Abstract
Regulation of bacterial iron homeostasis is often controlled by the iron-sensing ferric uptake repressor (Fur). The Bacillus subtilis Fur protein acts as an iron-dependent repressor for siderophore biosynthesis and iron transport proteins. Here, we demonstrate that Fur also coordinates an iron-sparing response that acts to repress the expression of iron-rich proteins when iron is limiting. When Fur is inactive, numerous iron-containing proteins are down-regulated, including succinate dehydrogenase, aconitase, cytochromes, and biosynthetic enzymes for heme, cysteine, and branched chain amino acids. As a result, a fur mutant grows slowly in a variety of nutrient conditions. Depending on the growth medium, rapid growth can be restored by mutations in one or more of the molecular effectors of the iron-sparing response. These effectors include the products of three Fur-regulated operons that encode a small RNA (FsrA) and three small, basic proteins (FbpA, FbpB, and FbpC). Extensive complementarity between FsrA and the leader region of the succinate dehydrogenase operon is consistent with an RNA-mediated translational repression mechanism for this target. Thus, iron deprivation in B. subtilis activates pathways to remodel the proteome to preserve iron for the most critical cellular functions.
Iron is an essential element for nearly all cells but is often of limited availability in natural environments (1). Bacteria have evolved sophisticated mechanisms to scavenge iron by the elaboration of high-affinity iron-chelating agents, siderophores, and their transport systems (2, 3). Expression of these iron uptake pathways is usually repressed under iron-sufficient conditions by an iron-sensing repressor. The paradigm for iron repression in bacteria is the ferric uptake repressor (Fur) protein as originally described in Escherichia coli (1). Fur orthologs are found in both Gram-negative and many Gram-positive bacteria, including Bacillus spp., Staphylococcus aureus, and Listeria monocytogenes (4). In Corynebacterium spp. and Mycobacterium spp., iron regulation is mediated instead by a functionally similar protein designated DtxR (5, 6) or IdeR (7, 8), respectively. In the Rhizobiales, iron regulation is mediated in large part by RirA, a member of the Rfr2 family of repressors (9).
Despite differences in the nature of the regulator, there are many common themes in the bacterial responses to iron starvation. Notably, in most well studied systems, there is an induction of siderophore biosynthesis and uptake or other pathways to increase iron assimilation. Bacillus subtilis expresses uptake systems for its endogenous siderophore bacillibactin, ferric citrate, and for a variety of siderophores made by other soil bacteria (10). In pathogenic bacteria, uptake pathways may also involve hemolysins, heme-binding proteins, and transferrin-binding proteins (2, 11, 12). B. subtilis can also use an elemental Fe uptake system YwbLMN (10), which is homologous to Saccharomyces cerevisiae Fet3p/Ftr1p and E. coli EfeUOB systems (13, 14). As a result of derepressed transport, fur mutants typically have elevated levels of chelatable iron within their cytosol (15) and are hypersensitive to H2O2, which reacts with free iron to generate hydroxyl radicals (16).
Despite the constitutive expression of iron uptake pathways, E. coli fur mutants have significantly reduced levels of total cellular iron (17). This apparent paradox is now understood as indicative of a small RNA (sRNA)-mediated repression of numerous iron-containing proteins (18, 19). In the absence of Fur, the RyhB sRNA represses the synthesis of succinate dehydrogenase (Sdh), aconitase, fumarase, and iron superoxide dismutase (20). As a result, total cellular iron is decreased, despite a significant increase in the pool of chelatable, cytosolic iron.
The repression of iron-rich enzymes when iron is limited is undoubtedly adaptive and has evolved multiple times. This repression, which we here refer to as an iron-sparing response, allows the cell to prioritize iron usage at times of limited availability. Similar RyhB-mediated responses have been described in Shigella flexneri and Vibrio cholerae (19). In Pseudomonas aeruginosa, two functionally analogous, Fur-regulated sRNAs were described in ref. 21. In Saccharomyces cerevisiae, iron deprivation leads to the activation of Aft1, which controls expression of iron uptake functions and Cth2, an RNA-binding protein that targets for degradation the mRNAs for aconitase, Sdh, and perhaps dozens of additional proteins (22). Iron-sparing responses are also likely to be present in Gram-positive bacteria. In Corynebacterium spp., the iron sensor DtxR represses the RipA protein, which, in turn, is a repressor of iron-using proteins (23). Analysis of the S. aureus proteome as a function of iron availability also suggests the presence of an iron-sparing response, but the mechanism is not yet known (24).
