Abstract
The El Tor biotype of Vibrio cholerae O1, causing the current seventh pandemic of cholera, has replaced the classical biotype, which caused the sixth pandemic. The CTX prophages encoding cholera toxin in the two biotypes have distinct repressor (rstR) genes. Recently, new variants of El Tor strains that carry the classical type (CTXclass) prophage have emerged. These “hybrid” strains apparently originate through lateral gene transfer and recombination events. To explore possible donors of the CTXclass prophage and its mode of transfer, we tested environmental V. cholerae isolates for the presence of CTXclass prophage and mobility of the phage genome. Of the 272 environmental V. cholerae isolates tested, 6 were found to carry the CTXclass prophage; all of these belonged to the O141 serogroup. These O141 strains were unable to produce infectious CTXclass phage or to transmit the prophage to recipient strains in the mouse model of infection; however, the CTXclass prophage was acquired by El Tor strains when cultured with the O141 strains in microcosms composed of filtered environmental water, a chitin substrate, and a V. cholerae O141-specific bacteriophage. The CTXclass prophage either coexisted with or replaced the resident CTXET prophage, resulting in El Tor strains with CTX genotypes similar to those of the naturally occurring hybrid strains. Our results support a model involving phages and natural chitin substrate in the emergence of new variants of pathogenic V. cholerae. Furthermore, the O141 strains apparently represent an alternative reservoir of the CTXclass phage genome, because the classical V. cholerae O1 strains are possibly extinct.
Keywords: hybrid Vibrio cholerae strain, toxigenic Vibrio cholerae
Cholera caused by toxigenic Vibrio cholerae is a major public health problem confronting many developing countries, in which outbreaks occur frequently and are closely associated with poverty and poor sanitation (1, 2). Seven distinct pandemics of cholera have been recorded since the first pandemic in 1817. The current seventh pandemic, which originated in Indonesia in 1961, is the most extensive in terms of geographic spread and duration. The causative agent is V. cholerae O1 of the El Tor biotype. The sixth pandemic and presumably the earlier pandemics were caused by V. cholerae O1 of the classical biotype (2).
Classification into biotypes is based on a set of phenotypic traits that include susceptibility to polymixin B, hemagglutination of chicken erythrocytes, hemolysis of sheep erythrocytes, the Voges–Proskauer test (which measures the production of acetylmethylcarbinol), and susceptibility to phages (1, 2). In toxigenic V. cholerae, the genes encoding cholera toxin (ctxAB) are located in the CTX prophage (3). Although most other parts of the CTX phage genomes are similar in the two biotypes of V. cholerae O1, the repressor genes (rstR genes) carried by CTX phages differ in CTXETφ and CTXclassφ (4, 5). Two other varieties of the rstR gene carried by CTXCalcφ and CTXenvφ also have been reported (6, 7). This diversity of rstR provides the molecular basis for heteroimmunity among the different CTX phages (4).
The El Tor biotype of V. cholerae O1 continues to cause cholera outbreaks in Bangladesh and other developing countries of Asia, Africa, and Latin America, whereas the sixth pandemic classical strains are no longer isolated from patients or the environment. The recent emergence of new variants of the El Tor strain, which carry the classical type (CTXclass) prophage, has been reported (8–10). These strains, commonly called “hybrid” variants, have been isolated from geographically distant locations, including Bangladesh and Mozambique. These strains display most of the typical traits of the El Tor biotype, but the resident CTX prophage in the strains is of the classical type.
Although the new hybrid variants presumably originated through lateral gene transfer and recombination events, the donor of the CTXclass prophage and its mode of transfer to El Tor strains in this evolutionary pathway are not clear. The aquatic habitats typically are ecosystems containing multiple microbial strains and species and high concentrations of phage and free DNA (11–13). These features, along with the recent observation that V. cholerae growing on a chitin substrate can take up and assimilate DNA from the environment (14), are presumed to promote horizontal gene transfer, leading to high levels of genomic diversity. Chitin, an insoluble polymer of GlcNAc, is abundant in the environment, particularly in the exoskeleton of marine crustaceans, and V. cholerae is known to attach to chitinaceous zooplankton (e.g., copepods) and metabolize chitin (15).
In the present study, we examined environmental isolates of V. cholerae for the presence of CTXclass prophage in an effort to identify possible donors of the CTXclass phage genome to the recently emerged hybrid variants of the El Tor strain. We also reproduced the environmental conditions under which these hybrid strains seem to have developed, allowing us to study the genetic mechanisms that likely led to their origination in the natural environment.
