Abstract
In Drosophila, the enzymatic activity of the glucan binding protein GNBP1 is needed to present Gram-positive peptidoglycan (PG) to peptidoglycan recognition protein SA (PGRP-SA). However, an additional PGRP (PGRP-SD) has been proposed to play a partially redundant role with GNBP1 and PGRP-SA. To reconcile the genetic results with events at the molecular level, we investigated how PGRP-SD participates in the sensing of Gram-positive bacteria. PGRP-SD enhanced the binding of GNBP1 to Gram-positive PG. PGRP-SD interacted with GNBP1 and enhanced the interaction between GNBP1 and PGRP-SA. A complex containing all three proteins could be detected in native gels in the presence of PG. In solution, addition of a highly purified PG fragment induced the occurrence not only of the ternary complex but also of dimeric subcomplexes. These results indicate that the interplay between the binding affinities of different PGRPs provides sufficient flexibility for the recognition of the highly diverse Gram-positive PG.
Keywords: Gram-positive bacteria sensing, pattern recognition receptors
Genetic analyses in Drosophila have led to a detailed understanding of innate immune recognition by invertebrates, a process that is to some extent conserved in higher animals (1). In flies, Toll and Imd (for Immune deficiency) have been shown to be the major pathways mediating host defense. Gram-positive bacteria and fungi strongly activate the Toll pathway, whereas Imd is deployed primarily against Gram-negative bacteria (2, 3). The mechanisms of pathogen discrimination that lead to triggering of these pathways have been attributed to structural differences in Gram-positive and Gram-negative peptidoglycan (PG) (4). PG is a heteropolymeric structure composed of a glycan strand cross-linked by short peptides. The glycan strand is made up of alternating N-acetylglucosamine and N-acetyl muramic acid in β-1,4 linkage. In contrast to the uniform structure of the glycan backbone, there is considerable variety in the linking peptides. At the third amino acid, l-Lysine is found in the majority of Gram-positive bacteria, whereas meso-diaminopimelic acid (mDAP) is present in Gram-negative bacteria and Gram-positive bacilli (5). Indeed, it has been shown that Lys-type PG is a potent activator of Toll, and DAP-type PG induces primarily the Imd pathway (4, 6–10).
Compared with the relative uniformity of Gram-negative PG, the structure and composition of Gram-positive PG defies easy categorization (11). There is considerable variability in both the interpeptide chains (a further link between the short peptides) and the mode of its cross-linking. This heterogeneity has hindered efforts to fit Gram-positive bacteria into a coherent model of pattern recognition. However, recent studies using highly purified PG components have revealed a clearer picture of inflammatory pathways in the fly (12). Gram-positive PGs are detected by two peptidoglycan recognition proteins (PGRPs), PGRP-SA and PGRP-SD (13, 14). Genetic studies indicate distinct roles for PGRP-SA and PGRP-SD, although there is also evidence for functional overlap (14). Flies deficient in PGRP-SA failed to activate the expression of antimicrobial peptides (AMP) after infection by Micrococcus luteus. These flies also exhibited a markedly reduced response after challenge by Enterococcus faecalis or Staphylococcus aureus (14), defects that were exacerbated in PGRP-SA; PGRP-SD double mutants. By contrast, AMP gene expression induced by infection with Staphylococcus saprophyticus was largely independent of PGRP-SA (14).
An additional player in Gram-positive bacterial sensing is the glucan-binding protein 1 (GNBP1) (15, 16). Recent biochemical studies revealed a presentation mechanism for the sensing of Gram-positive bacteria in flies (17). GNBP1 partially hydrolyzed Gram-positive PG to release free reducing ends of the muramic acid moieties for PGRP-SA binding. Thus, it is conceivable that the structural diversity of the Gram-positive bacterial cell wall requires a more flexible host defense characterized by the involvement of additional pattern recognition receptors such as PGRP-SD.
To investigate the biochemical basis of these flexible responses, we have purified functional recombinant PGRP-SA, -SD, and GNBP1 proteins expressed by insect cell lines. The presence of PGRP-SD significantly increased the binding ability of GNBP1 and in one instance PRGP-SA to Gram-positive PG. We also show that PGRP-SD interacted with GNBP1 and that the presence of PGRP-SD enhanced the interaction between PGRP-SA and GNBP1. Addition of a tetrameric muropeptide from S. aureus resulted in the formation of a ternary complex and different dimers. Finally, the presence of PGRP-SD in vivo augmented the signaling capacity of the GNBP1/PGRP-SA complex: there was a higher level of AMP gene expression when all 3 proteins were coexpressed compared with GNBP1/PGRP-SA alone. Our results are discussed in terms of the need for flexible host receptor partnerships to fight off Gram-positive bacterial infection.
Results
Characteristics of rGNBP1 and rPGRP-SA Binding to PG in the Presence of rPGRP-SD.