Here, we present evidence for an iron-sparing response in B. subtilis mediated primarily by a Fur-regulated sRNA (FsrA). The action of FsrA is independent of an Hfq-like protein encoded by B. subtilis but depends, in part, on three small, basic proteins designated as Fur-regulated basic proteins (Fbp) A–C. These basic proteins are postulated to function as RNA chaperones. We show that together these regulators contribute to many of the pleiotropic effects noted in fur mutant strains, including poor growth on a variety of media.
Results and Discussion
Fur Regulates the Expression of a sRNA (FsrA) and Three Small, Basic Proteins.
Genes repressed by Fur in B. subtilis are associated with Fur boxes (25). Classically, the Fur box is represented as a 19-bp inverted repeat, but we have noted that this sequence actually comprises two, overlapping operator sites with a 7-1-7 minimal recognition element (26). Although most Fur boxes are associated with operons encoding siderophore biosynthesis or uptake functions, or other proteins implicated in iron homeostasis, some sites are not closely apposed to annotated ORFs. Here, we identify three Fur-regulated transcription units that collectively encode one small, noncoding RNA (sRNA) and three small, basic proteins postulated to function as RNA chaperones.
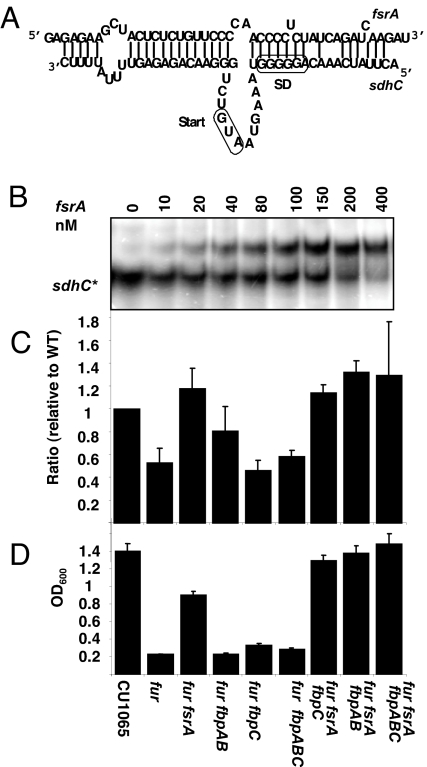
Anaysis of sequences downstream of one strongly predicted Fur box (17/19 match to consensus) identified a Fur-regulated sRNA (fsrA) highly conserved between three closely related Bacillus strains [supporting information (SI) Fig. S1]. The fsrA gene is unable to encode a protein product because it lacks any potential start codons. As expected, the FsrA sRNA is strongly up-regulated in a fur mutant strain (Fig. 1B) and in iron-starved wild-type cells (data not shown). As predicted, Fur binds to the fsrA regulatory region with high affinity (Fig. 1C). These results suggest that FsrA is a Fur-regulated sRNA.
Fig. 1.
Molecular components of the B. subtilis iron-sparing response. (A) Organization of the fsrA fbpAB, and fbpC transcription units. FB indicates a Fur box. (B) Northern blot analysis of fsrA expression in different strains. 23S rRNA was used as an internal loading control. (C) Binding of purified Fur to the promoter region of fsrA (0.1 nM) as detected by electrophoretic mobility shift assay. (D) Detection of FLAG-tagged FbpA, FbpB, and FbpC by Western blot with anti-FLAG antibodies. (E) Amino acid sequences and calculated pI of the FbpA, B, and C proteins.
The Fur-regulated ydbN gene encodes a small, 59-aa protein of unknown function (25). The predicted ydbN transcript initiates 161 nucleotides upstream of ydbN and encodes a second small protein (53 aa) also conserved in B. subtilis, Bacillus amyloquefaciens, and Bacillus licheniformis (Fig. S2). The pattern of base substitutions argues that these are protein-coding regions: many substitutions are in the degenerate third codon position. Both proteins are detected as FLAG-epitope tagged products in B. subtilis (Fig. 1D), and after expression in E. coli (data not shown). Here, we rename these proteins as Fur-regulated basic proteins (FbpA) and FbpB (formerly YdbN) (Fig. 1 A and E).
