Abstract
Nucleosome core particles in eukaryotes are linked by a stretch of DNA that is usually associated with a linker histone. Here, we show in yeast, that the presence of yeast linker histone Hho1p represses expression of a pol II transcribed gene (MET15) embedded in the rDNA. In vivo deletions of Hho1p sequences showed that the second globular domain is sufficient for that repression, whereas the presence of the N terminus is required for its derepression. In contrast, a run-on assay confirmed by a ChIP experiment showed that Hho1p is required for maximal pol I processivity during rDNA transcription. Psoralen accessibility experiments indicated that Hho1p is necessary for normal rDNA compaction. DNA array expression analysis comparing RNA transcripts in wild-type and hho1 strains before and after a heat-shock showed that Hho1p is necessary to achieve wild-type mRNA levels of transcripts that encode ribosomal components. Taken together, our results suggest that Hho1p is involved in rDNA compaction, and like core histones, is required for efficient rDNA transcription by pol I.
Keywords: chromatin, rDNA
The core histones of the nucleosome are among the most phylogenetically conserved proteins known (1). DNA (146 bp) are wrapped around the histone core in nearly two turns. In most eukaryotes, there is a stretch of DNA ranging from <20- to ≈70-nt bp, linking the nucleosome core particles (2). An additional histone, termed linker histone or histone H1, is usually associated with this DNA. When either native or reconstituted chromatin is treated with micrococcal nuclease, limited DNA digestion results in DNA fragments 146 bp long, whereas in the presence of linker histone, an additional 20 bp are protected (3), indicating that the linker histone interacts with DNA that extends beyond the nucleosome core.
Sequence and structural comparisons between known linker histones (4) have shown that most linker histones contain a globular domain having a winged helix-turn-helix motif (5–7). This domain has been shown to asymmetrically bind the DNA as it enters and exits the nucleosome. Specific sites on the globular domain have been identified that are required for the interactions between the histone and the DNA (7, 8). Although it is largely unstructured, the C terminus has been shown to facilitate the formation of a stem structure of the DNA entering and exiting the nucleosome core (9, 10). Neither the N nor the C termini sequences are conserved (11), although both are heavily basic, and their amino acid composition is similar (12).
The temporal transcription of histones, including linker histones, is very tightly cell-cycle-regulated, with transcription occurring during the S phase of the cell cycle (13). As is the case in other organisms, the yeast linker histone gene, HHO1, is transcribed during the S phase of the cell cycle (14). Furthermore, in a high-throughput chromatin immunoprecipitation experiment, of all 32 proteins tested (all involved in regulating various stages of gene regulation), none generated a ChIP pattern that was more similar to the four core histones than Hho1p (15). In two immunoprecipitation experiments, Hho1p was identified in a complex together with the core histones (16, 17). Together, these data argue strongly that yeast HHO1 encodes a functional homologue to linker histones in other species.
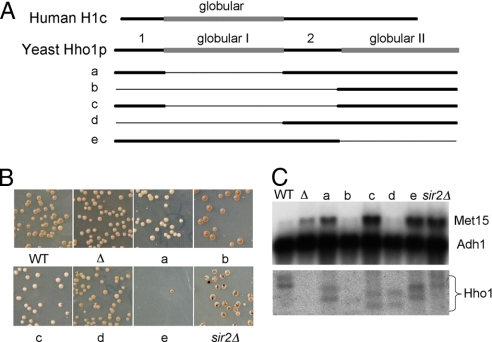
Despite its similarity to other linker histones, sequence comparison of Hho1p with other linker histones showed that it contains an additional globular domain (18), and that both globular domains conform to the canonical winged helix structure (19, 20). The protein sequences flanking the first globular domain resemble the N and C termini of typical linker histones. Fig. 1A shows a schematic comparison of human histone H1c and the yeast linker histone Hho1p.
Fig. 1.
An HHO1 disruption results in increased MET15 expression. (A) Schematic diagram illustrating hho1 deletions a-e. Human H1c is drawn as a reference (drawn to scale). (B) Mutant and wild-type strains were plated on Pb+ indicator plates. The lighter the color of the colony, the more MET15 expression, and the lower the presumed rDNA compaction. Sectors resulting from homologous recombination in the rDNA and loss of the MET15 gene are most numerous in the sir2Δ strain (JS218). Colonies were plated at 30°C for 5 days and then at 4°C for 1 additional week before photography. (C) Northern blot monitoring the expression of MET15 in the different mutants of Hho1 and sir2. Fragments of MET15 and ADH1 were used simultaneously as probes. Identical amounts of total cell RNA were loaded in each lane. A parallel gel (shown below) was probed with an Hho1 fragment. There are consistently two different-sized transcripts for Hho1. The differing sizes of the Hho1 transcripts in the different lanes correspond to the deletions made in the Hho1 gene by the mutagenesis. The hho1Δ deletion strain shows no hho1 transcript. Quantitation of the transcripts, normalized to the Adh1 probe, is presented in Table S1).
