Abstract
In vitro studies of pure tubulin have suggested that tubulin heterodimers in cells assemble into B-lattice microtubules, where the 8-nm dimers in adjacent protofilaments are staggered by 0.9 nm. This arrangement requires the tube to close by forming a seam with an A-lattice, in which the protofilaments are staggered by 4.9 nm. Here we show that Mal3, an EB1 family tip-tracking protein, drives tubulin to assemble in vitro into exclusively 13-protofilament microtubules with a high proportion of A-lattice protofilament contacts. We present a three-dimensional cryo-EM reconstruction of a purely A-lattice microtubule decorated with Mal3, in which Mal3 occupies the groove between protofilaments and associates closely with one tubulin monomer. We propose that Mal3 promotes assembly by binding to freshly formed tubulin polymer and particularly favors any with A-lattice arrangement. These results reopen the question of microtubule structure in cells.
INTRODUCTION
Microtubules are dynamic polymers1 of 8-nm αβ-tubulin heterodimer subunits assembled into polar protofilaments. Lateral contacts may be either αβ, making an A-lattice, or αα and ββ, making a B-lattice2. In vitro, pure tubulin in standard assembly buffers assembles into the B-lattice3, with an A-lattice-like ‘seam’, which led to the widespread assumption that the B-lattice predominates in vivo4.
Microtubules are most dynamic at the plus end, making this site an important target for regulatory proteins5-8, called plus end-tracking proteins (+TIPs). Among them, EB1 is a highly conserved protein apparently found in all eukaryotic organisms. It has been shown to stabilize microtubules in vivo7-10, but how it does this is unclear. One recent study on EB1 in vitro found that it had no effect on microtubule growth rate, with suppression of catastrophe and shrinkage rates11. Another found that EB1 can increase growth rate, suppress shrinkage and increase both catastrophe and rescue frequencies12. These authors also found that EB1 promotes the assembly of microtubules with 13 protofilaments, the number found almost universally in vivo. Mal3, the Schizosaccharomyces pombe homolog of EB1, has been shown to track the tips of growing microtubules in vitro9 and to decorate the seams of B-lattice brain microtubules that had been stabilized with Taxol for study by EM10. Here we show that Mal3 can drive tubulin to assemble into microtubules that, as well as 13-protofilaments, also have the A-lattice arrangement. These results reopen the question of which microtubule structure predominates in cells and have important implications for the in vivo structure and dynamics of microtubules.
RESULTS
Effect of full-length or truncated Mal3 on microtubule assembly
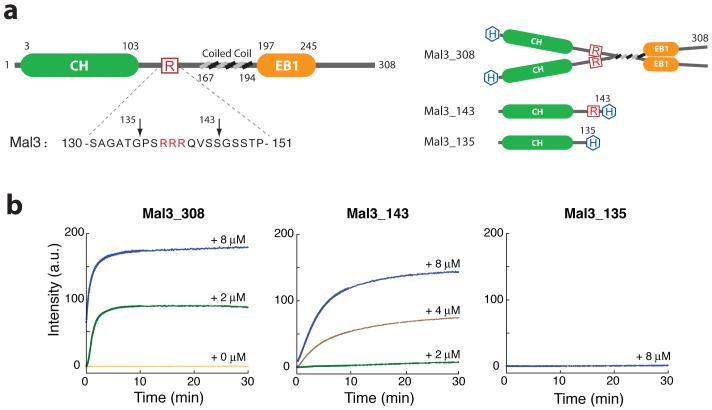
To study the structural mechanism by which EB1 family members stabilize microtubules, we made recombinant Mal3 constructs (Fig. 1a) and tested their effect on purified S. pombe tubulin. First, we examined their influence on polymerization using a light-scattering assay. We found that, even when the tubulin concentration was below the critical value for self-assembly, full-length Mal3 (Mal3-308) strongly promoted assembly (Fig. 1b). Microtubule assembly was also promoted by the first 143 N-terminal amino acids alone (Mal3-143; Fig. 1a), that is, the globular Calponin homology (CH) domain plus a 40-residue tail, predicted to be mainly unstructured13. This truncated molecule is monomeric by gel filtration (Supplementary Fig. 1), showing that dimerization of Mal3 is not essential for promoting microtubule assembly. The full-length protein is nonetheless a more potent nucleator of assembly (Fig. 1b). Deletion of a further eight residues from the C terminus of Mal3-143 (to create Mal3-135) abolishes its polymerization-enhancing activity. Notably, this eight-residue region contains a three-residue polyarginine sequence, suggesting a possible electrostatic role for this functionally crucial region.
Figure 1.
Mal3 promotion of microtubule assembly in vitro. (a) Left, domain structure of Mal3. CH and EB1-like C-terminal motif domains are indicated by green and orange, respectively. Boundaries for each domain are indicated by residue numbers. The amino acid sequence for residues 130-150 is shown below and the three arginine residues are highlighted in red. Right, domain structure for the constructs used in this study. 6×His-tags are highlighted in blue. (b) Microtubule-polymerization assay by 90° light scattering at λ = 350 nm. All assays contained 4 μM S. pombe tubulin (heterodimer concentration). Colored lines indicate the added Mal3 monomer concentration; orange, 0 μM; green, 2 μM; brown, 4 μM; blue, 8 μM.