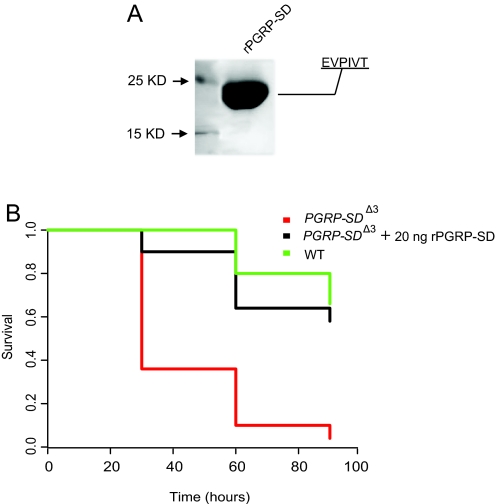
To study the role of PGRP-SD in pathogen sensing at the molecular level, we expressed recombinant PGRP-SD fused with a C-terminal 6xHis tag in insect cell culture using the baculovirus system. The protein was isolated by nickel-nitrilotriacetic acid affinity chromatography from Hi5 cell suspension culture and purified further by size-exclusion chromatography. N-terminal sequencing identified PGRP-SD with the signal peptide removed (Fig. 1). The functionality of the recombinant protein was tested in flies carrying a deletion of the pgrp-sd locus. Injection of 20 ng of rPGRP-SD protein before infection restored the ability of mutant flies to survive after immune challenge with S. aureus to a level comparable to that of infected WT adults (Fig. 1B). rPGRP-SA and GNBP1 were also expressed, purified, identified, and functionally tested, as described in ref. 17. Using these purified and functional proteins, we sought to determine how individual receptors and their combinations bound to different Gram-positive bacterial PGs, using PG from M. luteus (Ml), S. aureus (Sa), and S. saprophyticus (Ss).
Fig. 1.
Production of functional recombinant PGRP-SD in insect cells. (A) Purified PGRP-SD (predicted molecular mass: 20 kDa) was analyzed on a 15% SDS-reducing gel, indicating a single protein species. Protein was detected by Coomassie blue staining. Further identification by N-terminal sequencing (EVPIVT) showed a mature protein with the N-terminal signal peptide cleaved. (B) PGRP-SDΔ3 mutant flies are highly susceptible to S. aureus infection compared with WT adults. Injecting 20 ng of rPGRP-SD before infection, however, rescued lethality, producing a survival pattern comparable to that in WT animals. Survival patterns were plotted using Kaplan–Meier analysis. Statistical comparisons (log-rank test) were as follows: WT vs. SD rescue, P = 0.256; SD rescue vs. PGRP-SDJ3, P < 0.0001; WT vs. PGRP-SDJ3, P < 0.0001.
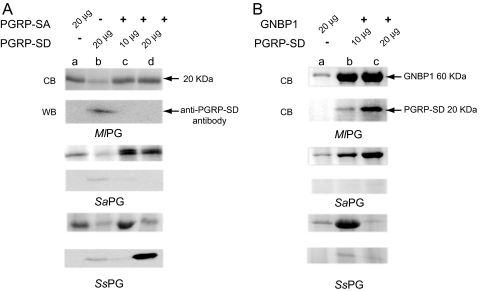
Binding of GNBP1 to PG from Ml, Sa, and Ss was greatly enhanced in the presence of PGRP-SD (Fig. 2B). The addition of 10 μg of PGRP-SD (reflecting a molecular ratio of 1:1) increased binding to GNBP1 by all PGs, whereas 20 μg of SD further increased the capacity of GNBP1 to bind to Sa PG (ratio of 1 GNBP1 to 3 PGRP-SD molecules; compare lanes b and c in Fig. 2B). However, a further increase in the concentration of PGRP-SD in the mix prevented binding of GNBP1 to Ss PG (compare lanes b and c in Fig. 2B, lower panel). This inhibition of GNBP1 binding to PG in the presence of high PGRP-SD concentrations was puzzling and could have 2 interpretations: (i) high concentrations of PGRP-SD were preventing GNBP1 binding by sequestering it away from, or interfering with, its interaction with Ss PG; or 2) high levels of PGRP-SD were aiding GNBP1 to hydrolyze Ss PG as in the case of the GNBP1/PGRP-SA complex (17), depleting the PG available for binding. We analyzed the supernatant of the binding reactions with PG from S. saprophyticus (Fig. 2B) by HPLC but found no evidence of PG hydrolysis (data not shown). However, we detected GNBP1 and PGRP-SD in the supernatant rather than the pellet, indicating that they were not bound to PG [see supporting information (SI) Fig. S1 and data not shown]. We observed that, by augmenting PGRP-SD, the amount of GNBP1 present in the supernatant was initially diminished but then rose again when 40 μg of PGRP-SD was added (Fig. S1). We therefore favor hypothesis 1, at least for this in vitro assay. Taken together, these data hint at an interaction between GNBP1 and PGRP-SD (see next section; note that this interaction could not be seen in the experiment of Fig. S1 because of the acetone precipitation treatment). In addition, the different behavior in terms of receptor binding to Sa and Ss PG may imply differences in PG structure other than muropeptide composition (e.g., the nature of cross-linking; see SI Text).
Fig. 2.