Results
V. cholerae O141 as a Reservoir of Classical CTX Prophage.
The classical biotype of V. cholerae O1, which probably is extinct, typically carried the CTXclass prophage. We investigated whether any other strain in the environment also could carry the CTXclass prophage, allowing the CTXclass genome to persist in nature despite the disappearance of the classical biotype of V. cholerae O1. In this study, all toxigenic V. cholerae O1 isolates from surface water were found to carry the CTXET prophage; however, analysis of 254 non-O1, non-O139 strains identified 11 CTX-positive strains, of which 6 carried the CTXclass prophage [see supporting information (SI) Table S1]. The CTX prophage in these six strains had the rstRclass gene, whereas that of the remaining five strains had rstRCalc or rstRenv genes, as determined by specific polymerase chain reaction (PCR) assays for these genes. These 11 CTX-positive non-O1, non-O139 strains also were positive for the toxin coregulated pilus (TCP) pathogenicity island and carried the tcpA allele of the classical biotype. Further analysis revealed that all six strains that carried the CTXclass prophage belonged to the O141 serogroup. Thus, the O141 strains appear to be a reservoir of the CTXclass phage genome. It is not clear how the O141 strains acquired the CTXclass prophage, but given that these strains are potentially pathogenic and coexisted with the classical strains, they are likely to have interacted and exchanged genetic material in either the human intestinal milieu or the aquatic environment.
The CTXclass Prophage of V. cholerae O141 May Be Acquired by Other Strains.
Toxigenic V. cholerae of the El Tor biotype is known to produce infectious CTXETφ particles. Thus, the propagation of the phage genome to recipient strains occurs most commonly through infection of the strains with cell-free phage particles. However, strains carrying CTXclass prophage do not produce infectious CTXclass φ (5, 10), and thus the mode of transfer of the CTXclass genome to recipient strains is unclear. Because CTXclass prophage has been detected in strains other than the classical biotype of V. cholerae O1 (8–10), the phage genome possibly can be acquired by these nonclassical strains through one or more alternative mechanisms. To track the transfer of the CTXclass phage genome from the toxigenic O141 strains, we introduced a selectable marker in the prophage resident in several O141 strains (Fig. 1). This genetic marking was done by introducing a PCR-amplified kanamycin-resistance determinant (KmR) marker (flanked by CTX phage genes) from strain O395NT (16) into toxigenic O141 strains through chitin-induced transformation (14).
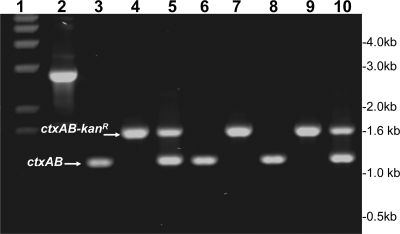
Fig. 1.
Analysis of genetically marked V. cholerae O141 strains carrying a KmR marker in the resident CTXclass prophage. These strains were produced by recombination with a PCR amplicon derived from strain O395NT using the chitin-induced transformation protocol (see text for details). Lane 1 shows molecular size markers corresponding to 1-kb DNA Ladder Plus (Invitrogen); lane 2, the PCR amplicon spanning zot and ctxB genes derived from O395NT, comprising the KmR marker flanked by CTX genes; lane 3, O141 strain V51 (native); lane 6, O141 strain 2615 (native); lane 7, a derivative of strain 2615 carrying the KmR marker; and lane 8, O141 strain 2634 (native). Lanes 4 and 5 show derivatives of V51 carrying the KmR marker in the CTX prophage, and lanes 9 and 10 show derivatives of strain 2634 carrying the KmR marker.
The chitin induction enhanced the uptake and recombination of the KmR-marked DNA into the CTXclass prophage carried by toxigenic O141 strains. These events were further verified by appropriate PCR assays to amplify the ctxAB region of the recombinants. Because the donor strain O395NT had a KmR gene inserted into the ctxAB operon, the PCR product derived from the KmR recombinants were larger than those from the wild-type strains (Fig. 1). When the wild-type toxigenic O141 strains carried two copies of CTXclass prophage, either one or both copies of the CTXclass prophage recombined with the incoming KmR-marked DNA (Fig. 1). The resulting KmR-labeled O141 strains were later used as donors to monitor the transfer of the CTXclass phage genome. V. cholerae O1 El Tor strains were previously shown to produce infectious CTXETφ particles, and transduction of recipient strains was found to occur efficiently inside the intestines of infant mice (3, 17). In the present study, analyses of the KmR-labeled O141 strains indicated that none of the strains produced CTXclassφ and that the phage genome could not be transferred to potential recipient strains inside the mouse intestine; however, recipient V. cholerae strains could be transformed at a frequency of ≈10−7 using total genomic DNA derived from the genetically marked O141 strains in the presence of chitin (Table 1). Subsequent analysis found that the KmR derivatives of the recipient strains carried KmR-labeled CTXclass prophage (Fig. 2).