The third Fur-regulated basic protein, FbpC, is encoded by the first identified member of the Fur regulon, metal-regulated gene C (mrgC) (27). An mrgC-lacZ transcriptional fusion is induced by the iron chelator EDDHA or by growth under iron-limiting conditions (27). With the completion of the B. subtilis genome sequence in 1997 (28), we noted that the mrgC ORF terminates after only 29 codons (Fig. 1E). The nearest annotated ORF (ypbR) lies 296 base pairs downstream of the mrgC transcript initiation site and is not regulated by Fur or by iron (25). The presence of a Rho-independent terminator predicts a ≈188-nt RNA product from this region. As noted for fbpA and fbpB, comparison of this region between sequenced Bacillus genomes reveals a pattern of base substitutions consistent with a protein coding region (Fig. S3). Moreover, addition of a FLAG epitope allows visualization of the corresponding protein (Fig. 1D).
Our working hypothesis is that these three small proteins function together with FsrA to direct repression of selected target genes. However, as shown below, FsrA can function at some targets independent of these proteins. Moreover, the fbpAB and fbpC operons also have functions independent of FsrA and these may be mediated by the encoded peptides, the RNA transcripts, or both.
fur Mutant Is Growth Impaired.
Previous observations indicate that a fur null mutation has pleiotropic effects on growth consistent with the hypothesis that Fur mediates the iron-dependent synthesis of iron-using enzymes of central metabolism. Indeed, transcriptome analyses revealed that a fur mutant had decreased levels of transcripts for several iron-containing enzymes and cytochromes and heme biosynthesis pathways (Table S1) (25). Similarly, microarray analyses of iron-starved B. subtilis have shown significantly decreased levels of mRNAs for both glutamate synthase and amino acid biosynthetic enzymes (29). The physiological consequences of these changes were observed directly in fluxome analyses. Fischer and Sauer (30) compared growth rates and metabolic fluxes in wild-type and 137 null mutants of B. subtilis. Most mutations led to little net change in metabolic fluxes, indicating that B. subtilis has a robust central metabolism. Of the strains tested, only one (cysP) had a slower growth rate than fur. Under the conditions used (M9 glucose minimal medium), the fur mutant grew at a rate one-half that of wild-type and there was a notable perturbation in the relative flux through the TCA cycle (reduced by 23%), and the absolute flux (citrate synthase activity) was reduced nearly 3-fold (30). This reduced flux is likely due to down-regulation of TCA cycle enzymes (see below). Motivated by these prior observations, we here initiated a detailed analysis of the phenotypes of a fur null mutant and isogenic derivatives lacking one or more of the molecular effectors postulated to mediate this iron-sparing response.
Iron-Sparing Response Reduces Cellular Iron Demand and Is Adaptive Under Severe Iron Limitation.
In E. coli, cells that lack an iron-sparing response are unable to prioritize iron utilization and are at a competitive disadvantage during growth under severe iron limitation (31). To determine whether the B. subtilis iron-sparing response is also adaptive under iron limitation, we monitored growth in medium containing a concentration of the iron chelator EDDHA sufficient to reduce yield of the wild-type strain by one-half. Under these conditions, a fur mutant grew significantly better than wild-type (Fig. 2A, lane 2 vs. lane 1), presumably because of the constitutive expression of the iron-sparing response in the precultures. Note that, under these conditions, even the wild-type strain derepresses the Fur regulon, so both strains should have a similar complement of iron uptake pathways. However, these strains are sfp0 (32) and are therefore unable to synthesize bacillibactin, which is necessary to access iron chelated by EDDHA (10).
Fig. 2.
Growth effects of mutants altered in the iron-sparing response. (A) The iron-sparing response is adaptive under severe iron limitation. Cell density after overnight growth under severe iron limitation (Chelex-treated glucose minimal medium with 1 μM EDDHA). Under these conditions, the fur mutant grows better than wild-type, but a fur mutant additionally lacking either fsrA or all three protein chaperones grew poorly. (B) Cell iron content (in micrograms of Fe per milligram of soluble protein) as determined by using spectrophotometric analysis of ferrozine-iron complexes.
To determine whether the fsrA, fbpAB, or fbpC operons were important for these growth effects, we extended this analysis to a panel of mutant strains. In contrast with the fur mutant, a double fur fsrA mutant strain is severely growth impaired in this medium (Fig. 2A, lane 3). We infer that the inability of these cells to prevent the synthesis of iron-consuming proteins hinders adaptation to these iron-limiting conditions. A severe growth defect was also noted in a fur mutant strain that still contains FsrA but lacks all three basic proteins (the fur fbpABC quadruple mutant; Fig. 2A, lane 6). These results suggest that the iron-sparing response requires both FsrA and one or more of these small proteins postulated to function as RNA chaperones. The fbpAB and fbpC operons, however, appear to be at least partially redundant in function. There appears to be a sufficient degree of iron-sparing in strains containing either FbpC alone (Fig. 2A, lane 4) or FbpAB (Fig. 2A, lane 5), to allow reasonably good growth (relative to wild-type cells) under these conditions.
