Abstract
Molecular recognition by the adaptive immune system relies on specific high-affinity antibody receptors that are generated from a restricted set of starting sequences through homologous recombination and somatic mutation. The steroid binding antibody DB3 and the catalytic Diels–Alderase antibody 1E9 derive from the same germ line sequences but exhibit very distinct specificities and functions. However, mutation of only two of the 36 sequence differences in the variable domains, LeuH47Trp and ArgH100Trp, converts 1E9 into a high-affinity steroid receptor with a ligand recognition profile similar to DB3. To understand how these changes switch binding specificity and function, we determined the crystal structures of the 1E9 LeuH47Trp/ArgH100Trp double mutant (1E9dm) as an unliganded Fab at 2.05 Å resolution and in complex with two configurationally distinct steroids at 2.40 and 2.85 Å. Surprisingly, despite the functional mimicry of DB3, 1E9dm employs a distinct steroid binding mechanism. Extensive structural rearrangements occur in the combining site, where residue H47 acts as a specificity switch and H100 adapts to different ligands. Unlike DB3, 1E9dm does not use alternative binding pockets or different sets of hydrogen-bonding interactions to bind configurationally distinct steroids. Rather, the different steroids are inserted more deeply into the 1E9dm combining site, creating more hydrophobic contacts that energetically compensate for the lack of hydrogen bonds. These findings demonstrate how subtle mutations within an existing molecular scaffold can dramatically modulate the function of immune receptors by inducing unanticipated, but compensating, mechanisms of ligand interaction.
Keywords: antibody–antigen complex, modulation of receptor specificity, molecular recognition, protein engineering, x-ray crystallography
Molecular recognition of antigens by the immune system is challenging because this process requires fulfillment of two opposing criteria. First, the repertoire must be able to recognize the vast universe of foreign antigens. Second, specificity and selectivity for any given antigen must be ensured to avoid self-reactivity and autoimmune diseases, such as lupus, rheumatoid arthritis, type I diabetes, or multiple sclerosis. How the immune system balances these factors remains incompletely understood despite decades of study. The adaptive immune response evolves immune receptors through recombination from a limited, but still substantial, arsenal of germ line precursors that are then optimized by class switching and affinity maturation. However, there are far fewer germ line precursors [≈108 different antibody sequences (1)] than potential antigens. Thus, a restricted number of antibody scaffolds must suffice for recognition of all possible ligands, including synthetic compounds that are not likely to be encountered in microbial infection or disease.
An interesting example that highlights the limitations of antibody specificity is the steroid binding antibody DB3 (2). DB3 was raised against a progesterone derivative to examine the role of progesterone during pregnancy in mice, but it cross-reacts with a configurationally diverse set of steroids with nanomolar affinity (3, 4). These compounds differ in the configuration of their A ring relative to the B, C, and D rings going from essentially planar to being bent out of the plane by almost 90° (Fig. 1 B and C). Structural analysis of DB3 complexes with different steroids has revealed that alternative binding modes rather than conformational changes of the protein largely account for its antigen cross-reactivity (4, 5). Depending on the steroid configuration, the B, C, and D rings are sandwiched by TrpH50 and TrpH100 in either a “syn” or an “anti” mode, as defined by the relative disposition of the two methyl groups of progesterone relative to TrpH50 (4), whereas the A ring can alternatively occupy two different pockets on the surface of the antibody (6). In each orientation, the specificity of the recognition is focused on the steroid D ring, which is deeply buried in a hydrophobic cavity and accepts a hydrogen bond from AsnH35 to the keto group at C17 or C20 (6). The C3 keto group either hydrogen bonds with HisL27d (for steroids in the syn orientation), or with a water molecule (for steroids in the anti orientation).
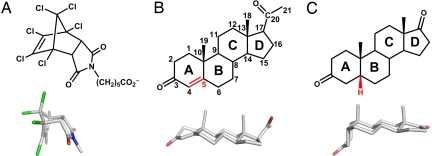
Fig. 1.