The linker DNA in yeast is unusually short (21–23). Because the globular domain of linker histones has been shown to bind DNA entering and exiting the nucleosome, it was proposed that each globular domain might individually bind an adjacent nucleosome, where the short linker distance is regulated by other factors (18, 20, 23). Alternatively, it was proposed that Hho1p could be responsible for the short distance between nucleosomes by binding adjacent nucleosomes simultaneously (18).
A number of NMR studies have investigated the structure of Hho1p with the goal of understanding the nature of the two domains (19, 20, 24). Each domain has been shown to have the typical winged helix-turn-helix motif. However, whereas the first globular domain is relatively structurally stable, the second one is much less so (19, 20).
Although linker histones appear to be ubiquitous in all eukaryotes and they have been intensively studied for many years, a clear understanding of the precise function of these proteins remains elusive. This stems largely from the absence of a clear phenotype in cells lacking linker histones. In lower eukaryotes, knockouts of linker histones had little measurable effect on phenotype. In Tetrahymena linker histone, knockouts in the macronucleus had little effect on RNA levels of different transcripts, although the volume of the nucleus increased by about a factor of two (25). In yeast, Hho1p disruption resulted in a small measurable increase in homologous recombination and a small shortening of life span (26). In higher eukaryotes, the presence of linker histones is required for proper embryogenesis (27, 28).
An earlier study in our laboratory showed, using chromatin immunoprecipitation, that Hho1p is preferentially localized to the rDNA (29). In this article, we describe experiments that functionally analyze yeast linker histone Hho1p, using a model system developed by the Boeke laboratory to monitor rDNA chromatin structure (30). In that system, MET15 is embedded in one of the rDNA repeats, and its expression is monitored in various genetic backgrounds. We constructed yeast strains that express only portions of Hho1p and tested the expression of the MET15 transcript embedded in the rDNA in each of these strains. Here, we show evidence that the second globular domain of Hho1p alone is sufficient to repress the expression of the MET15, whereas the presence of the N terminus of Hho1p promotes its transcription. By measuring psoralen accessibility to the rDNA we demonstrate that yeast linker histone is required for normal compaction of the chromatin at the rDNA locus. Finally, we show that normal processivity of RNA polymerase I transcription of rDNA requires the presence of HHO1.
Results
Hho1p Impedes Expression of a Pol II Transcript Embedded in rDNA.
To address the question of whether we could detect Hho1p function in the rDNA, we used a system developed by the Boeke laboratory to investigate the function of other proteins known to bind the rDNA. In that system, the transcription of a MET15 gene embedded in the NTS2 of one rDNA repeat is monitored by examining the color of the colonies when plated on Pb+ plates (30). Colonies expressing high levels of Met15p are white, whereas low levels of Met15p result in colonies having various shades of brown color that correlate with MET15 expression. When the MET15 gene product is completely absent, the colonies are very dark, approaching black. When MET15 is lost as a result of recombination, colonies form very dark brown sectors.
To test the effect of Hho1p on MET15 expression embedded in the rDNA, we created a strain in which the entire HHO1 coding sequence was deleted. We then compared the colors of the colonies when plated on Pb+ containing medium. As seen in Fig. 1B, hho1Δ colonies were significantly whiter than wild-type colonies that developed a brown color. This indicated that MET15 expression was higher in the hho1Δ strain than in the wild type. We verified these results by performing a Northern blot using RNA extracted from exponentially growing cells, probing with a MET15 probe. Deletion of HHO1 resulted in a significant increase in MET15 RNA levels, as can be seen in Fig. 1C. No significant change was seen in the sectoring of the colonies, indicating that homologous recombination was not drastically changed in the rDNA by deletion of HHO1. These data indicate that Hho1p serves to repress the RNA polymerase II transcription of MET15 when embedded in the rDNA.
The Second Globular Domain of Hho1p Is Sufficient to Partially Repress MET15 Expression, Whereas the Hho1p N Terminus Relieves This Repression.
As described above, yeast linker histone Hho1p contains two globular domains. Although the two globular domains are similar, they have been shown to have some structural differences (19, 20, 24, 31). The first globular domain (closer to the N terminus) is structurally more stable than the second globular domain, raising the possibility that only the first domain is functional. To address these and other questions, we used the “delitto perfetto” strategy (32) to express only parts of the Hho1p molecule in yeast strains harboring the MET15 gene embedded in their rDNA. Details of these constructs (mutants a-e) can be seen in Fig. 1A. Primers used for their construction are listed in Table S2. In each of these strains, the mutant hho1p was expressed from its native chromosomal location without any additional modifications (e.g., insertion of a selectable marker).
Using the MET15 assay described above, we monitored the color and sectoring of colonies formed when the mutant strains that lacked one or more Hho1p domains were plated on Pb+ indicator plates. In addition, we performed Northern analysis on RNA extracted from each of the cultures grown in rich media, hybridizing with a MET15 radiolabeled sequence. Our results clearly showed that the level of MET15 mRNA detected by colony color and Northern analysis depended on the hho1 mutant, as can be seen in Figs. 1 B and C.