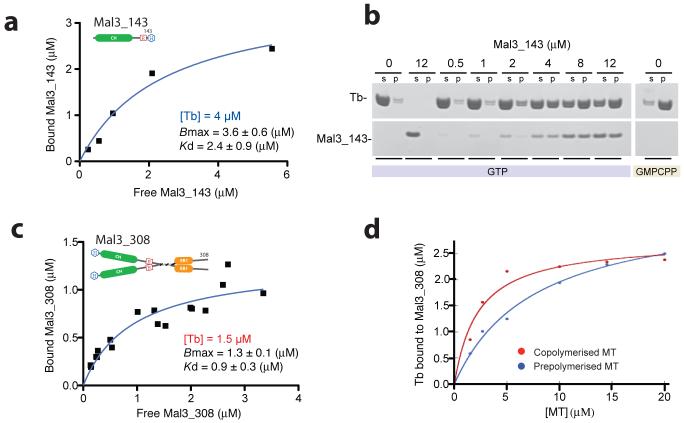
In copelleting assays with S. pombe tubulin (Fig. 2), we found that Mal3-143 drives polymerization, with the rate and extent of polymerization being dependent on the concentration of Mal3 (Fig. 2a,b and Supplementary Fig. 2a). We measured the affinity and stoichiometry of binding in two different ways, either by adding Mal3 to preformed microtubules or by polymerizing tubulin in the presence of Mal3. Pelleting of preassembled, GMPCPP-stabilized S. pombe microtubules in the presence of Mal3-143 showed stoichiometric binding to the lattice, with saturation at a ratio of ∼1:1 Mal3-143 monomer: tubulin heterodimer and a fitted Kd of 2.4 ± 0.9 (S.E.) μM. (Fig. 2a). Adding Mal3-143 to GTP tubulin and allowing assembly to proceed (Fig.2b and Supplementary Fig. 2), produced a similar final occupancy. The binding curve for Mal3-308 (Fig. 2c) was consistent with a ratio of ∼1:1 Mal3 dimer: tubulin heterodimer at full saturation. The curve fitted to the data in Figure 2c gives a single Kd value (0.9 μM) that may reflect a mixture of single-headed and double-headed binding; at subsaturating levels of Mal3-308, both heads of the dimer seem to occupy binding sites (see EM section).
Figure 2.
Mal3 binding to microtubules in vitro. (a) Binding of Mal3-143 to GMPCPP-stabilized S. pombe microtubules, as quantified by SDS-PAGE. The graph shows a single experiment and gave a fitted Kd of 2.4 ± 0.9 (±S.E.) μM. (b) Typical gel run for a pelleting assay. In this case, Mal3-143 (added concentrations, 0-12 μM, are indicated above the lanes) and 8 μM of S. pombe tubulin (Tb) heterodimers were copolymerized in the presence of 1 mM GTP or GMPCPP. s, supernatant fraction; p, pellet fraction. (Curves plotted for the samples with GTP are shown in Supplementary Fig. 2a.). (c) Binding of Mal3-308 to GMPCPP-stabilized S. pombe microtubules. The graph contains all the data points from three independent experiments. For the fitted curve, Bmax 1.3 ± 0.14 (±S.E.) μM and Kd 0.93 ± 0.28 (± S.E.) μM (giving 95% confidence intervals of 0.98-1.6 μM for Bmax and 0.35-1.5 μM for Kd). (d) Binding of 2.5 μM Mal3-308 to pig brain microtubules (MT), either after coassembly with tubulin (red) or after being mixed with preassembled GMPCPP-stabilized microtubules (blue). The data points shown are the result of one experiment. Two other experiments, using either GMPCPP or Taxol to stabilize the preassembled microtubules, gave similar differences from coassembled samples.
These data indicated that there is a binding site for Mal3 on every tubulin heterodimer. To relate them to previous work10 in which prestabilized brain microtubules were used, we titrated soluble brain tubulin or preformed brain microtubules against a fixed concentration of Mal3-308 (Fig. 2d). Polymerizing brain tubulin in the presence of Mal3-308 yielded up to approximately three-fold tighter binding of Mal3-308 compared to adding Mal3-308 to preassembled brain microtubules. We obtained similar results whether the preassembled microtubules were Taxol or GMPCPP stabilized. Several equivalent experiments with brain tubulin and Mal3-143 gave consistent results. The difference was less obvious for S. pombe tubulin, but preassembled S. pombe microtubules did need a higher concentration of Mal3 to saturate their binding sites (Supplementary Fig. 2). We concluded that assembly driven by Mal3 produced microtubules that, for some unknown reason, could bind Mal3 more efficiently.
EM of microtubules assembled with Mal3
We used EM to look at the effect of Mal3 on microtubule structure. Because our aim was to reconstruct a three-dimensional image of a microtubule fully decorated with Mal3-143, we sought 15- or 16-protofilament microtubules that might provide helical symmetry. We examined several hundreds of microtubules, and all seemed to have 13 protofilaments. This was unexpected because, in the absence of Mal3, S. pombe tubulin assembles similar numbers of 13- and 14-protofilament microtubules and a smaller proportion of wider tubes. This fact indicates that, like EB1 (ref. 12), Mal3 drives strongly toward assembly of only 13-protofilament microtubules, as does doublecortin14, another lattice-stabilizing microtubule-associated protein.