Enhanced binding of PGRP-SA and GNBP1 to PG and cell wall in the presence of PGRP-SD. (A) 20 μg of PGRP-SA were incubated with PG from Ml (top panel), Sa (middle panel), and Ss (lower panel) (see Materials and Methods) with or without PGRP-SD. Note that the 2 bands observed in lane c (Sa panel) are both PGRP-SA with and without the 6xHis tag. In our hands, storage of rPRGP-SA results in partial loss of the tag (L.W., unpublished observations). The identity of the bands was confirmed by an Ab against PGRP-SA (marks both bands) and one against the His tag (marks the upper band solely; data not shown). (B) 20 μg of GNBP1 were incubated with PG as above in the presence or absence of PGRP-SD. GNBP1 binding was significantly enhanced by PGRP-SD in a molecular ratio of 1:1 (or 20-μg GNBP1 and 10 μg of PGRP-SD) for all cases (lanes b). All of the protein bands in SDS-PAGE gel were visualized by Coomassie blue (CB) staining. We verified the presence of PGRP-SD by using an antibody against the protein in western blots (WB).
Binding of PGRP-SA to PG in the presence or absence of PGRP-SD revealed a different relationship (Fig. 2A). Binding of PGRP-SA (added in the fixed amount of 20 μg) to PG from Ml was not significantly altered in the presence of PGRP-SD. We noted, however, that although PGRP-SD was able to bind to this PG on its own, PGRP-SA could effectively compete it out (compare lanes b and c, d in Ml PG Fig. 2A, lower panel). However the affinity of PGRP-SA for Sa PG was augmented in the presence of PGRP-SD at 20 μg (a molecular ratio of 1:1 (Fig. 2A, compare Sa PG lanes a and d). Interestingly, an equal ratio of PGRP-SD to SA (20 μg of each) inhibited binding of PGRP-SA to Ss PG (compare Fig. 2A lower with middle panel). This inhibitory effect suggests a competitive mechanism of binding in 1:1 molecular ratios for Ss PG. In contrast to Ml PG, PGRP-SD prevented PGRP-SA from binding Ss PG (compare lanes c and d in Fig. 2A, lower panel).
Because the 2 proteins have similar molecular masses, our results were confirmed by using an antibody against PGRP-SD (anti-PGRP-SD panels, Fig. 2). For example, although 10 μg of PGRP-SD was mixed with 20 μg of PGRP-SA in all lanes c (Fig. 2A), no signal was observed with the antibody, leaving PGRP-SA as the only possible protein bound to the different PGs used. Conversely, in the case of Ss PG (Fig. 2A, lower panel), when 20 μg of each protein was present (lane d) the major protein bound was PGRP-SD, showing effective competition with PGRP-SA.
In conclusion, we have determined that PGRP-SA binding to PG in the presence of PGRP-SD depends on the microbe used. For Ml, PGRP-SA was able to bind and compete successfully with SD. PGRP-SD could bind to Ss PG, preventing PGRP-SA from doing so. This suggests that competition for binding to similar sites displaces 1 of the 2 proteins. We also observed cooperativity of PGRP-SA/SD in Sa PG binding. These differences may be accounted for by differences in PG structure. The determinant differences involved may include the extensive cross-linking in Ss and Sa but not Ml, or the different nature of cross-linking (Ss vs. Sa; see also SI Text and Fig. S2).
rPGRP-SD Interacts with rGNBP1 and Stabilizes the Interaction Between rGNBP1 and rPGRP-SA.
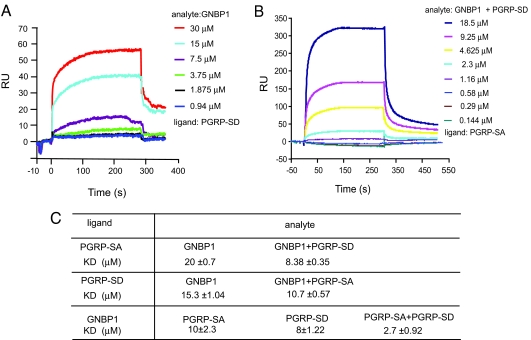
To address whether PGRP-SD interacts with both GNBP1 and PGRP-SD, we used surface plasmon resonance. Purified PGRP-SD was immobilized on the sensor chip (ligand), and rGNBP1 or rPGRP-SA (analyte) was injected over the chip. These experiments were performed in different orientations to confirm the interactions and were repeated at several different concentrations to produce Kd values (Fig. 3C). PGRP-SD binds to GNBP1 with a Kd of 15 μM (Fig. 3A). In contrast, the measured interaction with PGRP-SA has a Kd of 22 ± 2.5 μM that could only be quantified in one orientation (data not shown). This very weak association was confirmed by analytic ultracentrifugation (AUC; see Fig. S3 and text below). Therefore, the interaction of these proteins is unstable, or association requires the presence of both GNBP1 and a microbial ligand (see AUC experiments below and Fig. 4). Nevertheless, in the presence of PGRP-SD the interaction between GNBP1 and PGRP-SA was strengthened, as shown by a higher affinity of 8 μM compared with 20 μM using PGRP-SA alone as the ligand (Fig. 3B). When GNBP1 was used as ligand and PGRP-SA/PGRP-SD as analyte, the interaction recorded was 2.7 μM, compared with 10 μM for the single PGRP-SA/GNBP1 interaction (Fig. 3C).
Fig. 3.