Strikingly, hho1 mutants b and d that contain the second globular domain but do not contain the N terminus have low levels of MET15 expression, similar to wild type. However, like the sir2Δ strain, mutants a, c, and e, each of which contain the N terminus, express high levels of MET15. Our data also indicate that the second globular domain itself (mutant b) is sufficient to repress MET15 transcription embedded in the rDNA, whereas when the N terminus is present together with the second globular domain (mutant c), this repression is lost. Levels of ADH1 RNA expressed from its normal chromosomal location are unaffected by any of the hho1 mutants (Fig. 1C).
Processivity of RNA Polymerase I Depends on Hho1p.
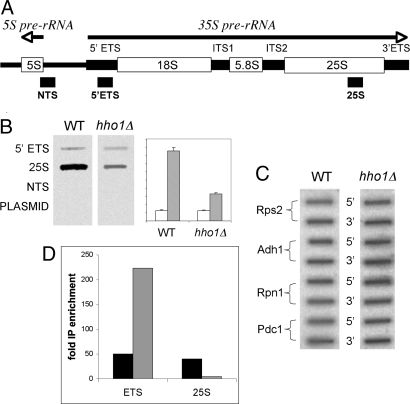
The experiments described above used a model system in which an RNA polymerase II transcribed gene (MET15) is used to monitor rDNA structure. Because this system is somewhat artificial and might not reflect native rDNA structure accurately, we asked how RNA polymerase I transcription in native rDNA is affected by the presence of Hho1p. To that end, we used a transcription run-on assay in which actively transcribing cells in the log phase of growth were labeled for a short time (10 min) with radiolabeled UTP. RNA was immediately extracted from wild-type and hho1Δ strains and hybridized with set amounts of short fragments of rDNA fixed to nitrocellulose in a slot blot. Each fragment represents a different region of the rDNA repeat. Three DNA fragments were chosen (see Fig. 2A) (33): (i) 5′ ETS close to the 5′ end of the 35S pre-rRNA transcript; (ii) 25S near the 3′ end of the transcript; and (iii) NTS, a sequence that is not transcribed. Each of the sequences is cloned in a plasmid vector.
Fig. 2.
Pol I processivity is defective in the hho1Δ mutant. Run-on transcription was performed as described in Methods. (A) Schematic diagram of the rDNA transcription unit. RNA polymerase I transcribes the 35S primary rRNA transcript. DNA sequences serving as probes in B and corresponding to the indicated segments of the nontranscribed spacer (NTS), the 5′ external transcribed spacer (5′ ETS), and the 25S rRNA are indicated. (B) Slot blot hybridization of the probes described in A with labeled RNA from wild-type and hho1Δ cells. Quantitation of two separate experiments with standard deviation bars on right (arbitrary abscissa units). Open and stippled bars represent ETS and 25S probes, respectively. (C) Slot blot hybridization of PCR amplified DNA fragments from 5′ and 3′ ends of genes transcribed by pol II. Primers are listed in Table S3. (D) Quantitation of ChIP experiment verifying defective RNA pol I processivity in hho1Δ. cross-linked, HA-tagged RNA pol I was immunoprecipitated from wild type and hho1Δ cells together with associated DNA. After cross-link reversal, quantitative PCR of ETS and 25S rDNA sequences were performed. Black and stippled bars represent enrichment of the DNA sequences relative to input DNA precipitated from wild-type and hho1Δ extracts, respectively.
As can be seen in Fig. 2B, the labeled RNA from both hho1Δ and wild-type strains hybridized to the 5′ ETS and the 25S rDNA fragments, whereas hybridization to the untranscribed DNA fragment and to the plasmid vector DNA was undetectable. Strikingly, however, significantly less labeled RNA isolated from the hho1Δ mutant hybridized to the 25S rDNA. Quantitation indicated there is about a 3-fold reduction in the hybridization to the 25S rDNA of the RNA isolated from the hho1Δ mutant relative to wild type.
Similar experiments with DNA fragments from the 5′ and 3′ of four different pol II transcribed genes (Fig. 2C) showed no difference in hybridization to the labeled RNA from either the wild-type or hho1Δ strains.
To verify these experiments a ChIP experiment was performed by using an HA-tagged RNA polymerase I, which replaced the native pol I. Probes from within the ETS and 25S were used to measure pol I occupancy during logarithmic growth. Although little difference was seen between pol I occupancy in the wild type, a 10-fold reduction was seen in the downstream 25S probe in the hho1Δ strain, confirming the transcription run-on experiment (Fig. 2D).
Rate of rRNA Synthesis Is Unaffected by the Presence of Hho1p.
To test the rate of rRNA synthesis, we pulse-labeled RNA in wild-type and hho1Δ cultures in midlog phase with 3H-uridine for 5 min. We then extracted the RNA and counted cpm/μg of RNA in a scintillation counter. All counts were TCA-precipitable, indicating that the measurement reflected incorporated 3H-uridine into RNA. No significant difference was observed between the strains in three parallel repeated experiments.
