricellulin, i.e., HA-delC or HA-delN, were expressed in Cld3delYVFlag L cells. Both deletion mutants colocalized with Cld3delYVFlag at cell–cell contacts, suggesting that the interaction between tricellulin and claudins does not require either of the cytoplasmic regions of tricellulin (Figure 2D). When a deletion mutant of tricellulin lacking both COOH and NH2 cytoplasmic domains was expressed in Cld3delYVFlag L cells, it resided in the endoplasmic reticulum (data not shown). Therefore, we cannot exclude the possibility that the COOH cytoplasmic domain and the NH2 cytoplasmic domain function redundantly to incorporate tricellulin into TJ strands.
From the observations described above, we conclude that tricellulin interacts with claudins in the plasma membrane independently from interaction with ZO-1. However, it still remains unclear how and when tricellulin becomes concentrated to tricellular contacts within claudin-based TJs during junction formation.
Knockdown of Occludin Leads to Mislocalization of Tricellulin to bTJs
The amino acid sequence comparison between tricellulin and occludin revealed a high degree of similarity in the COOH-terminal domains (∼32% identical). In addition, the genes encoding tricellulin and occludin are located in tandem on mouse chromosome 13 and human chromosome 5, indicating that these two proteins may have been duplicated during phylogenic evolution and that tricellulin and occludin are closely related molecules (Ikenouchi et al., 2005). Therefore, we reasoned that occludin might play a role in the localization of tricellulin at tTJs.
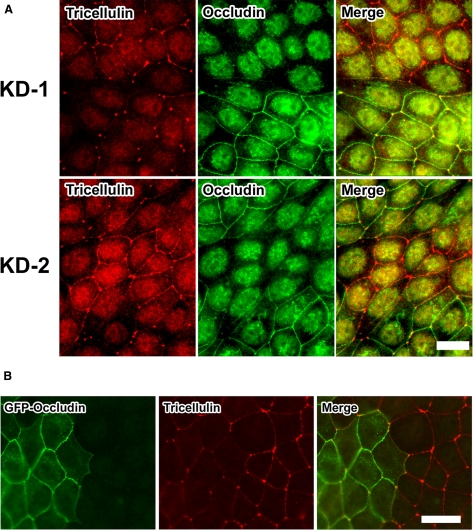
To gain insight into the role of occludin in the localization of tricellulin, we examined the localization of tricellulin in MDCK II cells in which the expression of occludin was suppressed by stably expressing short interfering RNA. Two independent MDCK II cell clones (KD-1 and KD-2) with suppressed occludin expression were established as described previously (Yu et al., 2005). In both clones, occludin protein expression was suppressed. Under confluent conditions, KD-1 and KD-2 cells were mixed and cocultured with parental MDCK II cells and double stained with antibodies against occludin and tricellulin. Interestingly, in occludin knockdown cells, the tricellulin signal at bTJs was significantly increased compared with that in parental cells. This finding suggests that occludin deficiency resulted in a redistribution of tricellulin from tTJs to bTJs (Figure 3A). mRNA or protein expression of tricellulin was not up-regulated in occludin knockdown cells (data not shown).
Figure 3.
Occludin deficiency leads to mislocalization of tricellulin. (A) Parental MDCK II cells were mixed and cocultured with occludin knockdown cells (KD-1 and KD-2, respectively) and double stained with anti-occludin pAb (green) and anti-tricellulin mAb (red). In parental cells (characterized by the presence of occludin), tricellulin is detected in tTJs, whereas in occludin-deficient cells, tricellulin is found in bTJs in addition to tTJs. Bar, 20 μm. (B) KD-1 cells exogenously expressing GFP-tagged mouse occludin were mixed and cocultured with KD-1 cells and double stained with anti-GFP mAb (green) and anti-tricellulin mAb (red). Localization of tricellulin at bTJs is abolished by the exogenous expression of mouse occludin. Bar, 20 μm.
Next, we established KD-1 cells exogenously expressing GFP-tagged mouse occludin. Mixed cultures of these cells and KD-1 cells revealed that this bTJ-directed localization of tricellulin was abolished by the exogenous expression of occludin (Figure 3B).