Table 1.
Horizontal transfer of CTXclass phage genome to recipient strains in the presence of a lytic phage and a chitin substrate
| Recipient strain | Presence of chitin | Donor strain | Presence of phage* | Transformation frequency |
|---|---|---|---|---|
| 2344–11 | No chitin | O141–2615-Km | No phage | 0 |
| Shrimp shell chitin | O141–2615-Km | No phage | 4.6 × 10−6 | |
| Shrimp shell chitin | O141–2615-Km | JSF-141Bφ | 2.0 × 10−4 | |
| 2344–16 | No chitin | O141–2634-Km | No phage | 0 |
| Shrimp shell chitin | O141–2634-Km | No phage | 2.7 × 10−7 | |
| Shrimp shell chitin | O141–2634-Km | JSF-141Bφ | 6.6 × 10−5 | |
| 2434–44 | No chitin | O141–2615-Km | No phage | 0 |
| Shrimp shell chitin | O141–2615-Km | No phage | 1.2 × 10−7 | |
| Shrimp shell chitin | O141–2615-Km | JSF-141Bφ | 4.7 × 10−5 | |
| C6706ΔhapA | No chitin | O141–2615-Km | No phage | 0 |
| Shrimp shell chitin | O141–2615-Km | No phage | 3.5 × 10−7 | |
| Shrimp shell chitin | O141–2615-Km | JSF-141Bφ | 2.9 × 10−4 | |
| 2749129 | No chitin | O141–2615-Km | No phage | 0 |
| Shrimp shell chitin | O141–2615-Km | No phage | 0 | |
| Shrimp shell chitin | O141–2615-Km | JSF-141Bφ | 2.0 × 10−7 | |
| 2749720 | No chitin | O141–2615-Km | No phage | 0 |
| Shrimp shell chitin | O141–2615-Km | No phage | 2.1 × 10−6 | |
| Shrimp shell chitin | O141–2615-Km | JSF-141Bφ | 9.6 × 10−4 | |
| 2756322 | No chitin | O141–2615-Km | No phage | 0 |
| Shrimp shell chitin | O141–2615-Km | No phage | 4.8 × 10−6 | |
| Shrimp shell chitin | O141–2615-Km | JSF-141Bφ | 7.3 × 10−4 | |
| 2684756 | No chitin | O141–2615-Km | No phage | 0 |
| Shrimp shell chitin | O141–2615-Km | No phage | 1.9 × 10−8 | |
| Shrimp shell chitin | O141–2615-Km | JSF-141Bφ | 1.2 × 10−6 |
*Phage JSF141Bφ specifically lyses the donor V. cholerae O141 strains.
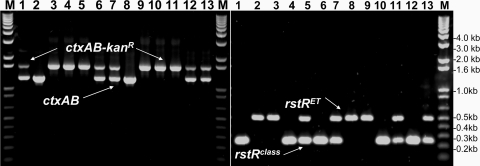
Fig. 2.
PCR analysis of the ctxAB genes (Left) and the rstR genes (Right) of CTX prophages carried by different derivatives of toxigenic V. cholerae O1 El Tor biotype strains that were subjected to transformation in microcosms in the presence of a genetically marked toxigenic O141 strain carrying a KmR marker in the resident CTXclass prophage, a lytic phage for O141 (JSF141Bφ), and pieces of sterile shrimp shell as a source of chitin. Lane 1 shows donor O141 strain 2615-Km; lane 2, wild-type V11–2615; and lane 8, strain C6706ΔhapA before transformation. Lanes 3–7 show different transformants of strain V11–2615 (toxigenic El Tor strain), and lanes 9–13 show different transformants of strain C6706ΔhapA. The markers in lanes marked “M” correspond to the 1-kb DNA Ladder Plus (Invitrogen). These findings demonstrate the origination of strains with diverse CTX genotype from the same parent strain. Although some of these retain the original CTX phage, others replace part or whole of the original prophage with the CTXclass phage genome.
Lytic Phages Enhance Chitin-Induced DNA Uptake in Microcosms.
The aquatic environment in Bangladesh is known to harbor various lytic phages active in V. cholerae, termed vibriophages (13). These vibriophages can disintegrate V. cholerae cells in the environment, resulting in release of their cellular DNA. In previous studies, we identified the presence of phages specific for different O1 and O139 strains. In the present study, we identified the JSF141B phage, which is specific for toxigenic V. cholerae O141 strains. To investigate whether this phage can enhance the chitin-induced acquisition of genetically marked CTXclass prophage by recipient strains, we set up experiments in microcosms composed of filtered environmental water, donor and recipient bacterial strains, pieces of sterile shrimp shell as a chitin source, and the lytic phage for V. cholerae O141 donor strains.