Complete rDNA Compaction Depends on Hho1p.
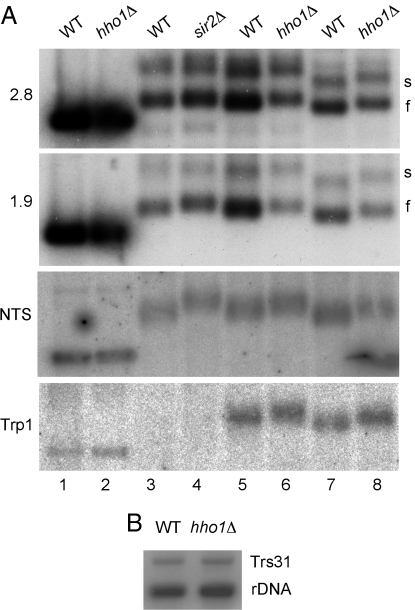
Accessibility to psoralen has been successfully used to distinguish between actively transcribing (“slow”) and quiescent (“fast”) copies of rDNA (34) and to evaluate the degree of chromatin compaction (30). As can be seen in Fig. 3A, deletion of Hho1p [like Sir2p (30)] results in increased accessibility of rDNA to psoralen that is detected by its reduced mobility in an agarose gel at all three loci tested (NTS, ETS, and 25S), indicating that Hho1p contributes to chromatin compaction. Interestingly, a similar result was obtained at TRP1. This is not surprising, because Hho1p is scattered throughout the chromatin (15). No significant difference in rDNA copy number was detected between wild-type and hho1Δ strains (Fig. 3B).
Fig. 3.
Deletion of Hho1 leads to increased chromatin accessibility to psoralen. (A) Psoralen cross-linking of rDNA. Wild-type, hho1Δ, and sir2Δ logarithmic cell cultures were exposed to 10 μg/ml psoralen and 365-nm UV light for 40 min. DNA was extracted, digested with EcoR1, electrophoresed on a 1.3% agarose gel, the cross-linking reversed and blotted onto a nitrocellulose membrane. The blot was probed with probes that recognize 1.9 and 2.8 kb fragments of the 35S rRNA transcript, the rDNA NTS, and the TRP1 gene. Strains used for the SIR2 control were deleted for TRP1. Blots were exposed to a phosphorimager screen. Cultures in lanes 1–6 were harvested at 0.3 OD600, lanes 7 and 8 at 0.9 OD600. (B) Copy number comparison of wild-type and hho1Δ strains. Extracted DNA was digested with EcoR1 and BamH1, electrophoresed, and blotted as above. The blot was probed with a mixture of two probes recognizing the 1.9-kb rDNA fragment and single-copy TRS31 DNA. Quantitation of the bands showed an rDNA/TRS31 ratio of 4.31 in the wild-type and 3.74 in the hho1Δ strain.
Hho1p Affects RNA Polymerase II Transcription at Sites Outside the rDNA Locus.
A recent article described changes in binding of a large number of proteins to yeast chromatin after heat shock using a “ChIP-on-chip” approach (15). Looking at the occupancy levels of quiescent and active RNA polymerase II promoters relative to non-promoter regions, the authors (15) showed that promoter occupancy by Hho1p is significantly lower at active promoters than at non-promoter regions, as is the case for core histones. Furthermore, upon heat shock, Hho1p is redistributed on the chromatin in a similar manner as core histones.
Against this background, we decided to revisit the question of polymerase II gene expression across the genome as a function of Hho1p presence and heat shock. We used a DNA array produced by Agilent on which 60-mer oligonucleotides are synthesized directly on the slides. This process yields DNA spots of very similar size, excellent uniformity, and small molar variability between spots. We expected that our results using this DNA array would be more reliable than results published earlier using a spotted DNA array (35). We extracted RNA from logarithmically growing wild-type and hho1Δ cells at both 25°C and 37°C after a 15-min heat shock. cRNA was synthesized from these RNA samples, labeling with Cy5 and Cy3, as described in supporting information (SI) Text.