Tricellulin Localizes at Both Edges of Bicellular Tight Junctions
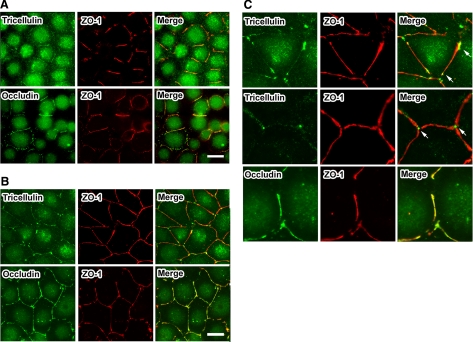
We next examined the behavior of tricellulin and occludin during the process of TJ formation by using a Ca2+ switch assay, according to the previously described protocol (Ando-Akatsuka et al., 1996). MTD1A cells were cultured in a low Ca2+ medium (containing 5 μM Ca2+) overnight under confluent conditions and then their polarization was initiated by transferring them to normal Ca2+ medium. At 0.5 h after Ca2+ repletion, both tricellulin and occludin began to localize at immature fragments of bTJs (Figure 4A). At 1 h after Ca2+ repletion, occludin colocalized with ZO-1 at entire developing bTJs, whereas tricellulin localized preferentially at both edges of elongating bTJs (Figure 4, B and C). From these results, we conclude that tricellulin localizes at the ends of bTJs during the formation of TJs and that this characteristic may explain why tricellulin localized at tTJs.
Figure 4.
Tricellulin localizes at both ends of bicellular tight junctions. (A) MTD1A cells were cultured in low Ca2+ medium (containing 5 μM Ca2+) overnight under confluent conditions and then polarization was initiated by transferring them to normal Ca2+ medium. After a 0.5-h incubation period, cells were stained with anti-occludin mAb (green) and anti-ZO-1 pAb (red) or anti-tricellulin mAb (green) and anti-ZO-1 pAb (red). Bar, 10 μm. (B) After an 1-h incubation period, cells were stained with anti-occludin mAb (green) and anti-ZO-1 pAb (red) or anti-tricellulin mAb (green) and anti-ZO-1 pAb (red). Bar, 10 μm. (C) Highly magnified images of cells in B that clearly show the localization of tricellulin at the edges of elongating bTJs (arrows).
DISCUSSION
In this study, we investigated for the first time how tricellulin is incorporated into TJs and concentrated at tricellular junctions. We found that tricellulin is incorporated into claudin-based TJs independently of binding to ZO-1. Unexpectedly, tricellulin affects the two-dimensional organization of TJ strands, implying that tricellulin may organize unique architectural characteristics of TJ strands in tTJs. At tricellular corners of epithelial cells, TJ strands are known to form cross-points between vertically oriented central sealing elements and horizontally oriented bicellular TJ strands (Staehelin, 1973). Tricellulin may be involved in the regulation of the two-dimensional organization of TJ strands at tTJs.
Tricellulin has at least two independent molecular mechanisms for tricellular localization. One is exclusion of tricellulin from bTJs by occludin. Previously, we reported that the deletion of the occludin gene from embryonic stem cells did not prevent differentiation of these cells into a polarized epithelium with clear TJs (Saitou et al., 1998). In contrast, occludin-deficient mice showed complex gross and histological phenotypes, such as chronic inflammation of the gastric epithelium, calcification in the brain, testicular atrophy, loss of cytoplasmic granules in striated duct cells of the salivary gland, and thinning of the compact bone, suggesting that deficiency of occludin caused moderate dysfunction of TJs or dysfunction of other cellular signaling pathways in which occludin is involved (Saitou et al., 2000; Schulzke et al., 2005). Recently, Yu et al. (2005) reported that the most important role of occludin may be in transducing signals from apoptotic cells through TJs to the actin cytoskeleton. Thus, the role of occludin in TJs has remained controversial.
In the present study, we found that the level of tricellulin in bTJs was significantly increased in occludin knockdown cells. In occludin knockout mice, tricellulin may compensate for some functions of occludin at bTJs and vice versa. To better understand the functions of tricellulin and occludin in the epithelial barrier, further generation of epithelial cells lacking tricellulin and occludin expression is required.
The other mechanism for tricellular localization of tricellulin is that it is targeted to both edges of elongating bTJs; however, the precise molecular interactions at the edges of bTJs and tTJs should be clarified in future studies. These findings are important first steps for understanding of tTJs.