The transformation frequency of various recipient strains in the presence of the phage was >100-fold higher than that in the absence of the phage (Table 1). Adding DNase to the culture reduced the transformation efficiency by ≈50-fold (data not shown), suggesting that phage-mediated release of cellular DNA from the donor cells was an important step in the transformation. However, as suggested previously (14), DNase may not completely abolish the transformation, because the conditions used in these assays may not have been optimum for DNase activity. Significantly, in the present study, V. cholerae O1 El Tor biotype strains from both clinical and environmental sources acquired the CTXclass prophage from V. cholerae O141 strains. These findings suggest that toxigenic V. cholerae O141 strains can act as a source of the CTXclass phage genome in the origination of hybrid El Tor strains (8) that, unconventionally, carry the CTXclass prophage.
The V. cholerae O141-specific JSF141B phage and related phages were detected and isolated from the water during the sampling period, and the phage concentration was sufficient to allow isolation with no need for pre-enrichment (data not shown). Given that both V. cholerae O1- and O139-specific phages also are found seasonally in high concentrations in surface water in Bangladesh (13), it seems possible that these phages could contribute to natural transformation by releasing DNA from the epidemic strain, particularly during the late stages of a cholera epidemic, when phage predation leads to the collapse of the epidemic (13, 18).
Arrangement of CTX Prophages in Transformed El Tor Strains.
Representative colonies of transformed V. cholerae O1 El Tor strains from the microcosms were grown in Luria broth (LB) containing kanamycin (50 μg/ml) and analyzed for the presence of CTXclass and CTXET phage genomes by specific PCR and hybridization assays. Analyses of multiple colonies of transformants derived from the same recipient strain showed different arrangements of the CTX phage genome. Whereas a proportion of the transformed cells retained their original CTXET prophage and also acquired the CTXclass prophage, in another population of cells, the CTXET prophage was replaced by the incumbent CTXclass phage genome (Fig. 2). These findings suggest that at least some of the naturally occurring hybrid El Tor strains may have originated from the seventh pandemic strain through a similar mechanism involving phages and chitin-induced transformation, as shown schematically in Fig. 3.
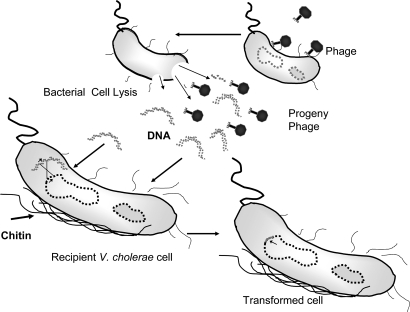
Fig. 3.
Role of lytic phages in chitin-mediated uptake of foreign DNA by V. cholerae in the aquatic environment. Lytic phages acting on V. cholerae cells release DNA from the donor cells. The free DNA fragments are taken up by recipient strains, aided by chitin-induced competence. Possible recombination between the foreign DNA and the host chromosome can cause genetic changes in the recipient cell.
Discussion
The emergence of genetic diversity among V. cholerae strains has been attributed to various factors, including the roles of mobile genetic elements, bacteriophages, and, more recently, the competence of V. cholerae to take up and assimilate free DNA from the environment while growing on a chitin substrate (1, 14). The results reported herein provide useful insight into how a combination of natural factors, including phages and chitin-induced transformation, can lead to the emergence of genetic variants among toxigenic V. cholerae strains.
An important recent development has been the emergence of hybrid strains of V. cholerae O1, which have traits of both the classical and El Tor biotypes (8, 9, 19). The most notable feature in some of these strains is the unusual presence of CTXclass prophage in an otherwise El Tor biotype background. Although previous studies suggested that these hybrid strains are the result of acquisition of the CTXclass prophage by El Tor strains, the donor of the CTXclass phage genome was unknown. Because the classical biotype of V. cholerae O1 now seems to be extinct, the identification of V. cholerae O141 strains carrying CTXclass prophage is significant, particularly in exploring the origin of the CTXclass prophage present in recently emerged variants of V. cholerae O1 El Tor strains. This finding also demonstrates that despite the disappearance of the classical biotype of V. cholerae O1, the CTXclass phage genome continues to thrive. Similar to classical biotype strains that inherently carry the CTXclass prophage, the toxigenic O141 strains carrying CTXclass prophage identified in this study are unable to produce infectious phage particles; however, we have demonstrated a mechanism through which El Tor strains may acquire the CTXclass prophage from toxigenic O141 strains independent of phage production.