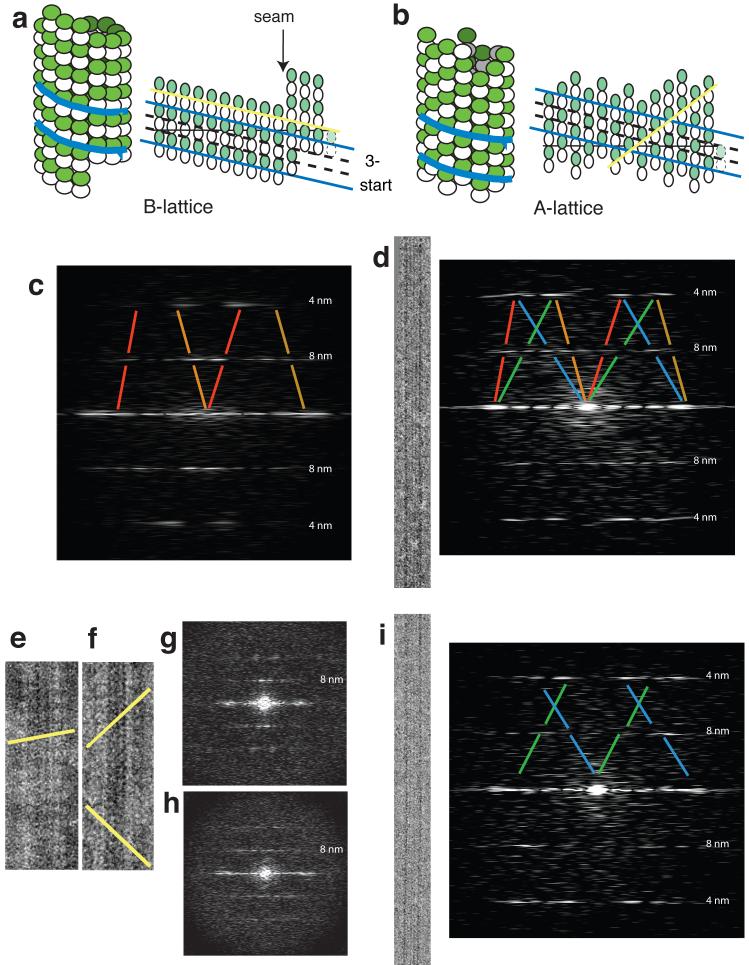
In a 13-protofilament microtubule, lateral contacts may produce a perfect A-lattice of 8-nm dimers or a B-lattice with one or more ‘seams’ (Fig. 3a,b). Images of undecorated microtubules seem to have a 4-nm rather than an 8-nm longitudinal repeat because α and β subunits of the tubulin heterodimer are indistinguishable at low resolution. When the microtubules are decorated with molecules that bind stoichiometrically to heterodimers, an 8-nm repeat becomes visible and provides a way to distinguish microtubules that have an A- or a B-lattice (Fig. 3 c-i). Self-assembled S. pombe tubulin, like brain tubulin3, forms B-lattice microtubules. After decoration with kinesin monomer, Fourier transforms of the images show a layerline with a spacing on the reciprocal lattice corresponding to 8nm, with strong peaks indicating predominantly a B-lattice (Fig. 3c, see peaks in gaps in the red and orange lines). Incubation of preformed microtubules with Mal3-143 instead of kinesin did not decorate sufficiently well to reveal the dimer lattices. However, when microtubule assembly was driven by adding Mal3-143, the computed diffraction patterns (Fig. 3d) did show an 8nm spacing. More notably, the 8-nm layerline showed a different pattern of peaks compared to kinesin-decorated microtubules. Instead of two strong peaks close to the central axis, peaks in this region were weaker, or even absent, and there were additional peaks further from the central axis. Some peaks could fit only an A-lattice arrangement of the tubulin heterodimers (Fig. 3d, i, peaks in gaps in the blue and green lines).
Figure 3.
Microtubules assembled with Mal3 show mixed lattices. (a,b) Diagrams of microtubules with A- and B-lattices of tubulin heterodimer subunits, represented both as three-dimensional microtubules and as opened-out sheets. White subunits, α-tubulin; green subunits, β-tubulin; blue lines, the direction of the 3-start family of helices, common to both lattices, that arises because subunits in adjacent longitudinal protofilaments are staggered by 0.9 nm; yellow lines, the directions of the decoration patterns, different on both lattices, as shown in e and f. (c) Diffraction pattern from a cryo-EM image of a pure tubulin microtubule decorated with kinesin head domains. Red and orange lines show the B-lattice contributions to the diffraction pattern. Red lines, contribution of the near side of the microtubule; orange lines, contribution of the far side. (d) Cryo-EM image of a microtubule copolymerized with Mal3 and its corresponding (mixed AB) diffraction pattern. Blue and green lines show the A-lattice contributions to the diffraction pattern. Blue lines, near-side contribution; green lines, far-side contribution. (e,f) Negative-stain EM images of microtubules decorated with kinesin, showing a pure tubulin microtubule (e) and a microtubule copolymerized with Mal3 (f). The yellow lines highlight the rows of kinesin on the microtubule walls (see a,b.) (g) Diffraction pattern of image e. (h) Diffraction pattern of image f. (i) cryo-EM image of a microtubule copolymerized with Mal3 and its corresponding A-lattice diffraction pattern.
The presence and intensities of peaks characteristic of A- or B-lattices varied greatly, with some rare coassembled microtubules showing only a B-lattice pattern and some showing only an A-lattice pattern (Fig. 3i). Overall, we saw A- and B-lattice patterns, either separately or in combination, in roughly equal proportions. To check that the A-lattice patterns really arose from the underlying tubulin lattice rather than from partial decoration with Mal3, we copolymerized tubulin with Mal3-143 and subsequently added monomeric kinesin, which bound stoichiometrically and displaced some of the Mal3-143, as determined by a pelleting assay (Supplementary Fig. 3). The pattern of kinesin decoration was first examined directly by negative staining. Microtubules assembled alone and decorated with monomeric kinesin clearly had the typical B-lattice, with lines of kinesins on the microtubule wall at about 10° to the horizontal axis (Fig. 3e). Microtubules coassembled with Mal3-143 typically show lines on the microtubule wall at a higher angle (Fig. 3f). The images’ Fourier transforms confirm that the former have a B-lattice and the latter mixed lattices (Fig. 3g,h), indicating that the mixed or A-lattice patterns are due to the underlying tubulin subunit arrangement. Such patterns are not restricted to microtubules assembled from S. pombe tubulin. Experiments with brain tubulin coassembled with S. pombe Mal3-143 also produced microtubules with mixed lattices (Supplementary Fig. 4).
We also looked at S. pombe microtubules assembled with Mal3-308. Diffraction patterns calculated from the images suggested that the microtubules were well decorated but, compared with microtubules assembled with Mal3-143, there were additional layerlines for longer periodicities than 8 nm. Unfortunately, these diffraction patterns were too variable to be analyzed in detail but could best be interpreted as arising from variable mixtures of molecules bound by one or two heads.