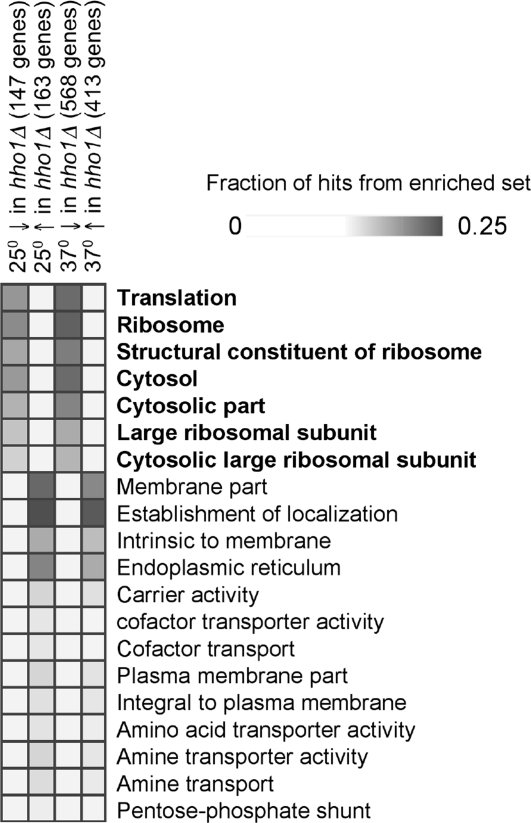
When comparing the wild type with the hho1Δ mutant grown at 25°C, examination of all genes except those with very low expression (4,285 genes with raw scores >500) showed that only 147 genes had RNA levels >1.3-fold higher in the wild type than in the hho1Δ mutant, whereas 163 genes had RNA levels >1.3-fold higher in the hho1Δ mutant than in the wild type. We found no correlation between the genes reported to be differentially expressed in the earlier publication (35) and our results. After a heat shock from 25°C to 37°C, 568 genes showed >1.3-fold lower expression levels in the hho1Δ mutant than in the wild type, whereas 413 genes showed >1.3-fold higher expression levels in the mutant than in wild type.
To characterize the gene sets that were differentially expressed (Datasets S1 and S2), we used Gene Set Enrichment software (36) (http://genie.weizmann.ac.il/genomica_web/gene_sets.jsp) that compares input gene sets with published gene sets derived from published DNA array experiments and from Gene Ontology (GO) annotations. When we compared the list of genes whose activity was higher in the hho1Δ mutant than in the wild type, either when cells were grown at 25°C or heat shocked to 37°C, we found no clear molecular function categories that were significantly enriched, although there was enrichment of genes that encode membrane bound proteins (Fig. 4). In contrast, when we compared the list of genes whose activity was lower in the hho1Δ mutant than in the wild type, we found that a large proportion of the genes whose products are essential for translation were significantly down-regulated. In particular, the RNA encoding most of the structural components of the ribosome was found in significantly lower amounts in the hho1Δ mutant than in the wild type, both at 25°C and after heat shock (see Fig. 4). These results complemented the results presented above that showed that rDNA is transcribed less efficiently in the hho1Δ mutant.
Fig. 4.
GO ontology of gene lists generated by expression microarray analysis. The fraction of hits from sets that are found in different GO ontology sets are displayed graphically (36). Here, we show only 20 of the categories. The analysis shown here was performed on gene sets after removal of genes expressed at a very low level (<500 units in the raw data). Above each column, the title relates to whether the gene set was higher (↑) or lower (↓) in the hho1Δ mutant than the wild type, and how many genes were in each set (in parentheses). GO ontologies that relate to translation are bolded. The complete figure and quantitative results can be found in Fig. S1.
Discussion
Linker histones are less evolutionarily conserved than core histones. Furthermore, it is clear that although linker histones have an important role in the dynamics of chromatin structure, they function in subtle ways (37). Functional studies have focused on reconstituted chromatin with mutant linker histones and on in vivo systems lacking one or more isoforms of the histone. The in vivo studies were limited by the lack of a convenient system in which mutant linker histone could be expressed from its own promoter at its native position in the chromosome without any additional modifications. Furthermore, there was not a convenient assay system to measure linker histone function.
Here, we address both these limitations of previous studies and show that they can be overcome. Using the “delitto perfetto” method (32), we constructed yeast strains whose mutant linker histone Hho1p was expressed from its natural promoter at its normal chromosomal location with no other modifications.
Because we had shown earlier that Hho1p concentrates in the rDNA (29), we sought an assay that would monitor Hho1p function in that context. Smith and Boeke (30) showed that the expression of MET15, which is transcribed by RNA polymerase II, is influenced by the local rDNA structure. Here, we showed (Fig. 1) that, as was the case in sir2 mutants (30), the partial silencing of MET15 embedded in the rDNA in the wild type strains was decreased in hho1Δ colonies, as seen by their losing their brown color on the Pb+ indicator plates. In addition, varying degrees of brown color were seen in hho1 mutants expressing only portions of the molecule. These data show that this experimental system can be used effectively to monitor yeast linker histone function in vivo. Previous experiments (30) using this system indicated that MET15 expression levels were a consequence of chromatin compaction at the rDNA locus. Our psoralen accessibility results confirm that Hho1p facilitates chromatin compaction of the rDNA, and at least one other locus (TRP1).
Linker histones typically contain a conserved globular domain that is preceded and followed by basic domains whose composition but not sequence is conserved. Hho1p has a similar structure, except that an additional globular domain is found at the N terminus. Sequence comparisons of Hho1p with other linker histones have shown that the first globular domain of Hho1p is more similar to other linker histones with respect to the presence of specific conserved lysine residues in putative DNA binding site II (K59 and K61) than the second domain, which lacks these lysines at homologous positions (N192 and G194). Structurally, this first globular domain is also more reminiscent of linker histones from other organisms than the second globular domain. The second domain has a less stable structure at physiological salt concentrations (19, 24). We decided, therefore, to test these domains individually in our MET15 assay system.