PGRP-SD interacts with both PGRP-SA and GNBP1. (A) rPGRP-SD was immobilized on a CM5 sensor chip, and serial dilutions of rGNBP1 were injected. The RU increased in accordance with the increasing concentration of the injected protein. (B) PGRP-SD stabilized the interaction between GNBP1 and PGRP-SA. rPGRP-SA was immobilized onto the CM5 sensor chip. rGNBP1 and rPGRP-SD were mixed at a 1:1 molar ratio and injected with a serial dilution from 18.5 μM to 0.144 μM. (C) The binding kinetics of all experiments (those described above and all reciprocal orientations) are presented. Ligand refers to the protein coated on the sensor chip. The term analyte is used for the protein injected.
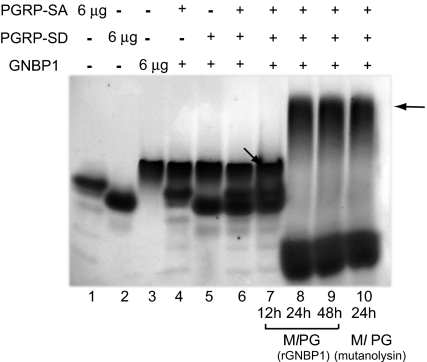
Fig. 4.
Ternary complex formation in the presence of PG. Processed PG shifted all of the three proteins into a higher-molecular-weight complex in native Tris-glycine PAGE gels. 6 μg of rPGRP-SA, rPGRP-SD, and rGNBP1 were used in single protein species controls (lanes 1–3). rGNBP1 mixed with either rPGRP-SA or rPGRP-SD as a 1:2 molar ratio is shown in lanes 4 and 5. The rPGRP-SA, rPGRP-SD, and rGNBP1 mix at 2:2:1 ratio is shown in lane 6. PG from Ml processed by rGNBP1 as described during 12 h (lane 7), 24 h (lane 8), or 48 h (lane 9) was incubated with the 3-protein mix (as defined in lane 6). Protein mix incubated with PG digested completely by mutanolysin (24 h) was used as control (lane 10). Arrows illustrate potential protein complexes. Lower bands in lanes 8, 9, and 10 represent PG fragments bound to PGRP-SA. In this form PGRP-SA will be running faster than the loading control of protein alone because AUC data indicate that it becomes more spherical upon microbial ligand addition (see Table S1, where S value increases from 2.0 to 2.2 upon muropeptide addition). Protein bands were visualized by Coomassie blue staining.
The results above show that all possible binary combinations of the 3 proteins lead to a measurable protein–protein interaction. The most stable was the PRGP-SD/GNBP1 association. Moreover, the data suggest that a stronger protein–protein interaction between PGRP-SA and GNBP1 occurs in the presence of PGRP-SD. However, surface plasmon resonance is a solid surface–based technology designed to quantify single protein–protein interactions rather than multiple and more complicated configurations. To explore the possibility of ternary complex formation in solution, we used Analytical Ultracentrifugation (AUC).
Ternary Complex and Heterodimer Formations in the Presence of Muropeptides or Peptidoglycan.
Deconvoluting the differently sedimenting species in a mixture of proteins is challenging; consequently, we began our analysis of their interactions by performing sedimentation velocity experiments on the isolated purified proteins PGRP-SD, PRGP-SA, and GNBP1. As shown in Fig. S3A and Table S1, PGRP-SA and PGRP-SD both seemed to be essentially mono-disperse, with similar sedimentation coefficients (s) of 2.2 ± 0.01S and 2.1 ± 0.01S, respectively. GNBP1 was also mostly mono-disperse (s = 3.7 ± 0.01S), with a very small amount of faster-sedimenting species (at 5.5 ± 0.5S) that may represent a dimer. Subsequent analysis of protein mixtures was cross-referenced to data for the pure proteins and reciprocal mixtures. Addition of highly purified tetrameric muropeptide from S. aureus at a concentration of 10 μM changed the behavior of binary and ternary mixtures of the proteins. Our detailed analysis for all possible combinations is presented in the accompanying SI Text and allowed us to interpret the behavior of the ternary mixture in the presence of muropeptide. In the latter case, up to 9 species were detected (Fig. S3H and Table S1). The peptide increased our ability to resolve individual sedimenting species in the sample, which included SA/SD, GNBP1, SA/SD-GNBP1, and the ternary complex SA-GNBP1-SD. There was a marked increase in the amount of species present at approximately 6–8S, the most abundant of which we have identified as the SA-GNBP1-SD-peptide complex (Fig. S3H). These results suggest the formation of a ternary complex in the presence of a microbial ligand.
A key point, however, was to strengthen our interpretation for the identities of sedimenting species visualized by g(s) analysis in Fig. S3. To this end, we performed a size-and-shape distribution analysis (c(s.fr), where fr is the frictional ratio (a sphere has a frictional ratio of 1 and more elongated species of >1). This allows of contour plots of c(s,M) to be generated, where M is the molecular mass of the protein. With this approach, we show that the sedimentation coefficients for the species we identify match their known molecular masses if the g(s) identifications were correct (see Fig. S4 and SI Text). Moreover, extending our analysis (Fig. S5), we are able to show the direct superposition of the c(s,M) plots in Fig. S4. Thus, the patterns of sedimenting species are consistent as between pure samples and mixtures, and, most importantly, the inclusion of the muropeptide allows the formation of novel complexes and increases the yield of others (for further analysis see SI Text and Fig. S5).