Supplementary Material
ACKNOWLEDGMENTS
This paper is dedicated to the memory of Dr. Shoichiro Tsukita. We thank all of the members of our laboratory (Department of Cell Biology, Kyoto University, Faculty of Medicine) for helpful discussions. We are indebted to Drs. J. M. Brandner, A. Nagafuchi, M. Umeda, and H. Niwa for continuous encouragement, helpful comments, and providing a reagent. This study was supported in part by a grant-in-aid for cancer research (to S. T. and M. F.) and a grant-in-aid for scientific research (A) (to S. T.) from the Ministry of Education, Science, and Culture of Japan.
Abbreviations used:
- AJ
adherens junction
- bTJ
bicellular tight junction
- TJ
tight junction
- tTJ
tricellular tight junction.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0530) on September 3, 2008.
REFERENCES
- Ando-Akatsuka Y., Saitou M., Hirase T., Kishi M., Sakakibara A., Itoh M., Yonemura S., Furuse M., Tsukita S. Interspecies diversity of the occludin sequence: cDNA cloning of human, mouse, dog, and rat-kangaroo homologues. J. Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M. G., Palade G. E. Junctional complexes in various epithelia. J. Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend D. S., Gilula N. B. Variations in tight and gap junctions in mammalian tissues. J. Cell Biol. 1972;53:758–776. doi: 10.1083/jcb.53.3.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita S. Claudin-1 and -2, novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 1998a;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Sasaki H., Fujimoto K., Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 1998b;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16:181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Hay E. D. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- Ikenouchi J., Furuse M., Furuse K., Sasaki H., Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol. 2005;171:939–945. doi: 10.1083/jcb.200510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J., Matsuda M., Furuse M., Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J. Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- Ikenouchi J., Umeda K., Tsukita S., Furuse M. Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J. Cell Biol. 2007;176:779–786. doi: 10.1083/jcb.200612080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter K., Balda M. S. Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- Nieto M. A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Saitou M., Ando-Akatsuka Y., Itoh M., Furuse M., Inazawa J., Fujimoto K., Tsukita S. Mammalian occludin in epithelial cells: its expression and subcellular distribution. Eur J. Cell Biol. 1997;73:222–231. [PubMed] [Google Scholar]
- Saitou M., Fujimoto K., Doi Y., Itoh M., Fujimoto T., Furuse M., Takano H., Noda T., Tsukita S. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J. Cell Biol. 1998;141:397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Furuse M., Sasaki H., Schulzke J. D., Fromm M., Takano H., Noda T., Tsukita S. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol. Biol. Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulzke J. D., Gitter A. H., Mankertz J., Spiegel S., Seidler U., Amasheh S., Saitou M., Tsukita S., Fromm M. Epithelial transport and barrier function in occludin-deficient mice. Biochim. Biophys. Acta. 2005;1669:34–42. doi: 10.1016/j.bbamem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Staehelin L. A. Further observations on the fine structure of freeze-cleaved tight junctions. J. Cell Sci. 1973;13:763–786. doi: 10.1242/jcs.13.3.763. [DOI] [PubMed] [Google Scholar]
- Staehelin L. A., Mukherjee T. M., Williams A. W. Freeze-etch appearance of the tight junctions in the epithelium of small and large intestine of mice. Protoplasma. 1969;67:165–184. doi: 10.1007/BF01248737. [DOI] [PubMed] [Google Scholar]
- Tsukita S., Furuse M., Itoh M. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- Turksen K., Troy T. C. Barriers built on claudins. J. Cell Sci. 2004;117:2435–2447. doi: 10.1242/jcs.01235. [DOI] [PubMed] [Google Scholar]
- Wade J. B., Karnovsky M. J. The structure of the zonula occludens. A single fibril model based on freeze-fracture. J. Cell Biol. 1974;60:168–180. doi: 10.1083/jcb.60.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. C., MacKenzie A., Hulbert W. C., Hogg J. C. A re-assessment of the tricellular region of epithelial cell tight junctions in trachea of guinea pig. Acta Anat. 1985;122:35–38. doi: 10.1159/000145982. [DOI] [PubMed] [Google Scholar]
- Yu A. S., McCarthy K. M., Francis S. A., McCormack J. M., Lai J., Rogers R. A., Lynch R. D., Schneeberger E. E. Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. Am. J. Physiol. Cell Physiol. 2005;288:C1231–C1241. doi: 10.1152/ajpcell.00581.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.