As can be seen in Fig. 1, mutants a–d lack the first globular domain. In addition, each contains one or both of the domains denoted as 1, at the N terminus, and 2, between the globular domains, in the figure. Strikingly, mutants a and c have considerably higher MET15 RNA levels than the wild type or than mutants b and d. An isogenic sir2 strain had high MET15 RNA levels (30), which were similar to those in our a and c hho1 mutants.
The globular domain of linker histones has been shown to be asymmetrically positioned at the entrance and exit of the DNA from the nucleosome core, presumably facilitating higher-order compaction of the chromatin (10, 38). In vitro studies showed that the C terminus is entwined with up to 30 bp of DNA entering and exiting the core particle, producing a stem structure (9, 10). In yeast, this domain may not be available for this function, because it is relatively short and is followed by a second globular domain. It is tempting to speculate that, during evolutionary time, the duplication of the globular domain in yeast might have facilitated the ability of yeast to deal with reduced linker DNA length by preventing the DNA stem formation, thereby permitting a shorter stretch of DNA between the nucleosome cores. The poor growth phenotype of mutant e, which contains only the first globular domain was reminiscent of reports that sea urchin histone H1 expressed in yeast is toxic (39, 40). Because the overall structure of mutant e (globular domain with flanking sequences) is similar to linker histones carrying only one globular domain, it may be that the C terminus of the mutant prevents normal yeast nucleosome spacing.
In vitro studies suggest that the linker histone N terminus might somehow be able to interfere with the binding of the globular domain, thus decreasing the wrapping of DNA around the core histones (9). If the N terminus of Hho1p functions similarly, it might assist pol II in displacing nucleosomes during transcription. This idea is supported by our data that show that the N terminus (domain 1 in Fig. 1A) is required for efficient MET15 transcription. Mutants b and d each lack that domain and thus have reduced MET15 transcription, whereas mutants a, c, and e each have the N-terminal domain, and each efficiently transcribes the MET15 gene. Although the Hho1p wild type contains the N terminus, it also has two globular domains, presumably making it more difficult for the polymerase to disrupt nucleosomes, thereby resulting in low MET15 transcription.
Schäfer et al. (41) showed that both globular domains of Hho1p can bind four-way junction DNA simultaneously, implying that one molecule of Hho1p is capable of binding two nucleosomes. We propose that the globular domains each bind linker DNA and thus repress pol II transcription, whereas the N terminus of Hho1p is used by components of the transcription and/or chromatin remodeling machinery to disrupt those interactions. Such a model might explain why the wild-type yeast that contain two globular domains in Hho1p expressed MET15 at a relatively low level.
The MET15 expression assay monitors transcription of a pol II transcript embedded in the rDNA. Recent data have indicated that, whereas a nucleosomal context impedes pol II transcription, pol I transcribes more efficiently in the presence of core histones (42). We asked, therefore, how transcription of the 35S pre-RNA would be affected by the lack of Hho1p. As described above, a transcription run-on experiment showed that, as in the case of core histones, yeast linker histone is required for efficient rDNA transcription. In our assay, we used two probes within the 35S pre-RNA template, one near the start of transcription and one near its 3′ terminus. After a 10-min labeling of wild-type cells with 32P UTP, the hybridization signal using the 3′ probe was about three times more intense than the hybridization signal using the 5′ probe. However, when using an RNA probe made from the hho1Δ mutant, the intensity of the bands was the same. We interpret these data as evidence that the processivity of pol I transcription in the hho1Δ mutant is substantially lower than in the wild type. This interpretation is supported by our ChIP experiment in which RNA pol I occupancy is ≈10-fold greater at the start of the 35S pre-RNA template than near its end.
The rates of transcription appear to be unaffected by Hho1p, because a 5-min pulse of 3H-uridine labeled RNA to the same specific activity in wild-type and hho1 strains. These results support the notion that nucleosomes serve to maintain a high transcription rate of the pol I complex rather than to inhibit its progress as they do for pol II transcription. They may also be a reflection of data that indicate the pol I complex transcribes through dynamic nucleosomal chromatin (37, 43). In our cultures, lowered pol I processivity in hho1 mutants is not a limiting factor of yeast growth rate or a determinant of rRNA levels, because neither is reduced in an hho1Δ strain. Curiously, our ChIP experiment shows an ≈4-fold greater RNA pol I occupancy at the start of the 35S pre-RNA template in the hho1Δ strain than in the wild type. This might indicate a stall in the pol I downstream of the ETS probe in the hho1Δ mutant.
Although Hho1p is more abundant at the rDNA locus than at other loci (29), it is distributed throughout the genome (15). Although it was shown earlier that expression levels of only a minority of genes was affected by the disruption of HHO1 (35), we revisited this question using higher-quality DNA arrays (Agilent) and using RNA that was extracted from isogenic wild-type and hho1Δ yeast before and after heat shock. As can be seen in Fig. 4, a significant number of genes were affected by the disruption of HHO1. Our results at 25°C were in agreement with previous Northern blot results of a small number of transcripts (29). Classification of genes whose expression changed >1.3-fold according to GO annotations showed that a large proportion of the genes whose products are involved in translation are down regulated in the hho1 mutant. Although it is possible that the down-regulation is a direct consequence of the absence of Hho1p at these genes, it is more likely that it is a result of general down-regulation of rRNA transcription. It is well known that synthesis of all of the ribosomal components is highly regulated, and it has been recently shown that RNA levels encoding ribosome components depends on rRNA transcription (44, 45).