The above results confirmed the data shown in Fig. S3, which in turn indicated the formation of various dimeric configurations and a ternary complex of the 3 proteins with the addition of a microbial ligand. We sought to determine, therefore, whether such a ternary complex could be detected in native gels. Fig. 4 shows representative results from these experiments. When Ml PG processed by rGNBP1 at 24-h and 48-h time points (lanes 8 and 9) or mutanolysin at 24 h (lane 10) was added to the mix of the three proteins, we observed a higher molecular weight complex, representing the association of GNBP1-PGRP-SA and PGRP-SD. This result was verified by Western blots using antibodies against the 3 proteins (data not shown). The molecular ratio for the formation of the complex was 2PGRP-SD: 2PGRP-SA: 1GNBP1. Our interpretation for the lower band observed in lanes 8, 9, and 10 is that it is PGRP-SA/PGRP-SD or PGRP-SA in complex with PG fragments. Clustering of PGRP-SA has been proposed by the Lee group as a model of complex initiation in Tenebrio molitor (24). Our AUC data predict a PGRP-SA/PGRP-SD interaction in the presence of a microbial ligand (Fig. S3). The fact that the band of the presumed complex or the PGRP-SA dimer is lower than the bands representing individual proteins may be explained again by the AUC data. There, increased sphericity of the 2 proteins was observed in PGRP-SA/PGRP-SD complexes in the presence of ligand (Fig. S4). In control experiments in which GNBP1 or mutanolysin-treated PG was run in the absence of PGRP-SA and PGRP-SD, only 1 band was observed corresponding to GNBP1 (Fig. S6). This indicates that the bands seen in these experiments were protein complexes and not products of PG degradation.
PGRP-SD Increases the Signaling Capacity of GNBP1/PGRP-SA In Vivo.
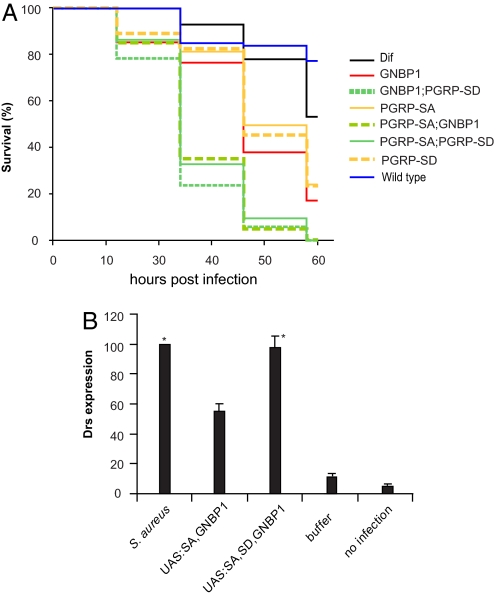
The biophysical and biochemical data presented above indicated the formation of a ternary complex for the reception of some Gram-positive bacteria. Genetic experiments have provided evidence that both PGRP-SA and PGRP-SD are important for the reception of Sa (14). To reconcile the genetic data with events at the molecular level, we tested the survival of double mutants (PGRP-SA; PGRP-SD or PGRP-SA; GNBP1 or GNBP1, PGRP-SD) against the single mutants and WT flies after Sa infection. Our results are shown in Fig. 5A. All double mutants were extremely susceptible to infection, with 80% of flies dying in 24 h. Single mutants were less sensitive to infection (only 30% of flies died in 24 h), although their survival was markedly reduced compared with WT adults. This signifies the importance of all 3 proteins in the defense against Sa. As pointed out by Bischoff et al. (14), dif flies were less sensitive to Sa challenge than the host receptor mutants, suggesting additional Toll-independent functions for fighting off this bacterial infection.
Fig. 5.
PGRP-SD, PGRPSA, and GNBP1 during host receptor signaling in vivo. (A) Kaplan–Meier analysis of survival experiments after Sa infection using double or single mutants of host receptors were carried out. Survival tests were performed in three independent experiments. Log-rank tests comparing the three experiments for each genotype indicated that the experiments were homogeneous (all P values very high and in the range of 0.7–0.98). Thus, it was valid to pool these experiments and for each genotype to analyze the data for all flies used. Because there were 35 pairwise tests between different genotypes there was an issue of multiplicity of tests. To address that, we used the Bonferroni correction for multiple tests (0.05 divided by the number of tests). This placed P values for significance at or <0.0014. The log-rank tests for the comparisons of the 8 genotypes presented in the graph gave P values that clearly placed the WT and Dif flies in one group (WT vs. Dif, P = 0.214), the single mutants in a second group, and the double mutants in a third group, whereby the statistical difference between all single vs. double mutant tests was significant (P < 0.0005) as judged by: GNBP1 vs. GNBP1, PGRP-SD, P < 0.0005; GNBP1 vs. PGRP-SA; GNBP1, P < 0.0005; PGRP-SA vs. PGRP-SA; PGRP-SD, P < 0.0005; PGRP-SA vs. PGRP-SA; GNBP1, P < 0.0005; PGRP-SD vs. PGRP-SD, GNBP1, P < 0.0005; and PGRP-SD vs. PGRP-SA; PGRP-SD, P < 0.0005. All single or double mutant combinations vs. the WT gave P < 0.0005. Differences between the single mutants were not statistically significant (GNBP1 vs. PGRP-SA, P = 0.003; GNBP1 vs. PGRP-SD, P = 0.033; PGRP-SA vs. PGRP-SD, P = 0.428). Finally, differences between double mutants were also not statistically significant (GNBP1, PGRP-SD vs. PGRP-SA; GNBP1, P = 0.008; GNBP1, PGRP-SD vs. PGRP-SA; PGRP-SD, P = 0.007; and PGRP-SA; GNBP1 vs. PGRP-SA; PGRP-SD, P = 0.512). (B) Concomitant overexpression of PGRP-SA and GNBP1 through the GAL4/UAS system results in activation of the AMP gene drosomycin (drs) used as a read-out for activation of the Toll pathway. This activation occurs in the absence of any immune challenge and amounts to 40% of drs induction after infection. Expressing PGRP-SD through a UAS-transgene at the same time as PGRP-SA and GNBP1 induces drs at the level seen by Gram-positive bacterial infection (S. aureus). Columns represent the percentage mean value of three independent experiments (corrected against the loading control RP49) with standard deviation represented as error bars. Asterisks indicate values that are not statistically different (P > 0.005) from each other. All other differences are statistically significant (P < 0.005).