It is well recognized that genetic exchanges between divergent bacterial lineages can contribute to the success of a species in complex and fluctuating environments, such as those in which V. cholerae may reside. Several studies also have pointed to such exchanges as important factors in V. cholerae population genetics and evolution (20–22). The presence of the CTXclass prophage in the O141 strains may be another such example. The finding that all O141 strains, which carry the CTXclass prophage, also carry the classical allele of tcpA gene (Table S1), further points to extensive genetic exchanges between the O141 and O1 classical strains, unless these two groups of strains are derived from a common ancestor. It is known that the CTX phage uses the TCP as a receptor for infecting susceptible V. cholerae cells (3). The existence of both CTX and TCP in these strains also suggests that the O141 strains may have acquired the CTX in a TCP-dependent manner, that is, by infection with a CTXclass phage. Thus, the CTXclass prophage, which is now mostly defective and unable to produce infectious particles, presumably was able to produce phage particles in the recent past.
The classification of CTX phages based on the diversity of the rstR gene reflects the type of CTX phage normally found in the El Tor and classical biotype strains; however, the significance of this classification on the epidemiology of the disease is not well understood. Although cholera toxin (CT) encoded by CTXclass and CTXET phages is almost similar, there are minor but presumably important differences in the primary structure of the B subunit of CT encoded by strains of the two biotypes harboring these phages. Immunologically, the B subunits of El Tor and classical CT share major epitopes but also have one or more weaker biotype-specific epitopes (23). Two related but not identical epitypes and three genotypes of CT are currently recognized (Fig. S1). CT1 is produced by classical biotype strains and by the U.S. Gulf Coast strains, whereas CT2 is produced by the El Tor biotype and O139 strains (24). The three genotypes of ctxB are based on three nonrandom base changes, resulting in changes in the deduced amino acid sequence. Genotype 1 is found in strains of the classical biotype worldwide and on the U.S. Gulf Coast, genotype 2 is found in El Tor biotype strains from Australia, and genotype 3 is found in El Tor biotype strains from the seventh pandemic and the Latin American epidemic (24). Thus, V. cholerae O1 El Tor biotype of the ongoing seventh pandemic produces CT of the CT2 epitype and genotype 3, whereas CT produced by the classical biotype belongs to the CT1 epitype and genotype 1. The emergence of El Tor strains producing classical CT appears to suggest that a target of the natural selection of various toxigenic V. cholerae strains may have been the type of toxin production, despite the observation that the classical biotype strains likely have become extinct, due to differences in ecological fitness between the classical and El Tor biotypes. Consequently, the recent emergence of El Tor biotype strains producing classical CT perhaps indicates an evolutionary optimization of the seventh pandemic strain. Continuing studies have shown that the El Tor strains that produce classical CT also are becoming the dominant type in India and Zambia (G.B.N., unpublished work).
Besides the hybrid El Tor strains that carry the CTXclass prophage and thus the rstRclass gene, another variant of V. cholerae O1 currently emerging in Bangladesh carries a CTX prophage with the El Tor type rstRET gene; however, the ctxB gene in the prophage has an identical nucleotide sequence to that of the typical classical ctxB gene (19). In the present study, a PCR-generated small DNA fragment (≈1.6 kb) from strain O395NT was successfully recombined in the resident CTX prophage of El Tor biotype strains in chitin-induced transformation (Fig. S2). Mechanistically, we presume that a similar event involving fragments of CTXclass DNA liberated by phage-mediated lysis of V. cholerae cells or other natural causes may have replaced the CT-B subunit and flanking genes of the originally resident CTXET prophage in these strains. Thus, the CTX prophages carried by these strains indeed may be hybrids of the CTXclass and CTXET phage genomes.
As mentioned earlier, El Tor strains carrying CTXclass prophage have now been isolated in several countries, including Bangladesh, India, Zambia, and Mozambique (8–10, 19). Previously, we analyzed the Mozambique strain and its CTX prophage, and the chromosomal phage integration sites, to investigate the origin of the strain (10). Our findings suggested that the Mozambique strain is derived from a progenitor similar to the seventh pandemic strain, involving multiple recombination events. In this pathway, an El Tor strain lost the CTXET phage and the TLC and RS1 sequences through a loop-out recombination event between related att sites, and subsequently acquired a CTXclass phage genome to evolve as the Mozambique strain. In the present study, we were able to convert multiple isolates of the seventh pandemic strain into derivatives carrying CTXclass phage and retaining the RS1 sequence and, occasionally, the originally resident CTXET phage. The CTX profiles of these transformants were more like hybrid strains referred to as the “Matlab variants” isolated from patients in Bangladesh (9). Thus, it appears that El Tor strains that carry the CTXclass prophage may belong to several clones originating in different regions through more than one evolutionary pathway.