We hypothesized that these effects of the FsrA sRNA and the FbpABC proteins might be explained by their role in reducing cellular iron demand. To measure the amount of iron needed to support cell growth we monitored iron content in wild-type and mutant cells grown under iron sufficient conditions (Fig. 2B). The amount of cellular iron (after washing to remove surface-associated ions) as measured by using ferrozine (33) was reduced ≈30–40% in the fur mutant (Fig. 2B, lane 2). Significantly, this effect was largely reversed in the fur fsrA and fur fbpAB strains (Fig. 2B, lanes 3 and 4). Surprisingly, this effect was exacerbated in the fur fbpC double mutant (Fig. 2B, lane 5). These results suggest that FsrA and the FbpAB proteins may function in the same pathway in determining iron content and that FbpC has an opposing effect. The imperfect correlation between the effects of these mutations on growth and on iron content hints at the complex nature of this stress response: FsrA clearly plays a major role in helping prioritize iron utilization but the effects of the FbpABC proteins are complicated and vary among the different protein targets.
Transcriptome Analyses Identify Genes Positively Regulated by Fur.
In E. coli, RyhB inhibits expression of ≈18 operons by RNA-RNA interaction (19). Like other sRNAs, RyhB affects steady state mRNA levels because of degradation of the resulting RNA-RNA duplexes, or simply because of the decreased stability associated with the lack of translation (34, 35). Regardless of mechanism, the effects of transacting sRNAs can be visualized, to a first approximation, by monitoring their effects on the transcriptome.
We hypothesized that the FsrA-mediated iron-sparing response should lead to specific changes in the transcriptome. In a previous microarray analysis, we demonstrated that, under iron-replete conditions, Fur negatively regulates the expression of ≈20 operons (by direct binding to associated operators) and functions as a positive regulator for numerous operons, including several encoding iron-containing enzymes and cytochromes (25). Down-regulation of some of these same genes was noted in a recent transcriptome analysis of B. subtilis under conditions of iron deprivation (29).
To identify candidate target operons for FsrA, we focused on the ≈240 genes that were decreased in mRNA level at least 4-fold in the fur mutant (25). Many of these genes (e.g., encoding ribosomal proteins and translation factors) are likely expressed at a lower level as a result of the slower growth rate of the mutant. Others encode iron-containing enzymes important for key metabolic pathways including enzymes of the TCA cycle, respiratory cytochromes, and amino acid biosynthesis (ILV, Cys, and Glu) (Table S1). Many of these same mRNAs are also down-regulated in cells starved for iron as measured 15 min after treatment with the iron chelator 2,2′-dipyridyl (DP) (Table S1).
FsrA sRNA Represses Expression of Succinate Dehydrogenase.
By analogy with RyhB, we hypothesized that FsrA might act as an antisense RNA for some or all of these target genes. Indeed, we identified several possible targets for FsrA by searching the B. subtilis genome, using TargetRNA (36) (Table S1). Among the strongest matches was the leader region of the operon encoding Sdh (Fig. 3A), a common target of iron-sparing responses in several organisms. In our previous microarray analyses, we noted that the sdhA, B, and C transcripts were down-regulated ≈3-fold in the fur mutant. This decrease is largely reversed in the fur fsrA double mutant (data not shown). This suggests that the decrease in transcript level is due to increased degradation subsequent to formation of the FsrA:sdhC duplex. In vitro, FsrA and the sdhC leader region form an RNA-RNA complex as judged by RNA mobility shift (Fig. 3B).
Fig. 3.
FsrA regulation of succinate dehydrogenase expression. (A) Predicted pairing between FsrA and the 5′-leader region of sdhC, the first gene of the sdhCAB operon. (B) Binding of the FsrA to the 5′ leader region of sdhC (200 nM) as detected by EMSA, using radioactively labeled sdhC (200 nM) and cold fsrA transcripts. (C) Level of SdhA protein as detected by proteomics (see Fig. S4 and S5). (D) Growth (OD600 after overnight growth at 37°C) in minimal medium containing succinate as carbon source.