Structures of ligands bound by 1E9, 1E9 LeuH47Trp/ArgH100Trp (1E9dm), and DB3. (A) The endo hexachloronorbornene derivative is a transition state analog of the Diels–Alder cycloaddition catalyzed by 1E9. (B and C) Progesterone (B) and 5β-androstane-3,17-dione (C) are structurally distinct steroids. Their A rings assume different orientations relative to the rest of the steroid skeleton (B, C, and D rings) because of the substitution at carbon C5 (indicated in red). Progesterone is C5-unsaturated (sp2 hybridization) with an ≈35° bent A ring. 5β-androstane-3,17-dione is 5β-substituted (sp3) and its A ring is almost perpendicular to the rest of the steroid skeleton.
Interestingly, DB3 shares 91% sequence identity to another antibody, 1E9, which efficiently catalyzes the cycloaddition between tetrachlorothiophene dioxide and N-ethylmaleimide (7). 1E9 was raised against a stable analogue of the bicyclic reaction intermediate (Fig. 1A) to which it binds with nanomolar affinity. Structural analysis and theoretical calculations have attributed the catalytic efficiency of 1E9 to enthalpic stabilization of the reaction intermediate, near-perfect shape complementarity of the hydrophobic binding site for the transition state, and a strategically placed hydrogen bond (8–10).
Although the steroid binding DB3 and the Diels–Alderase 1E9 antibodies derive from the same germ line sequences (VGAM3.8 and Vκ5.1 for the variable heavy and light chain gene segments, respectively), their variable domains exhibit 36 sequence differences. Six of these amino acids, namely residues L89, L94, H47, H97, H100, H100b, are located in the combining site, providing an explanation for their different functions (10) and weak cross-reactivity (11). Interestingly, some of the key combining site residues are identical; the hallmark residue of the VGAM3.8 gene family, AsnH35, is crucial for ligand binding by DB3 and catalysis by 1E9, and TrpH50 contributes significantly to ligand binding in both cases (4, 10). Otherwise, for DB3, specific interactions between the cavity-lining residues and the steroid skeleton, particularly with the D ring, are essential (4, 5), whereas, for efficient catalysis by 1E9, shape complementarity combined with a few specific interactions are most important (8, 9).
Piatesi et al. (12) showed by site-directed mutagenesis and binding studies that only two mutations are needed to interconvert the binding specificities of 1E9 and DB3. The LeuH47Trp/ArgH100Trp 1E9 double mutant (1E9dm) both binds steroids with nanomolar affinity and recapitulates the binding specificity of DB3 for a panel of structurally and configurationally distinct molecules. By structural analysis, we now investigate on an atomic level how these two mutations enable a restricted antibody scaffold to fulfill such diverse functions as catalysis of a Diels–Alder cycloaddition and high-affinity steroid binding. We uncovered unexpected steroid binding modes for 1E9dm that imply that the ligand-binding properties of structurally homologous protein-binding sites may evolve via unanticipated intermediates rather than directly. Our findings indicate that subtle changes in predefined binding sites can dramatically modulate selectivity and affinity by creating novel interaction mechanisms.
Results
1E9dm Fab Crystal Structures.
The crystal structure of the 1E9dm Fab was determined for the unliganded protein and in complex with two configurationally distinct steroids (13, 14), progesterone (Fig. 1B), and 5β-androstane-3,17-dione (Fig. 1C) (Table 1). Despite their high solvent content (≈75%, VM = 4.7 Å3/Da for 1 mol/ASU), the crystals diffracted to resolutions of 2.05 Å (apo protein), 2.40 Å (5β-androstane-3,17-dione complex), and 2.85 Å (progesterone complex). The Fab molecules are packed in a honeycomb lattice with large solvent channels (≈100 Å in diameter) that run parallel to the threefold axis. 1E9dm resembles typical Fab structures (15) and steroid binding does not induce conformational changes other than in the binding site (RMS deviation of 0.23–0.27 Å for superposition of all backbone atoms).
Table 1.