Chitin induces the expression of a 41-gene regulon involved in colonization and metabolism of chitin that includes genes predicted to encode a type IV pilus assembly complex (25). It has been suggested that during interepidemic periods, toxigenic V. cholerae exists in an unexplained ecological association with chitinaceous zooplanktons (e.g., copepods), and that a bloom of these crustaceans can eventually lead to cholera outbreaks (15). However, it should be clarified that chitin is an abundant source of carbon, nitrogen, and energy for many marine microorganisms. Although the attachment of V. cholerae O1 with copepods has been suggested, the genetic program for “chitin utilization” (25) is presumably shared by different Vibrio species and various strains within the species and is not confined to toxigenic V. cholerae O1.
Many Vibrio species that live in aquatic environments can use chitin as a carbon source, and the expression and regulation of genes involved in chitin utilization and the mechanism by which the Vibrio species attach to and colonize chitin surfaces are under investigation (25). The association with crustaceans is unlikely to have any direct implications in the enrichment of epidemic V. cholerae strains per se; however, the finding that chitin promotes DNA uptake by V. cholerae may facilitate the generation of diversity among the species. Our results demonstrate the role of a combination of two natural factors—phages and chitin induction—in the transformation of V. cholerae. Whereas chitin promotes natural transformation, vibriophages acting on susceptible bacteria can provide a ready source of DNA liberated from these bacteria (Fig. 3). This observation may have profound significance in understanding the interactions of multiple factors promoting V. cholerae evolution and producing genetically diverse pathogenic strains.
Materials and Methods
Bacterial Strains.
The environmental V. cholerae strains were isolated from surface water in Bangladesh. Clinical strains used as recipients in the transformation experiments were obtained from our culture collection. The genetically marked strains used either were obtained from our collection or were constructed during the present study. The relevant strains are described in Table S2.
Isolation of V. cholerae from Water.
Water samples were cultured to isolate V. cholerae as described in ref. 26. In brief, 25-ml aliquots of water were added to 225 ml of alkaline peptone water (peptone 1% [wt/vol] NaCl 1% [wt/vol], pH 8.5) and incubated at 37°C for 6 h. Dilutions of this enriched culture were plated on tauracholate tellarite gelatin agar plates (27) and grown overnight at 37°C. Suspected Vibrio colonies were picked and subjected to biochemical and serologic tests to identify V. cholerae belonging to the O1, O139, and non-O1, non-O139 serogroups (28). Relevant non-O1, non-O139 strains were later tested with O141 serogroup-specific antiserum to identify O141 strains. Environmental water samples are routinely tested in our laboratory for the presence of V. cholerae under a surveillance system in which weekly samples from 11 surveillance sites in Dhaka, Bangladesh (18) are tested.
O141 Serogrouping.
Antiserum was produced against a strain of V. cholerae O141 (strain V51). Bacteria were grown in brain heart infusion broth (Difco) for 20 h with continuous shaking. The bacteria were pelleted by centrifugation, washed, and reconstituted in physiological saline to 109 cfu/ml. Adult New Zealand White rabbits were immunized intravenously at 6-day intervals with 0.2-, 0.5-, and three 1.0-ml doses of bacterial suspension. A booster dose of 2 ml was administered 20 days after the last injection. The rabbits were killed 7 days after the last dose. The antiserum to V. cholerae O141 was made specific by absorbing the antiserum with a rough strain of V. cholerae (strain CA385) and strains representing several serogroups, including O1, O139, O22, and O155, as described in ref. 29. Selected non-O1, non-O139 strains were tested by slide agglutination assay to detect O141 strains.
Isolation of V. cholerae O141-Specific Phage from Water.
V. cholerae O141 strains V47, V48, V49, and V51 were used as potential indicator strains to detect and isolate V. cholerae O141-specific phage from environmental water using the plaque assay as described in ref. 13. Phages from representative plaques were purified further, and the specificity of each phage was tested using a panel of strains belonging to different species and serotypes.
Probes and PCR Assays.
The presence of virulence genes was investigated using specific DNA probes or PCR assays as described in ref. 26, with relevant PCR amplicons used as probes for hybridization whenever appropriate. Probes for the Vibrio seventh pandemic islands (VSP-I and VSP-II) (30) were generated by PCR from V. cholerae strain N16961 using primers based on published sequence of the respective genes (31). Probes were labeled using [α-32P]-deoxycytidine triphosphate (3000 Ci/mmol; Amersham Biosciences), and Southern blot hybridizations were conducted following standard methods (32).
Transformation Protocol.