Three-dimensional image reconstruction
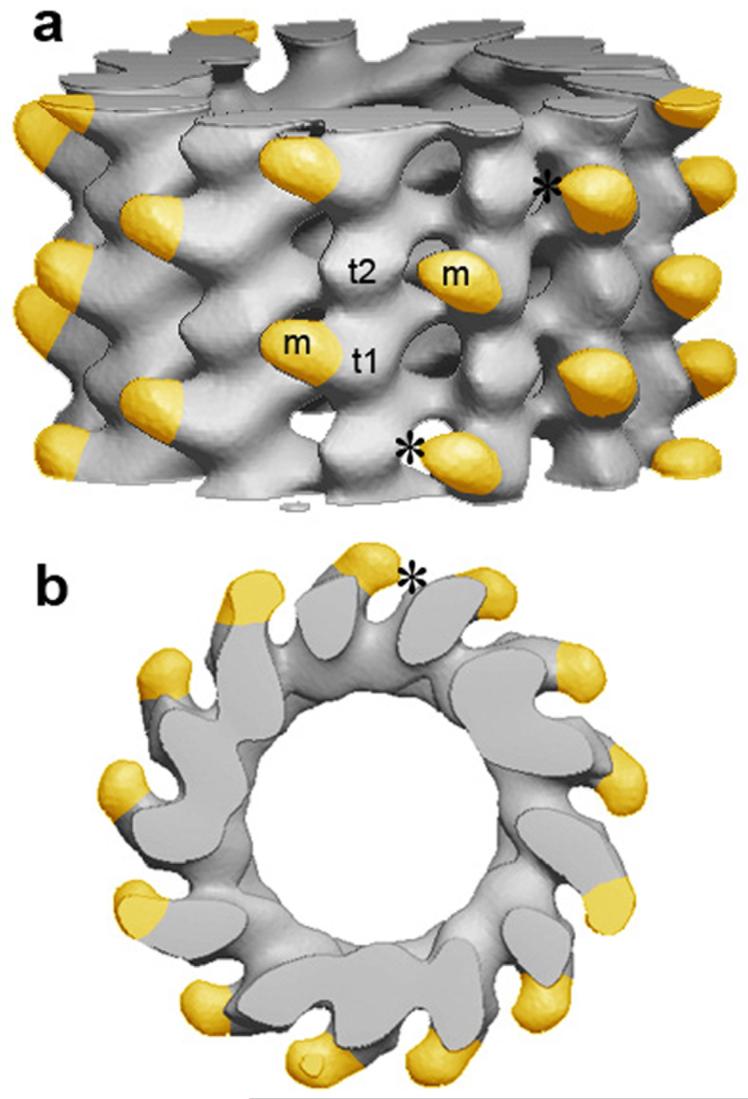
Fortuitously, 13-protofilament S. pombe microtubules have a slight helical twist (Supplementary Fig. 5), which allows analysis of the EM images by helical reconstruction methods. The three-dimensional map of a decorated A-lattice microtubule obtained in this way (Fig. 4) shows the Mal3 microtubule binding domain making an extensive contact with one tubulin subunit and occupying the groove between protofilaments. This site is consistent with metal-shadowed EM images showing Mal3 in a similar location10. We tentatively assign the tubulin subunit making the predominant contact as α-tubulin, because an alanine scanning study of Saccharomyces cerevisiae α-tubulin indicated extensive interaction with Bim1C, the S. cerevisae homolog of Mal3 (ref. 15). We see no contact between Mal3-143 and the neighboring protofilament at this resolution (2 nm), but they are sufficiently close that the predominantly unstructured C terminus of Mal3's microtubule binding domain could bridge across, potentially allowing the three arginines at the end of this flexible domain (Fig. 1a) to interact with the acidic E-hook16,17 of a neighboring tubulin subunit. This interaction would tend to mask the excess negative charge of the E-hook and might have a similar effect to high salt concentrations, which have also been shown to affect the microtubule lattice18. The role of this highly charged tubulin domain as a determinant of lattice geometry remains to be further tested.
Figure 4.
Three-dimensional reconstruction of Mal3-143-decorated A-lattice microtubule. (a) Side view of the microtubule. (plus end upward). (b) End-on projection seen from the plus end. Mal3 density is highlighted in yellow. Asterisks show where Mal3 (m) almost touches a tubulin subunit in the neighboring protofilament. Subunits t1 and t2 may represent α- and β-tubulin, respectively.
DISCUSSION
Effect of Mal3 on microtubule assembly in vitro
It was previously shown that Mal3 binds preferentially to the A-lattice seam of Taxol-stabilized B-lattice brain microtubules10. Our data now indicate that Mal3 not only binds preferentially to the A-lattice but can also drive the assembly of the A-lattice. We find that microtubules can be fully saturated either with Mal3 dimers or with single Mal3 heads (Fig. 2) and that both constructs drive tubulin assembly (Fig. 1). Cooperative binding by both heads of the dimer presumably contributes to the substantially higher polymerizing activity of the dimer compared with the monomer (Fig. 1b). A three-dimensional image (Fig. 4) shows that Mal3 heads occupy the interprotofilament grooves on the outer surface of a microtubule. We find that Mal3 specifies the assembly of exclusively 13-protofilament microtubules and can promote an estimated ∼50% of A-lattice contacts between the protofilaments. Pelleting assays indicated that Mal3 bound better to coassembled microtubules than to preassembled microtubules fixed in a B-lattice arrangement, especially in the case of brain tubulin (Fig. 2d) but, because coassembled microtubules did not consist of pure A-lattice, Figure 2d is likely to underestimate the difference in binding affinity for the A- and B-lattices. We also obtained better Mal3 decoration in our negative-stain and cryo-EM images when S. pombe tubulin was coassembled with Mal3 and consequently driven to make more A-lattice bonds. Our findings are not inconsistent with EM images showing Mal3 binding only to A-lattice seams10, if we assume that the specimen preparation for metal shadowing dislodged the more weakly bound molecules associated with B-lattice sites.