In addition to its participation in the sensing mechanism, we wanted to determine whether PGRP-SD potentiates the signaling ability of the GNBP1-PGRP-SA complex. Concomitant overexpression of PGRP-SA and GNBP1 through the UAS/GAL4 system and using a fat-body specific GAL4 driver (yolk-GAL4) was able to activate the pathway in the absence of infection as measured by the induction of the Toll-responsive AMP drosomycin (drs) (see also Gobert et al. (15)]. However, this activation was approximately 40% of that observed by injection of Sa. By expressing PGRP-SD along with PGRP-SA/GNBP1, activation of drs was comparable to infection. The data summarized in Fig. 5B indicated the cooperation of the 3 proteins in signaling and revealed a potential role for PGRP-SD in the signaling capacity of the ternary receptor complex.
Taken together, the data presented in Fig. 5 and previous results presented in this study detecting the formation of a PGRP-SA/GNBP1/PGRP-SD complex, correlate with the need for PGRP-SD in Sa recognition in vivo and in a ternary complex with PGRP-SA and GNBP1.
Discussion
In Drosophila, PG is the major inducer of AMP gene expression (4, 6–8). Results from genetic analyses show that PGRP-SA/GNBP1 and PGRP-SD act in a cooperative or partially redundant fashion to control activation of the Toll pathway and therefore survival after Gram-positive bacterial infection (14). In the present study, we further elucidate the molecular events during Gram-positive bacterial sensing in flies.
We have found that PGRP-SD provides flexibility to host receptor formation in vitro and in the presence of microbial ligands. Although PGRP-SD did not bind efficiently on its own, it significantly enhanced the binding of GNBP1 to PG from Ml, Sa, and Ss. However, in the case of Ml it was efficiently antagonized by PGRP-SA, whereas in the case of Ss PGRP-SD competed out PGRP-SA. Finally, in the case of Sa along with GNBP1, PGRP-SD enhanced the binding of PGRP-SA, indicating the possibility that in this case a complex with all 3 proteins may be formed. The potential of such formation was confirmed in solution with the addition of a highly purified tetrameric muropeptide from Sa. Finally, the facts that (i) all double mutant combinations are more sensitive to Sa infection than single mutants, and 2) overexpression of PGRP-SD potentiates the signaling capacity of the GNBP1/PGRP-SA complex, correlate with the notion of a ternary complex. Our current model of host receptor formation is presented in Fig. S7. We suggest that the complexity and variability of PG structure among the different species of Gram-positive bacteria used (11; see also SI Text and Fig. S2) may have led to a diversification of recognition strategies by the host. The ability to form a ternary complex and the formation of a variety of dimers may explain why fly mutants lacking pattern recognition receptors like PGRP-SA and SD respond differently depending on the type of Gram-positive bacteria. Thus, our results correlate well with corresponding genetic studies, indicating that sensing of certain species of Gram-positive bacteria like Ml is solely dependent on PGRP-SA/GNBP1, whereas recognition of Sa depends on all 3 molecules. Conversely, host recognition of some other Gram-positives (an example being Ss) is PGRP-SA independent (14).
The protein–protein interactions between receptors reported in this study are relatively weak (3–30 μM), and the relevance of these associations in vivo is not proven. Nevertheless, a comparison with other systems is informative. In mammalian adaptive immunity, interactions among cell-surface molecules in leukocytes are generally of low affinity (from 1 μM to 100 μM) (18, 19). For instance, T cell receptor (TCR) and peptide-MHC interactions range from 1 to 90 μM but are still the basis for the avidity hypothesis of negative and positive selection of T cells (20). This hypothesis emphasizes both the affinity of TCR to peptide–MHC and density of this complex on cell surfaces. The outcome of the avidity effect of complexes decides the fate of T cells. If the sum total of affinity of the complexes reaches the threshold to activate T cell maturation but remains weak enough to survive apoptosis, T cells are positively selected and develop further. The weak interactions of TCR with peptide–MHC complex on cell surfaces are essential to adjust between negative and positive selection in T cells.