Transformation of V. cholerae cells in the presence of chitin was done using the method described by Meibom et al. (14), with some modifications. Overnight cultures of the recipient V. cholerae strains were diluted 1:100-fold in LB medium and grown to an OD600 of ≈0.3. The bacteria were pelleted by centrifugation, washed, and resuspended in a 0.10 volume of filter-sterilized environmental water or 0.5% sterile sea salt solution (SS). Aliquots of a 2-ml bacterial suspension were dispensed into the wells of a 12-well tissue culture plate containing sterile pieces of shrimp shell. After incubation at 30°C, grown statically for 24 h, the planktonic phase was removed, and fresh water or SS was added. At the same time, 1–2 μg of the appropriate DNA was added to the wells. After 24 h, the shrimp shells were removed from the wells, washed in SS, and vortexed in SS to release any attached bacteria. The released bacteria were then plated onto LB plates without antibiotic and onto appropriate antibiotic-containing (75 μg/ml of kanamycin or 100 μg/ml of streptomycin) LB plates.
For transformation in the presence of phage, the donor and recipient strains grown and washed as above, were mixed together with a lytic phage specific to the donor cells (≈105 cfu/ml of each bacterial strain and 105 pfu phage per ml). Aliquots of the mixture were immediately dispensed into the wells of tissue culture plates containing pieces of sterile shrimp shell. After 24 h, the cells were released from the shrimp shells as before and plated on appropriate antibiotic plates to deselect the donor cells and select the transformants. Suspected transformants were further analyzed using PCR and hybridization assays to confirm the presence of the relevant genes. Total DNA or plasmid DNA was extracted from overnight cultures by standard methods (32) and purified using a microcentrifuge filter unit (Ultrafree-Probind; Sigma). The presence of chromosomally integrated CTX phage genomes or possible extrachromosomal replicative forms was analyzed by Southern blot analysis.
Labeling the CTXclass Prophage in V. cholerae O141 Strains.
The resident CTXclass prophages in several O141 strains (Fig. 1) were marked with a KmR marker, exploiting the chitin-induced transformation procedure (14). In brief, a 1.6-kb fragment between the ctxA promoter region and the end of ctxB gene, or a 2.7-kb region spanning parts of the zot and ctxB genes of V. cholerae strain O395NT (16), which carries the KmR marker in the CTX prophage, disrupting the ctxAB operon, was amplified by PCR. These amplicons, which contain the KmR marker flanked by CTX phage genes, were used to transform and recombine with the CTXclass prophage in toxigenic O141 strains. The recombinants were selected on LB-agar plates containing kanamycin (75 μg/ml) and further analyzed by PCR and Southern blot hybridization to confirm the acquisition of the KmR marker in the CTXclass prophages resident in these strains (Fig. 1).
Assay for CTX Phage Production.
To determine whether the toxigenic V. cholerae O141 strains or their KmR derivatives produced infectious CTXclassφ particles, the strains were analyzed both under laboratory conditions and inside the intestines of infant mice as described in ref. 17. In brief, strains were grown in LB at 30°C up to an absorbance at 540 nm (A540) of 0.2. The cells were collected by centrifugation, washed and resuspended in fresh LB. The suspension was divided into aliquots, to which mitomycin C (Sigma) was added to a final concentration of 20 ng/ml, and incubated for 6 h at 30°C. Supernatant fluids were filtered through 0.22-μm filters (Millipore), and the filtrates were analyzed for KmR-marked CTX phages by transduction assays (17).
For the in vivo assay, the same mixtures of phage and recipient cells, and freshly prepared mixtures of donor (KmR-marked) and recipient bacterial cells, were used to gastrointestinally inoculate groups of 5-day-old Swiss albino mice. The animals were killed after 16 h, and their intestinal contents were analyzed using appropriate antibiotic-containing plates to counterselect the donor cells and identify possible KmR derivatives of recipient cells. Strain SM44, an El Tor biotype strain carrying a KmR marker in the CTX prophage, was used as a positive control strain in the assays for phage production.
For unmarked wild-type strains, the possible presence of CTX phages in the culture supernatant was analyzed through detection of phage DNA. Culture filtrates were mixed with 0.25 volumes of a solution containing 20% polyethylene glycol (PEG-6000) and 10% NaCl and then centrifuged at 12,000 × g to precipitate possible phage particles. The precipitate was dissolved in a solution containing 20 mmol Tris-Cl (pH 7.5), 60 mmol KCl, 10 mmol MgCl, and 10 mmol NaCl and then digested with pancreatic DNaseI (100 units/ml) and RNase A (50 μg/ml) at 37°C for 2 h, to remove possible nucleic acids carried over from lysed bacterial cells. The solution was extracted with phenol-chloroform to disrupt possible phage particles, and the total nucleic acids were precipitated with ethanol. The nucleic acids were analyzed by enzymatic digestion and Southern blot hybridization using appropriate probes to detect the presence of CTX-phage DNA.
Supplementary Material
Acknowledgments.