The sequence complementarity between FsrA and sdhC predicts that FsrA will down-regulate translation of Sdh. Consistent with this expectation, the level of SdhA protein detected by 2D PAGE was significantly decreased in the fur mutant strain, but it was restored to wild-type levels in the fur fsrA mutant (Fig. 3C and Fig. S4). In addition, measurement of Sdh activity indicates an ≈8-fold decrease in the fur mutant strain but normal levels in the fur fsrA double mutant (data not shown). In general, there is a correlation between the level of expression of SdhA and the ability of the cells to grow on succinate as sole carbon source (Fig. 3D).
Many small regulatory RNAs require a chaperone, such as Hfq, which is important for the iron-sparing response in E. coli (18). Although the B. subtilis ymaH gene encodes a protein homologous to Hfq, this protein is not required for FsrA-mediated inhibition of Sdh expression because a fur ymaH double mutant does not regain the ability to grow on succinate. To date, the function of Hfq remains unclear in B. subtilis and it is apparently not needed for the function of other known sRNAs (37, 38). Perhaps because of the extensive sequence complementarity between FsrA and the sdhC mRNA (Fig. 3A), this binding interaction also does not require the postulated FbpAB and FpbC protein chaperones either in vitro (Fig. 3B) or in vivo (growth on succinate is not restored in a fur fbpABC mutant; Fig. 3D).
FsrA sRNA Represses Expression of Aconitase.
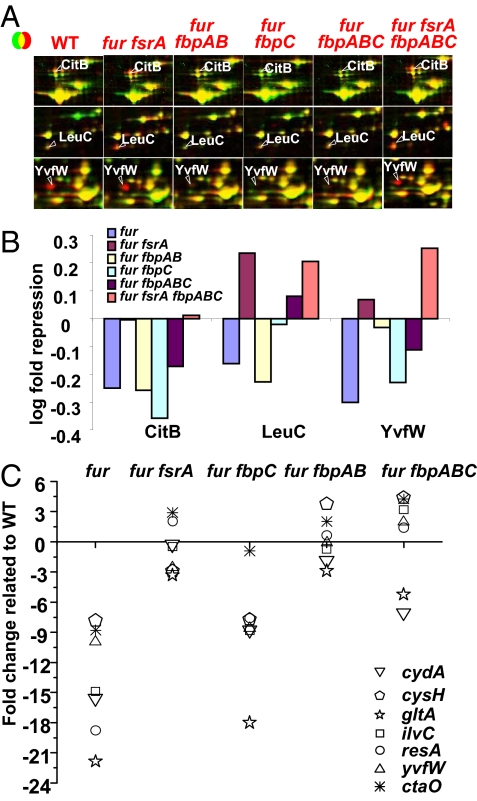
To futher explore the effects of FsrA and these Fur-regulated proteins on gene regulation, we have used 2D PAGE to monitor both cytosolic proteins and membrane-associated lipoproteins in our collection of mutant strains (Figs. S4-S6 and Table S2). As expected, those proteins directed repressed by Fur are up-regulated in all of the fur mutant strains. Among those proteins that are down-regulated in the fur mutant, many are increased in level in strains additionally carrying mutations in fsrA and/or the fbpAB or fbpC operons. Three examples of proteins with unique patterns of regulation are illustrated in Fig. 4A and quantified in Fig. 4B.
Fig. 4.
Remodeling of the proteome functions to reduce cellular iron utilization. (A) Selected examples of proteins positively regulated by Fur (CitB, LeuC, and YvfW) as visualized by using 2D PAGE (see Figs. S4–S6 for complete datasets). (B) Quantitation of protein expression levels for CitB, LeuC, and YvfW from A. (C) RT-PCR analysis of mRNA levels for representative FsrA target genes. Fold-change in mRNA expression levels (relative to wild type) was measured by using quantitative RT-PCR. Results shown are representative of experiments performed at least three times.
Aconitase (CitB) is decreased in intensity ≈2-fold in the fur mutant and this effect is reversed only in those strains lacking FsrA. Thus, like Sdh, the repression of aconitase requires FsrA but is independent of the fbpAB and fbpC operons. Aconitase contains a 4Fe–4S cluster and is a known target of iron-sparing responses in C. glutamicum (mediated by the RipA protein, ref. 23) and in S. cerevisiae (mediated by Cth2p; 22). It is interesting to note that B. subtilis aconitase, like its mammalian counterparts that function as iron-regulatory element binding proteins (IRE-BP), has an iron-regulated RNA-binding activity (39). Thus, under conditions of severe iron deprivation FsrA mediates repression of continued synthesis of aconitase and loss of iron from existing aconitase molecules may further reduce enzyme activity and activate its RNA-binding function. Although the roles of aconitase as an RNA-binding protein are not well understood in B. subtilis, is has been reported to bind internally to the feuABC mRNA (39), which encodes the major uptake system for the endogenous siderophore bacillibactin (10).