In an analogous manner, the weak protein–protein interactions we observe for GNBP1, PGRP-SA, and PGRP-SD may underlie a potential avidity effect if we take into account the polymeric structure of PG. The number of protein complexes bound to long and intact polymeric PG may dictate the strength of interaction with PG in a quantitative manner. With more protein complexes bound, the interactions could become additively stronger, capable of generating an interface and of recruiting other molecules, resulting in more PG digestion and activation of a downstream serine protease cascade. With shorter PG fragments, the interactions become weaker, and the coreceptors leave this interface. Eventually the signal generated is repressed, leading to reduced AMP expression. Evidence in support of this proposal is that (1) the immune response is proportional to the polymeric status of muropeptide (8); and 2) the tetrameric muropeptide is capable of stabilizing and forming a larger protein complex when GNBP1, PGRP-SA, and PGRP-SD are present (this study).
Materials and Methods
Fly Strains and Procedures.
The fly strains used were described GNBP1osi, UAS-GNBP1 (15), PGRP-SAseml, UAS-PGRP-SA (13), UAS-PGRP-SD, PGRP-SDΔ3, PGRP-SD, GNBP1 (14), yolkGAL4, and UAS-Dif (25). Infections for the functional assay were performed as described in ref. 17. AMP gene expression and quantification of Northern blots as described in ref. 8. A w isogenic strain was used as WT control.
Cloning of PGRP_SD Full-Length cDNA and Expression Construct in pFastBac1.
PGRP-SD full cDNA was obtained by RT-PCR using total RNA from WT flies (Oregon-R strain). Total RNA from 5 adults was extracted by TRIZOL reagent (Invitrogen) according to the manufacturer's instructions. Purified total RNA was subsequently reverse-transcribed to first-strand cDNA with the RETROscript kit (Ambion). Using a pair of gene-specific primers at the 5′ and 3′ termini of the PGRP-SD gene, the full-length cDNA was amplified from the first-strand cDNA and subcloned into pSTBlue-1 (Novagen). The PGRP-SD full-length cDNA construct was verified by DNA sequencing.
Expression and Purification of Recombinant Proteins.
The expression conditions for the 3 recombinant proteins were empirically determined at different time points in multiples of infection (MOI) in either Hi5 or Sf9 cells. Recombinant GNBP1 and PGRP-SA were best expressed in Sf9 cells and purified as described in ref. 17. Optimized protein expression conditions for PGRP-SD was confirmed by Western blot analysis using Petra anti-His-tag antibody conjugated with horseradish peroxidase (Qiagen) as in Hi5 cells at MOI of 10.0 48 h after infection. Viruses were amplified by infection of Hi5 cell suspension cultures with 2% FBS. PGRP-SD was thus purified from 4-liter suspension culture of Hi5 cell at MOI of 10.0 2 days after infection. After centrifugation, the supernatant was concentrated on a Centramate tangential flow system (Pall Filtron) with buffer exchanged to binding buffer (150 mM NaCl and 20 mM Tris·HCl, pH 7.5, 10 mM imidazole). After addition of EDTA-free protease inhibitors (Roche), PGRP-SD His6-tagged protein was purified by Qiagen Superflow NiNTA agarose and gel filtration on Superdex 75 or 200 columns (GE Healthcare) on an FPLC system (GE Healthcare). The identity of PGRP-SD was confirmed by N-terminal sequencing (PNAC facility, Cambridge University, Cambridge, U.K.).
Isolation of Bacterial Cell Wall, Peptidoglycan, and Muropeptides.
Peptidoglycan from M. luteus (reference strain DSM20030), S. saprophyticus (reference strain ATCC15305), and from the clinical isolate S. aureus strain COL, and muropeptide purification from the latter, was performed as described (8, 17).
Protein Binding Assays to Cell Wall, Peptidoglycan, Whole Bacteria, Generation of Antibodies, and Western blot analysis.
The binding activity of proteins to PG was performed by incubating proteins with 200 μg of PG in a 300-μl reaction with binding buffer (20 mM Tris·HCl, pH 8.0; 300 mM NaCl). For the binding enhancement experiment of PGRP-SD, PGRP-SA and GNBP1 were fixed as 20 μg with PGRP-SD added with increasing amounts as 10 μg, 20 μg, and 40 μg. After 3-h incubation on a shaking platform, PG was sedimented at 16,000 × g for 5 min. PG pellet was then washed three times with 1 ml washing buffer (100 mM Tris·HCl, pH 8.0; 500 mM NaCl and 0.02% Tween-20). Subsequently PG pellet was dissolved directly in 20-μl SDS sample buffer and subjected to an SDS-reducing PAGE gel. The gels were stained with SimplyBlue staining solution (Invitrogen) after electrophoresis.
To detect the protein compositional change in the supernatant after centrifuging to remove the PG pellet, protein in the supernatant was precipitated by adding 9× volume of ice-cold acetone. The acetone–supernatant mixtures were incubated at −20°C for 4 h and subjected to centrifuge at 14,000 rpm at 4°C for 15 min. The protein pellets were washed with 1 ml of ice-cold acetone for 5 min and centrifuged again at 14 000 rpm at 4°C for 15 min. Protein pellets were finally dissolved directly in 20 μL 2× SDS-PAGE running buffer and loaded onto a 15% SDS-PAGE directly. Bands were visualized using the SimplyBlue staining solution from Invitrogen.