This research was funded in part by National Institutes of Health Grants 2RO1-GM068851-5 and AI070963-01A1 under sub-agreements between Harvard Medical School and the International Centre for Diarrhoeal Disease Research, Bangladesh. The International Centre for Diarrhoeal Disease Research, Bangladesh is supported by countries and agencies that share its concern for the health problems of developing countries.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805560105/DCSupplemental.
References
- 1.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Morris JG, Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous bacteriophage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 4.Kimsey HH, Waldor MK. CTXφ immunity: Application in the development of cholera vaccines. Proc Natl Acad Sci USA. 1998;95:7035–7039. doi: 10.1073/pnas.95.12.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis BM, Moyer KE, Boyd EF, Waldor MK. CTX prophages in classical biotype of Vibrio cholerae: Functional phage genes but dysfunctional phage genomes. J Bacteriol. 2000;182:6992–6998. doi: 10.1128/jb.182.24.6992-6998.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis BM, Kimsey HH, Chang W, Waldor MK. The Vibrio cholerae O139 Calcutta bacteriophage CTXΦ is infectious and encodes a novel repressor. J Bacteriol. 1999;181:6779–6787. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE. Characterization of VPI pathogenicity island and CTXφ prophage in environmental strains of Vibrio cholerae. J Bacteriol. 2001;183:4737–4746. doi: 10.1128/JB.183.16.4737-4746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ansaruzzaman M, et al. Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis. 2004;10:2057–2059. doi: 10.3201/eid1011.040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nair GB, et al. New variants of. Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol. 2002;40:3296–3299. doi: 10.1128/JCM.40.9.3296-3299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque SM, et al. Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci USA. 2007;104:5151–5156. doi: 10.1073/pnas.0700365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki MT, et al. Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb Ecol. 2004;48:473–488. doi: 10.1007/s00248-004-0213-5. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque SM, et al. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci USA. 2005;102:1702–1707. doi: 10.1073/pnas.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meibom KL, Blokesch M, Dolganov NA, Wu C, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 15.Colwell RR, Huq A. In: Vibrio cholerae and Cholera: Molecular to Global Perspectives. Wachsmuth IK, Blake PA, Olsvik O, editors. Washington, DC: ASM Press; 1994. pp. 117–133. [Google Scholar]
- 16.Mekalanos JJ, et al. Cholera toxin genes: Nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 17.Faruque SM, et al. Analysis of environmental and clinical strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: Molecular basis for the origination of new strains with epidemic potential. Infect Immun. 1998;66:5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruque SM, et al. Self-limiting nature of seasonal cholera epidemics: Role of host-mediated amplification of phage. Proc Natl Acad Sci USA. 2005;102:6119–6124. doi: 10.1073/pnas.0502069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair GB, et al. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006;44:4211–4213. doi: 10.1128/JCM.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltran P, et al. Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol. 1999;37:581–590. doi: 10.1128/jcm.37.3.581-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bik EM, Bunschoten AE, Gouw RD, Mooi FR. Genesis of the novel epidemic Vibrio cholerae O139 strain: Evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubey RS, Lindblad M, Holmgren J. Purification of El Tor cholera enterotoxins and comparisons with classical toxin. J Gen Microbiol. 1990;136:1839–1847. doi: 10.1099/00221287-136-9-1839. [DOI] [PubMed] [Google Scholar]
- 23.Finkelstein RA, et al. Epitopes of the cholera family of enterotoxins. Rev Infect Dis. 1987;9:544–561. doi: 10.1093/clinids/9.3.544. [DOI] [PubMed] [Google Scholar]
- 24.Olsvik O, et al. Use of automated sequencing of polymerase chain reaction–generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol. 1993;31:22–25. doi: 10.1128/jcm.31.1.22-25.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meibom KL, et al. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faruque SM, et al. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera epidemic area. Proc Natl Acad Sci USA. 2004;101:2123–2128. doi: 10.1073/pnas.0308485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monsur KA. A highly selective gelatine-taurocholoate-tellurite medium for the isolation of Vibrio cholerae. Trans R Soc Trop Med Hyg. 1961;55:440–442. doi: 10.1016/0035-9203(61)90090-6. [DOI] [PubMed] [Google Scholar]
- 28.World Health Organization, Bacterial Disease Unit. Guidelines for the Laboratory Diagnosis of Cholera. Geneva, Switzerland: WHO; 1974. [Google Scholar]
- 29.Albert MJ, et al. Characterization of Aeromonas trota strains that cross-react with Vibrio cholerae O139 Bengal. J Clin Microbiol. 1995;33:3119–3123. doi: 10.1128/jcm.33.12.3119-3123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dziejman M, et al. Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci USA. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab Press; 1982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.