In addition to its role in mediating repression of Sdh and aconitase, FsrA appears to be the major factor determining several phenotypes characteristic of the fur mutant. In addition to an inability to grow on succinate, a fur mutant is unable to grow on fumarate as sole carbon source or on ammonium as sole nitrogen source (possibly reflecting regulation of the gltAB operon). In both cases, growth is restored in the fur fsrA double mutant, but not in the fur fbpAB fbpC mutant. Therefore, these growth phenotypes depend on FsrA, but not on the three protein-coding genes.
Repression of YvfW Requires Products of Both the fsrA and fbpAB Operons.
Expression of YvfW, an unknown function ferredoxin-like protein, is also repressed in a fur mutant strain (Fig. 4A and Table S1). However, in this case repression is alleviated by either mutation of fsrA or the fbpAB operon (Fig. 4). Regulation of YvfW by both FsrA and the fbpAB operon is confirmed by RT-PCR (Fig. 4C) and has also been seen in transcriptome comparisons of these same strains (G.T.S. and J.D.H., unpublished results).
Repression of Isopropyl Malate Dehydratase (LeuCD) Requires Products of the fsrA and fbpC Operons.
The expression of enzymes involved in branched chain amino acid biosynthesis was also strongly regulated by iron. This likely reflects the fact that isopropyl malate dehydratase, the product of the leuC and leuD genes, is an iron-sulfur containing enzyme. In B. subtilis, the ilv-leu operon encodes seven proteins that participate in branched chain amino acid biosynthesis and expression of this operon is subject to complex control (40). At least three different transcription factors regulate transcription of this operon, and the primary transcript is processed to yield several discrete mRNA species with differing stabilities (41). LeuC protein levels were reduced in a fur mutant, but, in this case, repression was largely alleviated by mutation of either fsrA alone or fbpC alone (Fig. 4) and completely relieved in multiply mutant strains. The leuCD genes were shown to be derepressed in a ripA mutant in C. glutamicum, indicating that this enzyme is also subject to the iron-sparing response in this organism.
fsrA, fbpAB, and fbpC Operons Function in Combination to Regulate Protein Synthesis.
To extend this analysis and determine whether these changes in protein levels were correlated with changes in mRNA levels, we used quantitative RT-PCR to monitor the transcript levels for several presumed target operons of the iron-sparing response (Fig. 4C). The fur mutant had a decreased level of mRNA corresponding to biosynthetic enzymes for branched chain amino acids (ilvC), cytochromes (cydA, resA, and ctaO), cysteine (cysH), glutamate (gltA), and the ferredoxin-like protein (yvfW). The levels of many of these RNAs were restored to near wild-type levels in the fur fsrA, fur fbpAB, or fur fbpAB fbpC mutant strains. As noted in the examples above, FsrA appears to have the strongest effect on expression and RNA levels were restored to near wild-type levels for each of these targets in the fur fsrA double mutant strain. Strong effects were also noted for the fur fbpAB mutant strain, whereas the fur fbpC strain displayed a full derepression for only one tested target gene, gltA (Fig. 4C). These results highlight the complexity of this regulatory system and suggest a model in which the FbpAB and FpbC proteins may function to modulate the activity or the targeting of the FsrA sRNA.
Constitutive Expression of the Iron-Sparing Response Is Maladaptive Under a Variety of Growth Conditions.
To explore the physiological role of this iron-sparing response, we have used an automated growth (optical density) monitoring system (BioScreen) to monitor our panel of mutant strains in LB and glucose minimal medium with and without supplementation with various amino acids (Table S3). Consistent with previous results, the fur mutant displayed both a reduction in the maximal rate of growth and a prolonged lag phase in a variety of conditions and was unable to grow with succinate or fumarate as sole carbon source. These effects were partially suppressed either by mutation of fsrA or, in some cases, by mutation of one or both of the fbpAB and fbpC operons. When the tabulated growth data were analyzed by using a numerical taxonomy approach (42), it became clear that the fur, fur fbpAB, and fur fbpC mutants all cluster together, indicating that the individual mutation of either the fbpAB or the fbpC operons did little to suppress the poor growth phenotypes under most conditions (Table S3). Conversely, the fur fsrA and the fur fbpAB fbpC mutants clustered with the wild-type strain indicating that mutation of either the sRNA or all three peptide-encoding genes suppressed many of the growth phenotypes. These results are consistent with the general notion that the fbpAB and fbpC loci are at least partially redundant in function, as noted for growth in medium supplemented with EDDHA (Fig. 2A).