The reconstructed three-dimensional image (Fig. 4) shows that each Mal3 head binds to one side of a protofilament, leaning into the groove, and it seems likely that the essential positively charged region of the linker connecting Mal3's CH domains interacts directly with a negatively charged tubulin tail on the next protofilament. As yet, there is no equivalent image of Mal3 or EB1 monomers bound to a B-lattice microtubule, but it seems reasonable to assume that it would look fairly similar to Figure 4, including the same broad contact with a subunit in one protofilament. A Mal3 head bound in a B-lattice environment might interact with the next protofilament through the charged polypeptide tails in a manner similar to that predicted for the A-lattice sites, but more weakly.
Model for assembly enhancement in vitro
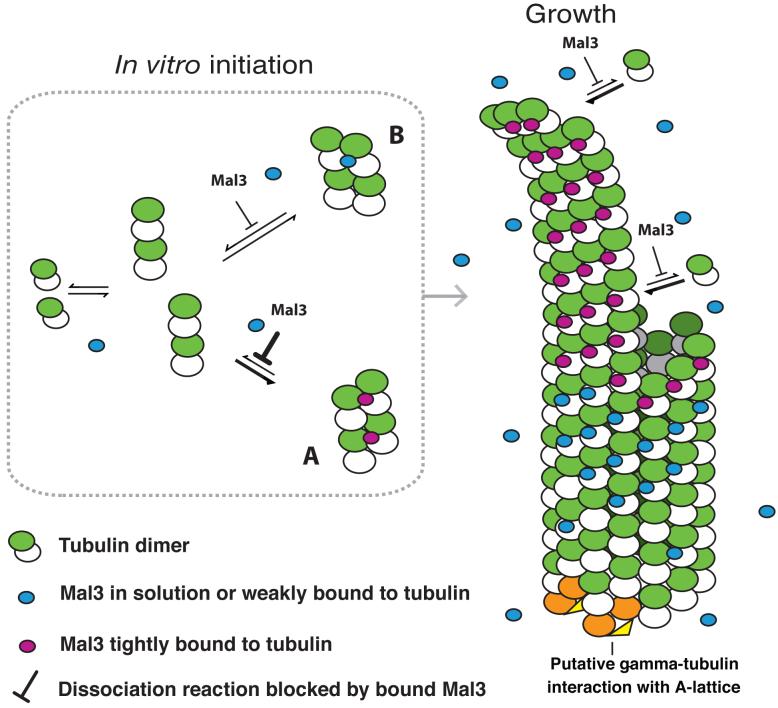
To account for Mal3's ability to drive tubulin to assemble into 13-protofilament, predominantly A-lattice microtubules, we propose the model shown in Figure 5. In this model, Mal3 exerts its effects on assembly by stabilizing otherwise-transient tubulin assembly intermediates. We envisage that tubulin heterodimers reversibly associate into both A- and B-lattice arrangements. In the absence of Mal3, the B-lattice arrangement is the more stable and B-lattice microtubules are consequently formed. In the presence of Mal3, the A-lattice arrangement is also stabilized, and assembly is also constrained to form 13-protofilament microtubules. The model figure shows a pure A-lattice microtubule, but in vitro only a minority of the microtubules assembled would have this lattice; the majority would have a variable number of seams.
Figure 5.
Model of Mal3 promotion of A-lattice assembly. In vitro tubulin heterodimers will form oligomers that may either continue to assemble or may dissociate again. We propose that Mal3 binds to and stabilizes A-oligomers better than B-oligomers, thus promoting their growth into larger complexes and seeding a future microtubule with a high proportion of A-lattice contacts. As a microtubule grows, Mal3 binds tightly to the tip, which might correspond to an extension with a similar structure to the oligomer seeds. Mal3 binds only weakly to the closed microtubule lattice. White subunits, α-tubulin; green subunits, β-tubulin. Stably bound Mal3 monomers are shown in magenta. More weakly bound Mal3 is shown in blue. γ-tubulin complexes (orange and yellow)27,28 may initiate pure A-lattice assembly in vivo by interacting with both α- and β-tubulin29.
In previous work, attention has focused on the mechanism by which Mal3 recognizes microtubule tips. Two main hypotheses have been advanced. The intense labeling of the tips of microtubules growing in the presence of Mal3 (ref. 9) suggests that the protein concentrates there by binding tightly, but gradually dissociates from the lattice as the tip moves away. As already argued9, it is possible that Mal3 binds with higher affinity to a particular structure, such as the sheet-like extension often observed at microtubule plus ends19. It has been suggested20,21 that the gentle outward curvature of such sheets, intermediate between the straight assembled protofilaments22 and the highly curved disassembled structure23, reflects a unique conformation during an early step in assembly. Alternatively, Mal3 may bind more tightly to a cap of tubulin subunits containing GTP or GDP.Pi (whilst the cleaved cleaved phosphate remains trapped in the nucleotide binding pocket) than it binds to GDP-filled subunits in the body of the microtubule. The 8-s lifetime of the bright aggregates, which is long compared with the time taken for the sheet to close up and be replaced9, favors the cap theory.
Our own model focuses on the in vitro nucleation activity of Mal3 (Fig. 1b). Because Mal3 does not interact detectably with soluble tubulin dimers and does not affect microtubule-elongation rate (refs 9, 11 and data not shown), we postulate that it lowers the critical concentration for assembly by interacting with and stabilizing transient assemblies. We hypothesize that oligomeric assemblies that seed microtubules in vitro might have a similar structure to the sheet-like structure present at the tip of a growing microtubule, as well as having GTP bound as in the cap of a growing microtubule.