Data collection and refinement statistics of the 1E9dm structures
| Apo | Progesterone complex | 5β-androstane-3,17-dione complex | |
|---|---|---|---|
| Data collection | |||
| Space group | P3121 | P3121 | P3121 |
| Unit cell dimensions | |||
| a, Å | 127.6 | 128.4 | 127.3 |
| b, Å | 127.6 | 128.4 | 127.3 |
| c, Å | 91.9 | 91.8 | 92.0 |
| Resolution, Å | 50.00–2.05 (2.29–2.05)* | 50.00–2.85 (2.92–2.85)* | 50.00–2.40 (2.49–2.40)* |
| Rsym,† % | 8.2 (52.5) | 11.1 (38.2) | 8.5 (52.9) |
| 〈I/σI〉 | 15.6 (2.4) | 9.6 (2.4) | 14.3 (2.7) |
| Completeness, % | 99.6 (100.0) | 96.4 (88.3) | 99.9 (99.9) |
| Unique reflections | 54,536 | 20,438 | 33,845 |
| Redundancy | 4.1 (3.9) | 3.9 (4.1) | 4.3 (4.3) |
| Refinement | |||
| Rwork‡/Rfree,§ % | 17.8 / 20.4 | 18.3 / 23.6 | 17.1 / 21.6 |
| Refined atoms | |||
| Protein | 3,536 | 3,351 | 3,416 |
| Ligand | 8 | 23 | 21 |
| Water | 278 | 65 | 175 |
| Sulfate | 65 | 70 | 125 |
| Average B values | |||
| Protein, Å2 | 48 | 46 | 47 |
| Ligand, Å2 | 58 | 35 | 46 |
| Water, Å2 | 52 | 33 | 47 |
| Sulfate, Å2 | 64 | 47 | 57 |
| rmsd | |||
| Bond lengths, Å | 0.017 | 0.015 | 0.015 |
| Bond angles, ° | 1.67 | 1.65 | 1.57 |
| Ramachandran plot | |||
| Allowed | 99.8 | 100.0 | 99.5 |
| Favored | 98.4 | 95.2 | 97.5 |
| Disallowed | 0.2 | 0.0 | 0.5 |
* Highest resolution shells are shown in parenthesis.
†Rsym = ΣhklΣi Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl)
‡Rwork = Σhkl||Fc(hkl)| − |Fo(hkl)||/Σ hkl|Fo(hkl)|.
§Rfree is calculated in the same manner as Rwork but from 5% of the data that was not used for refinement.
1E9dm Apo Structure.
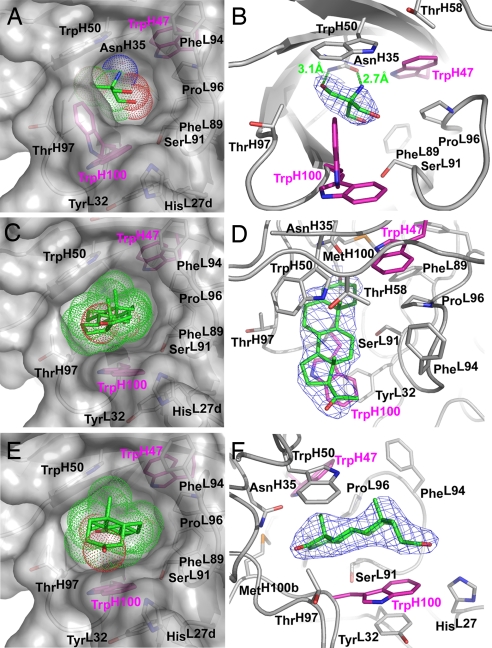
The majority of the residues in the 1E9dm binding pocket are contributed by the complementarity-determining regions (CDRs). The active site is lined by the hydrophobic side chains of PheL89, PheL94, ProL96, TrpH47, TrpH50, TrpH100, and MetH100b; the Cα and Cβ atoms of SerL91; the Cβ and Cγ2 atoms of ThrH58 and ThrH97; the Cδ2, Nε2 and Cε1 atoms of HisL27d; and the side chain of AsnH35. The latter residue forms two hydrogen bonds (2.7 and 3.1 Å) to a Tris molecule (Fig. 2 A and B) that originates from the protein storage buffer.
Fig. 2.
Structures of the 1E9dm Fab combining site with different bound ligands. (Left) Surface representations of the combining site residues with the bound ligands (green) in similar perspectives. The van der Waals radii of the ligands are indicated as dots to give an impression of their fit in the binding pocket. The leuH47Trp and ArgH100Trp mutations were introduced into WT 1E9 to generate 1E9dm (pink). (Right) 2Fo − Fc electron density maps (1σ level) in different views around the ligands (blue mesh) highlighting the quality of the maps. (A and B) 1E9dm Fab apo structure with the bound Tris buffer molecule. The open and closed conformations of TrpH100 are illustrated. (C and D) 1E9dm progesterone complex. (E and F) 1E9dm 5β-androstane-3,17-dione complex.