Conclusion
The results presented here establish the broad outlines of the iron-sparing response in B. subtilis. Like the enterobacteria, this response involves at least one sRNA (FsrA) that acts to down-regulate the synthesis of numerous iron-containing enzymes and iron-consuming biosynthetic pathways. However, FsrA-mediated regulation also involves the fbpAB and fbpC genes. As monitored by RT-PCR, down-regulation many RNA targets in the fur mutant requires both the FsrA sRNA and the fbpAB operon (Fig. 3A). In some, but not all, cases, fbpC is also involved. As monitored by proteomics, the effects are also complex and FsrA appears to be the most important effector for many of the protein targets.
The fbpAB and fbpC genes encode small, basic proteins that we propose function as dedicated Fur-regulated RNA chaperones (either alone or in heterooligomeric complexes). We do not exclude the possibility that the fbpAB and fbpC transcripts may also function as sRNAs to affect gene expression. Indeed, E. coli sgrS has recently been shown to regulate sugar transport by acting both as a sRNA and by encoding a protein product (43). Ongoing studies are directed at defining the complete suite of targets for FsrA-mediated regulation and the precise roles of the fbpAB and fbpC genes in what appears to be a complex combinatorial mechanism for regulation of gene expression.
Materials and Methods
Strain Construction and Growth Conditions.
B. subtilis strains were constructed by using long-flanking homology PCR as described in ref. 44. For selection, antibiotics were added as follows: 1 μg/ml erythromycin and 25 μg/ml lincomycin (for selecting for macrolide-lincosamide-streptogramin B resistance), 100 μg/ml spectinomycin, 10 μg/ml chloramphenicol, 15 μg/ml kanamycin, and 10 μg/ml neomycin. B. subtilis was grown in LB or in a Mops-based glucose minimal medium suitable for iron starvation studies (27). Unless otherwise indicated, liquid media were inoculated from an overnight preculture and incubated at 37°C with shaking at 200 rpm. E. coli DH5α was used for routine DNA cloning as described in ref. 45. Restriction enzymes, DNA ligase and DNA polymerases were used according to the manufacturer's instructions (New England Biolabs). Northern blot analysis and EMSA experiments were done by using standard techniques as described in SI Methods.
Overexpression and Detection of FbpA, B, and C Proteins.
Expression constructs for C-terminal FLAG-tagged fusions were constructed by PCR (46). The resulting products were cloned in pET16B and pDGG1664 (47) and expressed in E. coli and B. subtilis respectively. The fusions were detected by Western blot analysis, using anti-FLAG antibodies (Sigma).
Quantification of Target Genes Expression by Real-Time PCR.
RNA was extracted from strains grown in LB to mid-log phase, using an RNA extraction kit (Qiagen). cDNA was synthesized by using random hexamer primers and Taqman reverse transcription kit (Roche), and real-time PCR was carried out by using SYBR Green (Bio-Rad) and gene-specific primers according to the manufacturers' instructions.
Analysis of RNA–RNA Complex Formation.
Both target RNA (5′ 166 sdhC mRNA) and fsrA were synthesized in vitro from PCR-generated template fragments, using a T7 RNA polymerase kit (Ambion) and purified from 6% denaturing polyacrylamide gels according to the manufacturer's instructions. RNA-RNA complex formation was measured according to (38), using 6% native polyacrylamide gels containing 1× TBE. Dried gels were analyzed by PhosphoImaging.
Proteome Analysis.
B. subtilis strains were grown in Belitsky minimal medium (48) to an OD500 of 0.4 and harvested for the proteome analysis, using dual channel imaging of 2D PAGE gels as described in ref. 49 (Figs. S4 and S5 and SI Methods). To define the membrane-attached lipoproteome (Fig. S6), strains were grown in LB and harvested 1 h after entry into the stationary phase at OD540 = 3.0. Sdh activity was measured in cells grown in minimal succinate medium according to ref. 50.
Supplementary Material
Acknowledgments.
We thank E. Massé for helpful comments on this manuscript. This work was supported by National Institutes of Health Grant GM059323.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711752105/DCSupplemental.