Relationship to function in vivo
In summary, our in vitro experiments indicate that Mal3, a member of the EB1 tip-tracking protein family, functions as a cofactor or catalyst for assembly, driving tubulin to assemble, even at concentrations below the critical value for pure tubulin, into a 13-protofilament A-lattice arrangement. EB1 itself has already been shown to promote the assembly of 13-protofilament microtubules12. Thus, EB1 family members share doublecortin’s14 role of specifying protofilament number. Mal3 also promotes the formation of A-lattice in vitro, although not entirely countering (under our conditions) tubulin’s preference to assemble with a B-lattice. Considering the ubiquity of EB1 family proteins in eukaryotes, the question of which lattice is prevalent in cells must now be regarded as open. Microtubules assembled in vitro show considerable polymorphism, whereas in vivo microtubules with more or fewer than 13 protofilaments are rarely observed, showing that cells regulate microtubule diameters efficiently. It is likely that the microtubule lattice is equally well controlled, and it is also possible that the choice of tubulin heterodimer arrangement may hold useful information. We can speculate that, for example, the arrangement may provide a mechanism to define subpopulations of microtubules with distinct properties (and distinct microtubule-associated proteins), as already demonstrated for tubulin modifications24,25.
Mal3-deletion strains of S. pombe have short, stubby microtubules26. Mal3 is therefore not absolutely required for microtubule initiation but nonetheless has an important role in promoting growth. Every growing microtubule in normal cells apparently carries Mal3 at its tip, and the same seems to be true of EB1 family proteins in higher eukaryotes. Mal3/EB1 must work in conjunction with other microtubule-organizing proteins, including γ-tubulin at the minus ends of microtubules. The action of Mal3 in restricting protofilament number and in driving the formation of the A-lattice will therefore augment any action of γ-tubulin. It is not known whether microtubule-initiating complexes, such as the γ-tubulin ring complex27 or the γ-tubulin small complex28, promote the A-lattice, but this would explain why γ-tubulin was originally identified as a protein that interacts with β-tubulin29. This finding, indicating a direct interaction between β- and γ-tubulin, misleadingly suggested, at the time, that the αβ-tubulin dimer was oriented with β-tubulin toward the minus end. In the case of a B-lattice a complete ring of γ-tubulins would actually make only one contact with β-tubulin, whereas in an A-lattice each pair of γ-tubulins would make contact with αβ-tubulin (compare Figure 3a with Figures 3b and 5). Small γ-tubulin complexes (individual pairs) probably would not make any such contact at all.
Potentially, Mal3 in cells might distinguish between microtubules assembled from designated organizing centers and those assembled randomly from chance excesses of tubulin. The structural actions of Mal3 might then represent a mechanism for marking non-γ-tubulin-nucleated microtubules. However, the question of which lattice is favored in cells will be difficult to resolve. Although studies of microtubules derived from cells have reported a B-lattice4, there is a danger that Taxol, which was added to stabilize the microtubules, may have driven assembly of soluble tubulin present in the extracts into the B-lattice. Our work raises afresh the possibility that the A-lattice is the preferred lattice in the cell. We believe it will now be necessary to reassess the role of the microtubule lattice in cellular microtubule dynamics. An important aim is to address the technical challenges that must be overcome to determine the preferred microtubule lattice structure in living cells.
METHODS
Protein expression & purification
For tubulin, we created an S. pombe strain mmsp174 (h- ura4.d18 arg3.D4 atb2-::nda2+) containing only α1- and β-tubulin protein isoforms by replacing the atb2 (α2) gene protein-encoding region with the protein-coding region of the α1 gene nda2 using a DNA fragment that was PCR-amplified from genomic DNA using a previously described method30. Single isoform S. pombe α1- and β-tubulin was prepared by a modification of a previously described method31 to be described in detail elsewhere. Purified tubulins were desalted into K-PEM buffer32 (100 mM PIPES, 1 mM MgSO4 and 2 mM EGTA adjusted to pH 6.9 with KOH) containing 20 μM GDP before storage in liquid nitrogen. Protein concentrations were determined by UV-absorption scan of protein samples dissolved in 6 M guanidine hydrochloride, assuming full nucleotide occupancy and ε of 108,390 M-1 cm-1.
For Mal3-308, a DNA fragment encoding full-length Mal3 was PCR amplified from wild-type (strain 972) S. pombe genomic DNA and cloned into a pET17b vector (Novagen) with an N-terminal tag of MGSSHHHHHH. The DNA sequence was confirmed by DNA sequencing. The protein was expressed in Escherichia coli BL21 Star (DE3) strain (Invitrogen) following IPTG induction at 20 °C for 2 h and purified on a nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen) by a previously described method33. The Mal3-308-containing fraction from the Ni-NTA column was loaded on a HiLoad 16/60 Superdex 200 pg column (GE Healthcare) that was equilibrated with K-PEM buffer containing 100 mM NaCl. Fractions were frozen and stored in liquid N2. Mal3-308 concentration was determined by absorbance at 280 nm in 6M GuHCl using ε of 37,025 M-1 cm-1.
Fragments encoding Mal3-143 and Mal3-135 were PCR amplified from the pET17b-Mal3 vector and cloned into a pHis17 vector with a C-terminal tag of GSHHHHHH. Their sequences were confirmed by DNA sequencing. The proteins were expressed and purified in the same way as full-length Mal3 except that they were loaded on a HiLoad 16/60 Sephacryl column (GE Healthcare) equilibrated with 20 mM Tris (pH 7.2).
Kinesin-1 T93N construction, expression and purification were done as described previously34
Microtubule-polymerization assay
We mixed 4 μM of S. pombe tubulin and various concentrations of Mal3 constructs on ice in K-PEM buffer containing 1 mM DTT and 1 mM GTP, and transferred the mixture to a quartz fluorimeter cuvette, temperature-controlled to 25 °C by a Cary single-cell Peltier heater. Scattering of 350-nm light at 90° was recorded in a Cary Eclipse Fluorescence spectrophotometer. After a 30-min incubation, samples were immediately transferred on to poly(L-lysine)-coated slide glass and observed by video-enhanced differential interference contrast (DIC) microscopy using a charge-coupled device (CCD) camera C3077 and Argus video processor (Hamamatsu Photonics).