All residues in the binding site are well ordered, except for TrpH100 whose side chain is flexible as indicated by less well defined electron density beyond Cγ and elevated B values (average of 62 Å2 as compared with the average of 44 Å2 for other side-chains in the binding site). Different orientations of the indole ring can be discerned (Fig. 2B); “open” states are characterized by π-stacking of the TrpH100 side chain with the phenol ring of TyrL32, whereas the indole of TrpH100 rotates into the ligand binding pocket of “closed” states to minimize exposure of the hydrophobic surface (4).
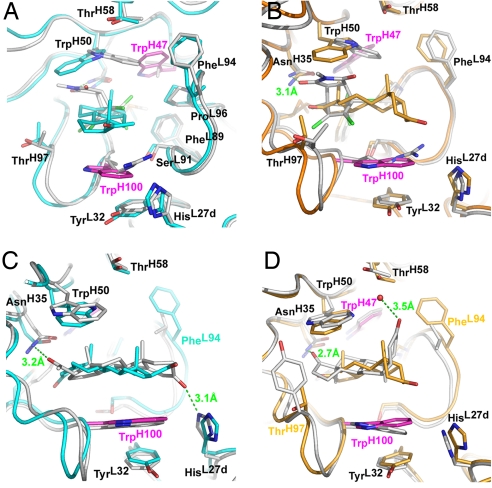
The LeuH47Trp and ArgH100Trp mutations significantly alter the shape of the 1E9 binding pocket such that the combining site of the double mutant now resembles a fusion between 1E9 and DB3 (Fig. 3). As in DB3, the TrpH100 side chain can either act as a surrogate ligand for the unliganded binding pocket, or in the open conformation, as a hydrophobic platform that can adapt to distinct ligands to provide van der Waals interactions. Although the ArgH100Trp mutation does not significantly change the affinity for the 1E9 TSA, it enhances the affinity for steroids (12). Residue H47, however, primarily acts as a specificity switch: Replacement of LeuH47 by the bulky TrpH47 leads to a complete reorganization of the 1E9 binding site and three orders of magnitude weaker TSA binding (12), because it sterically forces the TrpH50 indole to rotate ≈145° around its Cβ-Cγ bond [Fig. 4 A and B and supporting information (SI) Figs. S1 and S2] to assume approximately the same location as in DB3 (4, 5) (Fig. 3 B and C). However, the TrpH50 side chain is rotated ≈75° around χ1 and 180° around χ2 compared with DB3 (Fig. 4 C and D and Figs. S3 and S4), although no obvious spatial restrictions or interactions would favor a particular rotamer in either protein.
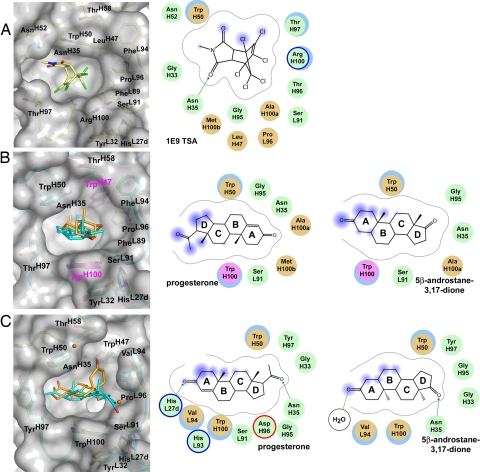
Fig. 3.
Ligand binding by Diels–Alderase 1E9 (A), 1E9dm (B), and the steroid-binding DB3 (C). (Left) Proteins are shown in the same orientation to demonstrate the distinct shapes of the combining sites and the different ways in which the respective ligands are bound. Light yellow, 1E9 TSA; cyan, progesterone; orange, 5β-androstane-3,17-dione and two ordered water molecules. (Right) 2D schemes of the ligand binding modes are shown. Green, polar residues; brown, hydrophobic; blue-lined circle, basic; red-lined circle, acidic; gray dashed line, proximity contour; fuzzy blue, ligand exposure; blue underlayed circle, receptor exposure; green arrow, side chain donor; olive line, solvent contact.
Fig. 4.