References
- 1.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Wandersman C, Delepelaire P. Bacterial iron sources: From siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 3.Faraldo-Gomez JD, Sansom MS. Acquisition of siderophores in gram-negative bacteria. Nat Rev Mol Cell Biol. 2003;4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 4.Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 5.Brune I, et al. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics. 2006;7:21. doi: 10.1186/1471-2164-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wennerhold J, Bott M. The DtxR regulon of Corynebacterium glutamicum. J Bacteriol. 2006;188:2907–2918. doi: 10.1128/JB.188.8.2907-2918.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez GM. Control of iron metabolism in Mycobacterium tuberculosis. Trends Microbiol. 2006;14:320–327. doi: 10.1016/j.tim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Ranjan S, Yellaboina S, Ranjan A. IdeR in mycobacteria: From target recognition to physiological function. Crit Rev Microbiol. 2006;32:69–75. doi: 10.1080/10408410600709768. [DOI] [PubMed] [Google Scholar]
- 9.Johnston AW, et al. Living without Fur: The subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals. 2007;20:501–511. doi: 10.1007/s10534-007-9085-8. [DOI] [PubMed] [Google Scholar]
- 10.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol. 2006;188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 12.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosse C, et al. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol. 2006;62:120–131. doi: 10.1111/j.1365-2958.2006.05326.x. [DOI] [PubMed] [Google Scholar]
- 14.Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol Microbiol. 2007;65:857–875. doi: 10.1111/j.1365-2958.2007.05802.x. [DOI] [PubMed] [Google Scholar]
- 15.Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: Protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdul-Tehrani H, et al. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilderman PJ, et al. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 23.Wennerhold J, Krug A, Bott M. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J Biol Chem. 2005;280:40500–40508. doi: 10.1074/jbc.M508693200. [DOI] [PubMed] [Google Scholar]
- 24.Friedman DB, et al. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2006;2:e87. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- 26.Baichoo N, Helmann JD. Recognition of DNA by Fur: A reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, James LP, Helmann JD. Metalloregulation in Bacillus subtilis: Isolation and characterization of two genes differentially repressed by metal ions. J Bacteriol. 1993;175:5428–5437. doi: 10.1128/jb.175.17.5428-5437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 29.Miethke M, Westers H, Blom EJ, Kuipers OP, Marahiel MA. Iron starvation triggers the stringent response and induces amino acid biosynthesis for bacillibactin production in Bacillus subtilis. J Bacteriol. 2006;188:8655–8657. doi: 10.1128/JB.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer E, Sauer U. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat Genet. 2005;37:636–640. doi: 10.1038/ng1555. [DOI] [PubMed] [Google Scholar]
- 31.Jacques JF, et al. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 32.Quadri LE, et al. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 33.Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem. 2004;331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 34.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tjaden B, et al. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvaggi JM, Perkins JB, Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heidrich N, Chinali A, Gerth U, Brantl S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol Microbiol. 2006;62:520–536. doi: 10.1111/j.1365-2958.2006.05384.x. [DOI] [PubMed] [Google Scholar]
- 39.Alen C, Sonenshein AL. Bacillus subtilis aconitase is an RNA-binding protein. Proc Natl Acad Sci USA. 1999;96:10412–10417. doi: 10.1073/pnas.96.18.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tojo S, et al. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol Microbiol. 2005;56:1560–1573. doi: 10.1111/j.1365-2958.2005.04635.x. [DOI] [PubMed] [Google Scholar]
- 41.Mader U, Hennig S, Hecker M, Homuth G. Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes. J Bacteriol. 2004;186:2240–2252. doi: 10.1128/JB.186.8.2240-2252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Litzner BR, Caton TM, Schneegurt MA. Carbon substrate utilization, antibiotic sensitivity, and numerical taxonomy of bacterial isolates from the Great Salt Plains of Oklahoma. Arch Microbiol. 2006;185:286–296. doi: 10.1007/s00203-006-0096-6. [DOI] [PubMed] [Google Scholar]
- 43.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butcher BG, Helmann JD. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 46.Kaltwasser M, Wiegert T, Schumann W. Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl Environ Microbiol. 2002;68:2624–2628. doi: 10.1128/AEM.68.5.2624-2628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 48.Stulke J, Hanschke R, Hecker M. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J Gen Microbiol. 1993;139:2041–2045. doi: 10.1099/00221287-139-9-2041. [DOI] [PubMed] [Google Scholar]
- 49.Tam le T, et al. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ Microbiol. 2006;8:1408–1427. doi: 10.1111/j.1462-2920.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- 50.Hatefi Y. Resolution of complex II and isolation of succinate dehydrogenase (EC 1.3.99.1) Methods Enzymol. 1978;53:27–35. doi: 10.1016/s0076-6879(78)53009-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.