Microtubule binding assays
For the prepolymerized microtubule binding assay of Mal3-143, 5 μM of purified S. pombe tubulin in 82 mM K-PIPES (pH 6.8), 0.12 mM MgSO4, 0.88 mM MgCl2, 1.1 mM EGTA, 1.25 mM DTT and 1.25 mM GMPCPP was incubated on ice for 15 min and then at 30 °C for 1 h to polymerize. We mixed 40 μl of prepolymerized microtubules with 10 μl of Mal3-143, giving a final concentration of 4 μM tubulin in 86 mM K-PIPES, pH 6.8, 0.3 mM MgSO4, 0.7 mM MgCl2, 1.3 mM EGTA, 1 mM DTT, 20 mM NaCl and 1 mM GMPCPP and incubated for 10 min at 30 °C.
For the copolymerized tubulin and Mal3-143 binding assay, 50 μl mixtures of 8 μM tubulin and Mal3-143 were copolymerized in 1 mM GTP, 88 mM K-PIPES, pH 6.8, 0.4 mM MgSO4, 0.6 mM MgCl2, 1.4 mM EGTA, 1 mM DTT and 20 mM NaCl. The samples were mixed on ice and incubated for 5 min, then at 30 °C for 30 min. All samples were centrifuged at 50,000 r.p.m. (90720 g), 30 °C for 5 min in a TLA100 rotor using an Optima TLX ultracentrifuge (Beckman). The pellet fractions were resuspended in 50 μl of BRB80 buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2 and 1 mM EGTA).
For the prepolymerized S. pombe microtubule binding assay of Mal3-308, Mal3-308 was incubated for 10 min at 25 °C in K-PEM, 30 mM NaCl, 1 mM GMPCPP, 1 mM AMPPNP-Mg2+ with 1.5 μM of prepolymerized GMPCPP microtubules. Samples (50 μl) were pelleted in a TLA100 rotor for 5 min at 50,000 r.p.m. and 25 °C. The pellet fractions were resuspended in 50 μl of K-PEM buffer. For the experiment with pig microtubules and Mal3-308, 2.5 μM Mal3-308 dimer was either coassembled with pig brain tubulin (35 °C 10 min then 25 °C for 20 min) or mixed with preassembled pig GMPCPP-stabilized microtubules (10 min 25 °C) in K-PEM supplemented with 10 mM NaCl, 1 mM DTT and 1 mM GMPCPP. Samples (50 μl) were pelleted in a TLA100 rotor for 5 min at 50,000 r.p.m. and 25 °C. The pellet fractions were resuspended in 50 μl of K-PEM buffer.
Pellet and supernatant fractions were analyzed by SDS-PAGE using NuPAGE 4-12% (w/v) Bis-Tris precast gels run in MES buffer (Invitrogen). Gels were stained with Sypro Red (Invitrogen) following the manufacturer’s instructions and the fluorescence images of the gels were recorded using a Molecular Imager FX (Bio-Rad). Protein bands were quantified using Quantity One software (Bio-Rad) within the linear signal intensity range, which was determined using standard curves of Mal3-143 or Mal3-308, respectively. The concentration of Mal3 bound to microtubules was plotted against the concentration of unbound Mal3.
To analyze Mal3-308 binding to prepolymerized S. pombe microtubules, the copelleting experiment was repeated three times, and all data points from the three independent experiments were included in the graph (Fig. 2c). For quantification of the Mal3-308 pellet and supernatant fractions, each SDS-PAGE gel contained a standard curve of purified Mal3-308. A plot of the standard curve data was fitted using linear regression and the amount of Mal3-308 in the pellet and supernatant samples was determined from the integrated fluorescence intensity of individual bands using the standard curve. The measured Mal3-308 amount (the sum of the amount in the supernatant plus the amount in the pellet) was similar to the total amount of Mal3 added to the assay. A 30% root-mean-square error in the amounts reflects all pipetting errors as well as any nonspecific losses. The measured amount of Mal3-308 in the pellet (bound Mal3) was plotted against the measured amount of Mal3-308 in the supernatant (free Mal3), and the data were fitted by the equation [bound Mal3 to microtubule] = (Bmax × [free Mal3]) / (Kd + [free Mal3]). The nonlinear fit of the data, Bmax (binding sites), Kd, their standard error and 95% confidence intervals were all calculated using the program PRISM (GraphPad software). The standard error and confidence interval were determined from the nonlinear fit of the data set.
To analyze Mal3-143 copelleting experiments, the amount of bound Mal3-143 was calculated from the relative Sypro Red staining (SRS) of the pellet and supernatant fractions and the total amount of Mal3-143 added using the relationship (SRS of Mal3-143 in pellet) * [total Mal3-143 added] / ((SRS of Mal3-143 in pellet) + (SRS of Mal3-143 in supernatant)).
The total amount (SRS pellet + SRS supernatant) of Mal3-143 was not appreciably different (∼10%) from the total amount present in a control where no microtubules were added. A plot of bound against free Mal3 was fitted, as described for Mal3-308, with R2 = 0.967.
Electron microscopy
We mixed 5 μM of S. pombe tubulin and 6 μM of Mal3-143 in BRB80 (80 mM K-PIPES (pH 6.8), 1mM MgCl2, 1 mM EGTA and 1 mM DTT) with 1 mM GTP, and incubated the mixture on ice for 5 min. We then induced polymerization by incubating the mixture at 30 °C for 15 min. The sample, applied on a copper grid coated with a holey carbon film, was rapidly frozen in liquid ethane. The grids were examined using a Gatan cold stage in a FEI F20 electron microscope operating at 200 kV. The images, at 50,000 magnification with a defocus of 1-3 μm, were photographed with Kodak SO-163 film and digitized in 6-μm steps using an MRC-KZA scanner.