Overlay of the combining sites of 1E9, 1E9dm, and DB3 with bound ligands. (A) 1E9 and its TSA (gray) superimposed with 1E9dm binding progesterone (cyan). This view is rotated around the z axis ≈90° compared with B–D to demonstrate the movement of the TrpH50 side chain in 1E9dm caused by the LeuH47Trp mutation. (B) The TSA bound by 1E9 (gray) and 5β-androstane-3,17-dione bound by 1E9dm (orange). (C) Progesterone bound by 1E9dm in the inverse head-to-tail binding mode with a buried A ring (cyan). The same steroid bound by DB3 (gray). (D) 5β-androstane-3,17-dione bound by 1E9dm (orange) and DB3 (gray).
Steroid Recognition by 1E9dm.
The remodeling of the 1E9 active site due to the LeuH47Trp mutation in 1E9dm allows it to snugly sequester both the slightly bent progesterone and the highly kinked 5β-androstane-3,17-dione (Fig. 2 C and E). Although structurally distinct, both steroids occupy essentially the same general location in the deep cavity in an orientation approximately perpendicular to the protein surface (Fig. 3B). Three rings of the steroid skeleton (78% of the steroid surface) are deeply buried in the pocket, whereas the ring proximal to the pocket entrance stacks with the more mobile and adjustable indole side chain of TrpH100. In 5β-androstane-3,17-dione, the D ring is buried in the protein interior and the perpendicular A ring packs against TrpH100 (Fig. 2F). In contrast, progesterone is bound with an inverse head-to-tail arrangement burying its A ring and packing the C and D rings, including the C20 substituent, against TrpH100 (Fig. 2D).
The majority of interactions between the steroids and 1E9dm are van der Waals contacts (46 of 49 and 42 of 42 for the progesterone and 5β-androstane-3,17-dione complexes, respectively). The heavy chain contributes many more interactions than the light chain (71.3% and 28.7% for VH and VL, respectively) in both complexes. More specifically, CDR H3 (41.1%), H2 (16.0%) and L3 (26.9%) provide the majority of the contacts, whereas H1 (8.4%), and L1 (1.8%), and some framework residues of the heavy chain (5.8%), make more modest contributions. Interestingly, 35% of all contacts are provided by only 2 of 18 interacting residues, namely TrpH50 (15%) and TrpH100 (20%). The indole rings of these residues engage in hydrophobic stacking and sandwich the steroid skeletons of progesterone (Fig. 2 C and D) and 5β-androstane-3,17-dione (Fig. 2 E and F).
Comparison of Steroid Binding by 1E9dm and DB3.
Even though 1E9dm and DB3 have comparable low nanomolar affinities for progesterone and 5β-androstane-3,17-dione (12), actual steroid binding and interactions with the antibody are accomplished differently. The principal axis of the steroid skeleton is rotated by ≈40° in 1E9dm compared with DB3 (Fig. 3 B and C). DB3 binds unsaturated C5 or 5α-substituted steroids (Fig. 1B) in a syn orientation and 5β-substituted steroids (Fig. 1C) in an anti binding mode (Fig. 3C) but, in both modes, its specificity is exclusively focused on the buried steroid D ring (4, 6). In contrast, 1E9dm binds both steroids in a syn orientation (Fig. 3B). As in DB3, 5β-androstane-3,17-dione is inserted so that its D ring is buried (Figs. 2F and 3B and Fig. S4). Most unexpectedly, progesterone is bound in 1E9dm with the A ring being most deeply buried (Figs. 2D and 3B). However, this progesterone binding mode does not translate to any significantly lower binding affinity. The Kd values of the progesterone complexes of 1E9dm and DB3 are both low nanomolar (12). The aforementioned ligand binding differences are caused by the discriminative space requirements of the corresponding H100b residues. 1E9 contains MetH100b, whereas DB3 features a bulky PheH100b that provides high shape complementarity to steroids and positions them closer to the opening of the DB3 combining site (Fig. 3C). The substitution of MetH100b in 1E9 versus PheH100b in DB3 reconfigures the binding pocket of 1E9dm and generates space in the base of the pocket that allows for deeper penetration of the ligands.