When our datasets were compared with datasets from other DNA array experiments, we found a significant correlation with ChIP-on-chip experiments that used RAP1 (46) and RPD3 (47). Rap1p targets the promoters of genes that encode ribosomal proteins, thus it is not surprising that the datasets overlap. Rpd3p (a histone deacetylase) is part of a complex that contains Sin3p and Sap30p and is involved in transcriptional silencing. It has been shown to be required for the silencing of rDNA genes entering stationary phase (48) and for maintenance of rDNA chromatin structure (49). The overlap of these datasets with ours suggests that Hho1p may functionally interact with Rap1p and Rpd3p at the rDNA to modulate its chromatin structure.
The assays demonstrated in this article now afford the opportunity to use the power of yeast genetics to functionally dissect Hho1p and to study its interactions with other proteins in the context of rDNA. As was found previously for H3 and H4 core histones, yeast linker histone Hho1p acts to inhibit transcription of a pol II transcript (MET15), whereas it is necessary for efficient transcription of a pol I transcript. It will be interesting in the future to continue dissection of the Hho1p molecule to better understand how linker histone functions in different chromatin contexts.
Methods
Yeast Strains and Growth Conditions.
The parent yeast strain for mutant constructs in this article was JS237 (MATα, his3Δ200, met15Δ1, ura3-167, leu2Δ1, RDN1::Ty1-met15) (30). This strain carries MET15 in the rDNA. JS218 is sir2::His-3. Experiments comparing wild-type and hho1Δ strains used BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and its derivative, Y02125 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YPL127C::kanMX4), respectively, obtained from Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/col_index.html). The parent strain carrying 6X HA-tagged RNA polymerase I was Y312 (MATa, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, trp1::hisG, can1::hisG, ade2::hisG, RPA42::HA-TRP1). A strain deleted in Hho1 was made by transformation with a PCR fragment carrying YPL127C::kanMX4.
Mutant strain construction is described in SI Text. Pb+ indicator plates were as described (30), except they contained 40 mg of adenine hemisulfate per l.
Transcription Run-On Assay.
The transcription run-on assay was performed essentially as described (33, 50). Details can be found in SI Text.
RNA Pulse–Chase.
To pulse-label with 3H-uridine, the cells were transformed with a plasmid containing URA3 (pUG35) and grown to midlog phase in ura− media. Cells were labeled with 5 μCi/ml uridine (41 Ci/mmol) for 5 min.
ChIP.
Wild-type and hho1Δ strains harboring an HA-tagged version of RNA pol I were grown to 0.3 OD600 in yeast extract/peptone/dextrose. Chromatin was cross-linked by formaldehyde, and ChIP was performed (51). Products were purified by the PCR Clean-Up System (Promega) and analyzed by quantitative radioactive PCR. Products of PCR were run on a polyacrylamide gel and quantified using a phophoimager and TINA software (Fuji).
Psoralen Cross-Linking.
Psoralen cross-linking and analysis were performed for 40 min essentially as described (52).
DNA Array Hybridization.
DNA array hybridizations were carried out on Agilent whole genome arrays, as described in SI Text.
Supplementary Material
Acknowledgments.
We thank the Boeke and Griesenbeck laboratories for yeast strains, the Baserga laboratory for rDNA probes, Ilya Freidkin for suggesting that this assay system could be used to study Hho1p and for initiating preliminary experiments in that direction, Irit Shooltheis for suggesting and initiating the psoralen experiments, Jennifer Gallagher for very helpful discussions, and Liat Moskovitz for assistance with some of the experiments. We are especially grateful to Itamar Simon, in whose laboratory the ChIP was performed. This work was supported by internal Mina and Everard Goodman Life Sciences Faculty funding at Bar Ilan University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9095).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709403105/DCSupplemental.