We carried out the kinesin-decoration experiment by coassembling 5 μM S. pombe tubulin with 10 μM kinesin in BRB80, 1mM AMPPNP and 1 mM GMPCPP at 30 °C for 15 min, or by coassembling 6 μM Mal3 with 5 μM S. pombe tubulin in BRB80, 1 mM AMPPNP and 1 mM GMPCPP at 30 °C for 15 min and subsequently adding 10 μM kinesin. Samples were then applied on a carbon-coated grid, washed with BRB80 and negatively stained with 1% (w/v) uranyl acetate.
Image analysis
Cryo-EM images were processed using the MRC package of programs35; images were boxed, floated and fast-Fourier transformed. Layerline data were selected (Supplementary Figs. 5 and 6) and averaged, and a three-dimensional map at ∼2-nm resolution was calculated by Fourier-Bessel transformation.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank J. Löwe and the members of his group for their support and comments. This work was supported by the Medical Research Council, Marie Curie Cancer Care and Cancer Research UK.
REFERENCES
- 1.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Amos L, Klug A. Arrangement of subunits in flagellar microtubules. J. Cell Sci. 1974;14:523–549. doi: 10.1242/jcs.14.3.523. [DOI] [PubMed] [Google Scholar]
- 3.Song YH, Mandelkow E. Recombinant kinesin motor domain binds to β-tubulin and decorates microtubules with a B surface lattice. Proc. Natl. Acad. Sci. USA. 1993;90:1671–1675. doi: 10.1073/pnas.90.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kikkawa M, Ishikawa T, Nakata T, Wakabayashi T, Hirokawa N. Direct visualization of the microtubule lattice seam both in vitro and in vivo. J. Cell Biol. 1994;127:1965–1971. doi: 10.1083/jcb.127.6.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuyler SC, Pellman D. Microtubule “plus-end-tracking proteins”: The end is just the beginning. Cell. 2001;105:421–424. doi: 10.1016/s0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- 6.Mimori-Kiyosue Y, Tsukita S. “Search-and-capture” of microtubules through plus-end-binding proteins (+TIPs) J. Biochem. 2003;134:321–326. doi: 10.1093/jb/mvg148. [DOI] [PubMed] [Google Scholar]
- 7.Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Morrison EE. Action and interactions at microtubule ends. Cell. Mol. Life Sci. 2007;64:307–317. doi: 10.1007/s00018-007-6360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 10.Sandblad L, et al. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 11.Manna T, Honnappa S, Steinmetz MO, Wilson L. Suppression of microtubule dynamic instability by the +TIP protein EB1 and its modulation by the CAP-Gly domain of p150glued. Biochemistry. 2008;47:779–786. doi: 10.1021/bi701912g. [DOI] [PubMed] [Google Scholar]
- 12.Vitre B, et al. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 2008;10:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- 13.Honnappa S, John CM, Kostrewa D, Winkler FK, Steinmetz MO. Structural insights into the EB1-APC interaction. EMBO J. 2005;24:261–269. doi: 10.1038/sj.emboj.7600529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moores CA, et al. Mechanism of microtubule stabilization by doublecortin. Mol. Cell. 2004;14:833–839. doi: 10.1016/j.molcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Richards KL, et al. Structure-function relationships in yeast tubulins. Mol. Biol. Cell. 2000;11:1887–1903. doi: 10.1091/mbc.11.5.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharyya B, Sackett DL, Wolff J. Tubulin, hybrid dimers, and tubulin S. Stepwise charge reduction and polymerization. J. Biol. Chem. 1985;260:10208–10216. [PubMed] [Google Scholar]
- 17.Sackett DL, Wolff J. Proteolysis of tubulin and the substructure of the tubulin dimer. J. Biol. Chem. 1986;261:9070–9076. [PubMed] [Google Scholar]
- 18.Dias DP, Milligan RA. Motor protein decoration of microtubules grown in high salt conditions reveals the presence of mixed lattices. J. Mol. Biol. 1999;287:287–292. doi: 10.1006/jmbi.1999.2597. [DOI] [PubMed] [Google Scholar]
- 19.Chretien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahadevan L, Mitchison TJ. Cell biology: powerful curves. Nature. 2005;435:895–897. doi: 10.1038/435895a. [DOI] [PubMed] [Google Scholar]
- 22.Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 23.Ravelli RB, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 24.Gundersen GG, Bulinski JC. Distribution of tyrosinated and nontyrosinated α-tubulin during mitosis. J. Cell Biol. 1986;102:1118–1126. doi: 10.1083/jcb.102.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- 28.Kollman JM, et al. The structure of the γ-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell. 2008;19:207–215. doi: 10.1091/mbc.E07-09-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oakley CE, Oakley BR. Identification of γ-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- 30.Erdeniz N, Mortensen UH, Rothstein R. Cloning-free PCR-based allele replacement methods. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis A, Sage CR, Wilson L, Farrell KW. Purification and biochemical characterization of tubulin from the budding yeast Saccharomyces cerevisiae. Biochemistry. 1993;32:8823–8835. doi: 10.1021/bi00085a013. [DOI] [PubMed] [Google Scholar]
- 32.Walker RA, et al. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browning H, Hackney DD. The EB1 homolog Mal3 stimulates the ATPase of the kinesin Tea2 by recruiting it to the microtubule. J. Biol. Chem. 2005;280:12299–12304. doi: 10.1074/jbc.M413620200. [DOI] [PubMed] [Google Scholar]
- 34.Krylyshkina O, et al. Modulation of substrate adhesion dynamics via microtubule targeting requires kinesin-1. J. Cell Biol. 2002;156:349–359. doi: 10.1083/jcb.200105051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.