Polyclonal antibodies against GNBP1, PGRP-SA, and PGRP-SD were raised in rabbits by Eurogentec using purified proteins with the His-tag removed. Standard Western blot analysis was carried out to confirm the functionality of the antibodies against purified proteins.
Surface Plasmon Resonance.
Surface plasmon resonance experiments were carried out on a Biacore 2000 instrument using CM5 sensor chips (Biacore) as described before (17).
Analytic Ultracentrifugation.
Sedimentation velocity experiments were performed in a Beckman Optima XL-I analytical ultracentrifuge, as described in ref. 21. Samples of PGRP-SA (20 μM), SD (20 μM), and GNBP1 (8 μM) alone and in combination were spun at 40 000 rpm and imaged using interference optics. Analysis by the g(s) method used SEDFIT (22). Single species display a Gaussian distribution in sedimentation coefficient, owing to the effects of diffusion, about a central point that is the sedimentation coefficient of the species. The Gaussian distributions were fitted using the program PROFIT, and the areas under individual peaks within the distribution were calculated using the equation A = σα[rad]2π[/rad], where A is the area, σ is the half width, and α is the height of each peak.
To calculate c(s,fr) distributions, 2D plots of the relationship between sedimentation coefficient and frictional ratio (fr) were determined, whereby diffusional broadening of the sedimenting protein boundaries was used to determine the frictional characteristics of the sample. The function c(s,fr) was then used to derive functions such as c(s,M), dependence of sedimentation coefficient and weight, and c(s,Rs), dependence of sedimentation coefficient and Stokes radius. These calculations were all performed using SEDFIT (22, 23). The superimposed plots shown in Fig. S5 were generated from the output of the c(s,fr) analysis using PROFIT.
Native PAGE Gels and Ternary Complex Detection.
Proteins were mixed as a molar ratio of 1 (GNBP1: 6 μg): 2 (PGRP-SA: 6 μg): 2 (PGRP-SD: 6 μg) with 2 μl of processed PG after 12 h, 24 h, and 48 h digestion with GNBP1. After incubation at room temperature for 30 min, 2× native gel loading buffer was mixed with the samples and subjected to 4–12% Tris-glycine gel under native running conditions (Invitrogen). Individual proteins and protein combinations without added PG were used as controls. The gel shown in Fig. 4 is a typical result. Four independent experiments were conducted.
Supplementary Material
Acknowledgments.
We thank Julien Royet, Dominique Ferrandon, and Jean-Marc Reichhart for fly strains and Yvonne Griffiths for help with statistical analysis. L.W. was supported by a Dorothy Hodgkin postgraduate award from Research Councils U.K. This work was supported in part by Grant POCI/SAU-IMI/56501/2004 from the Fundação para a Ciência e a Tecnologia (to SRF), a Royal Society International Collaboration Grant U.K./Portugal (to S.R.F. and P.L.) and a pump-priming grant from the EPA Cephalosporin Trust Fund (to P.L.). R.J.C.G. is a Royal Society University Research Fellow. Work in the United Kingdom was funded by the Medical Research Council through a Program Grant (to N.J.G.) and a Career Establishment Grant (to P.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710092105/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Ligoxygakis P. Pathogen recognition and signalling in the Drosophila innate immune response. Immunobiology. 2006;211:251–261. doi: 10.1016/j.imbio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre B, Hoffman JA. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 4.Leulier F, Parquet C, Pili-Flouri S, Ryu JH, Caroff M, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 5.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, et al. Monomeric and polymeric Gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 7.Stenbak CR, et al. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- 8.Filipe SR, Tomasz A, Ligoxygakis P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005;6:327–333. doi: 10.1038/sj.embor.7400371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim JH, et al. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- 10.Swaminathan CP, et al. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs) Proc Natl Acad Sci USA. 2006;103:684–689. doi: 10.1073/pnas.0507656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber JR, Moreillon P, Tuomanen EI. Innate sensors for Gram-positive bacteria. Curr Opin Immunol. 2003;15:408–415. doi: 10.1016/s0952-7915(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 12.Chaput C, Boneca IG. Peptidoglycan detection by mammals and flies. Microbes Infect. 2007;9:637–647. doi: 10.1016/j.micinf.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 13.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 14.Bischoff V, et al. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- 15.Gobert V, et al. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- 16.Pili-Floury S, et al. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J Biol Chem. 2004;279:12848–12853. doi: 10.1074/jbc.M313324200. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, et al. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. 2006;25:5005–5014. doi: 10.1038/sj.emboj.7601363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis SJ, Ikemizu S, Wild MK, van der Merwe PA. CD2 and the nature of protein interactions mediating cell–cell recognition. Immunol Rev. 1998;163:217–236. doi: 10.1111/j.1600-065x.1998.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 19.van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;4:217–224. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 20.Ashton-Rickardt PG, et al. Evidence for a differential avidity model of T cell selection in the thymus. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 21.Harkiolaki M, Gilbert RJC, Jones EY, Feller SM. The C-terminal SH3 domain of CRKL as a dynamic dimerization module transiently exposing a nuclear export signal. Structure. 2006;14:1741–1753. doi: 10.1016/j.str.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 23.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JW, et al. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc Natl Acad Sci USA. 2007;104:6602–6607. doi: 10.1073/pnas.0610924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rutschmann S, et al. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity. 2000;12(5):569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.