References
- 1.Baxevanis AD, Landsman D. Histone sequence database: A compilation of highly-conserved nucleoprotein sequences. Nucleic Acids Res. 1996;24:245–247. doi: 10.1093/nar/24.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 3.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 4.Kasinsky HE, Lewis JD, Dacks JB, Ausió J. Origin of H1 linker histones. FASEB J. 2001;15:34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- 5.Cerf C, et al. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: Full assignment, tertiary structure, and comparison with the globular domain of histone H5. Biochemistry. 1994;33:11079–11086. doi: 10.1021/bi00203a004. [DOI] [PubMed] [Google Scholar]
- 6.Clore GM, Gronenborn AM, Nilges M, Sukumaran DK, Zarbock J. The polypeptide fold of the globular domain of histone H5 in solution. A study using nuclear magnetic resonance, distance geometry and restrained molecular dynamics. EMBO J. 1987;6:1833–1842. doi: 10.1002/j.1460-2075.1987.tb02438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 8.Goytisolo FA, et al. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- 9.Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- 10.Sivolob A, Prunell A. Linker histone-dependent organization and dynamics of nucleosome entry/exit DNAs. J Mol Biol. 2003;331:1025–1040. doi: 10.1016/s0022-2836(03)00831-3. [DOI] [PubMed] [Google Scholar]
- 11.Van Holde KE. Chromatin. New York: Springer; 1989. [Google Scholar]
- 12.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 13.Schumperli D. Cell-cycle regulation of histone gene expression. Cell. 1986;45:471–472. doi: 10.1016/0092-8674(86)90277-1. [DOI] [PubMed] [Google Scholar]
- 14.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast. Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dilworth DJ, et al. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–965. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, et al. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landsman D. Histone H1 in Saccharomyces cerevisiae - a double mystery solved. Trends Biochem Sci. 1996;21:287–288. [PubMed] [Google Scholar]
- 19.Ali T, Thomas JO. Distinct properties of the two putative “globular domains” of the yeast linker histone, Hho1p. J Mol Biol. 2004;337:1123–1135. doi: 10.1016/j.jmb.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 20.Ali T, Coles P, Stevens TJ, Stott K, Thomas JO. Two homologous domains of similar structure but different stability in the yeast linker histone, Hho1p. J Mol Biol. 2004;338:139–148. doi: 10.1016/j.jmb.2004.02.046. [DOI] [PubMed] [Google Scholar]
- 21.Horz W, Zachau HG. Deoxyribonuclease II as a probe for chromatin structure. I. Location of cleavage sites. J Mol Biol. 1980;144:305–327. doi: 10.1016/0022-2836(80)90093-5. [DOI] [PubMed] [Google Scholar]
- 22.Lohr D, et al. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci USA. 1977;74:79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas JO, Furber V. Yeast chromatin structure. FEBS Lett. 1976;66:274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- 24.Ono K, et al. The linker histone homolog Hho1p from. Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy. Nucleic Acids Res. 2003;31:7199–7207. doi: 10.1093/nar/gkg931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen X, Yu L, Weir JW, Gorovsky MA. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 26.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 27.Fan Y, et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Freidkin I, Katcoff DJ. Specific distribution of the. Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin. Nucleic Acids Res. 2001;29:4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 31.Sanderson A, Stott K, Stevens TJ, Thomas JO. Engineering the structural stability and functional properties of the GI domain into the intrinsically unfolded GII domain of the yeast linker histone Hho1p. J Mol Biol. 2005;349:608–620. doi: 10.1016/j.jmb.2005.03.085. [DOI] [PubMed] [Google Scholar]
- 32.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 33.Gallagher JE, et al. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast. Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellauer K, Sirard E, Turcotte B. Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem. 2001;276:13587–13592. doi: 10.1074/jbc.M011196200. [DOI] [PubMed] [Google Scholar]
- 36.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 37.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 38.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linder C, Thoma F. Histone H1 expressed in Saccharomyces cerevisiae binds to chromatin and affects survival, growth, transcription, and plasmid stability but does not change nucleosomal spacing. Mol Cell Biol. 1994;14:2822–2835. doi: 10.1128/mcb.14.4.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miloshev G, Venkov P, Van Holde K, Zlatanova J. Low levels of exogenous histone H1 in yeast cause cell death. Proc Natl Acad Sci USA. 1994;91:11567–11570. doi: 10.1073/pnas.91.24.11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schäfer G, Smith EM, Patterton HG. The Saccharomyces cerevisiae linker histone Hho1p, with two globular domains, can simultaneously bind to two four-way junction DNA molecules. Biochemistry. 2005;44:16766–16775. doi: 10.1021/bi0511787. [DOI] [PubMed] [Google Scholar]
- 42.Tongaonkar P, et al. Histones are required for transcription of yeast rRNA genes by RNA polymerase I. Proc Natl Acad Sci USA. 2005;102:10129–10134. doi: 10.1073/pnas.0504563102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones HS, et al. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chédin S, et al. Is ribosome synthesis controlled by pol I transcription? Cell Cycle. 2007;6:11–15. doi: 10.4161/cc.6.1.3649. [DOI] [PubMed] [Google Scholar]
- 45.Laferté A, et al. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 47.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 48.Sandmeier JJ, et al. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 2002;21:4959–4968. doi: 10.1093/emboj/cdf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oakes ML, et al. Role of histone deacetylase Rpd3 in regulating rRNA gene transcription and nucleolar structure in yeast. Mol Cell Biol. 2006;26:3889–3901. doi: 10.1128/MCB.26.10.3889-3901.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elion EA, Warner JR. An RNA polymerase I enhancer in. Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2002;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 52.Dammann R, Lucchini R, Koller T, Sogo JM. Transcription in the yeast rRNA gene locus: Distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol Cell Biol. 1995;15:5294–5303. doi: 10.1128/mcb.15.10.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.