In contrast to DB3, hydrogen-bonding interactions do not appear to play a major role in steroid binding by 1E9dm. Because of the different location and arrangement of progesterone and 5β-androstane-3,17-dione in 1E9dm versus DB3, the steroid keto groups that are buried in the interior of 1E9dm do not engage in any strong hydrogen bonds, as any potential hydrogen bonding donors are either too distant (>3.5 Å) or in unfavorable geometry. Instead, the polar keto groups are either packed against the hydrogens of MetH100b Cγ and the AlaH100a peptide plane in the case of progesterone or against the hydrogens of AsnH35 Nδ2 and GlyH95 Cα in the case of 5β-androstane-3,17-dione. However, because of the formation of the crystal lattice, AsnL28 Nδ2 of a symmetry-related Fab hydrogen bonds weakly with O20 of progesterone or with O3 of 5β-androstane-3,17-dione, but these interactions would not occur in solution.
1E9dm does not hydrogen bond with either of the steroids, which is surprising because, in all DB3 steroid complexes, AsnH35 hydrogen bonds with the steroid keto group at C17 or C20 (Fig. 4 C and D and Figs. S3 and S4), thus orienting the steroid in the pocket and providing specific recognition (6). DB3 also hydrogen bonds with the C3 keto or hydroxyl groups of the syn or anti bound steroids with HisH27d Nε2 or a water molecule adjacent to ThrH58, respectively (Fig. 4 C and D and Figs. S3 and S4). DB3 thus provides hydrophobic interactions with the steroid skeleton and hydrogen bonding with either steroid keto or hydroxyl groups, whereas 1E9dm uses a more extended arsenal of hydrophobic interactions that are associated with deeper penetration of the ligands into the protein. Remarkably, despite the different binding mechanisms, DB3 and 1E9dm exhibit similar high affinities for these structurally distinct ligands (12)¶, indicating that the increased number of hydrophobic interactions between 1E9dm and the steroids energetically compensates for the lack of hydrogen bonds (16).
Discussion
To enhance our understanding of how subtle differences in highly homologous binding sites can modulate selectivity, affinity, and function, we investigated the evolution of the ligand recognition properties of two structurally related but functionally distinct antibodies, the steroid-binding DB3 and the Diels–Alderase 1E9, via crystallography. Piatesi et al. (12) recently showed that the specificity and function of 1E9 can be gradually switched to that of DB3 by mutation of only five residues (PheL89Ser/LeuH47Trp/ThrH97Tyr/ArgH100Trp/MetH100bPhe) in the combining site. As expected, because of its increased resemblance of DB3, 1E9 loses its ability to catalyze the Diels–Alder reaction or even bind the TSA, but gains steroid-binding properties. Residues at position H47 and H100 were identified as most crucial for discriminating between the Diels–Alder activity of 1E9 and steroid-binding by DB3. Remarkably, the introduction of only these two mutations (LeuH47Trp/ArgH100Trp) into the WT 1E9 binding site increases the affinity for steroids up to 14,000-fold and results in a 1E9 variant (1E9dm) that recapitulates the specificity of DB3 for a panel of structurally dissimilar ligands (12). Despite this functional mimicry, comparison of the 1E9dm and the DB3 crystal structures reveals structural differences that give rise to distinctively different shapes for their ligand binding sites and result in novel modes of steroid binding (Fig. 3). The fusion of the 1E9 framework with the DB3 mutations that confer altered ligand specificity in 1E9dm have created a binding site that accommodates steroids by unanticipated mechanisms and modes of binding without sacrificing affinity for a panel of steroids that differ in configuration at C5.
The ArgH100Trp mutation is significant for steroid binding by 1E9dm, because it provides first shell van der Waals interactions with the steroid skeleton, which act cooperatively with the LeuH47Trp mutation. In 1E9dm the TrpH100 side chain acts as a surrogate ligand for the apo binding site and undergoes a closed-to-open transition upon steroid binding just as in DB3. Slight adaptations of its side-chain conformation then facilitate hydrophobic stacking with different ligands. Although arginine and tryptophan have been found to be functionally equivalent at position H100 in some steroid-binding antibodies (17), the importance of the ArgH100Trp substitution for switching specificity is consistent with mutational studies on both 1E9 (12) and DB3 (18).
The nature of the second interaction shell, specifically the residue at H47, which does not make direct contact with the transition state analog in 1E9 and barely interacts with the steroids in 1E9dm or DB3, is critical for discrimination between the different classes of ligands bound by 1E9 and DB3. LeuH47 defines a pocket shape that is well suited for stabilizing the transition state of the Diels–Alder reaction, thus enabling catalysis, whereas TrpH47 is a prerequisite for steroid recognition and tight binding. The domino effect on other residues (namely TrpH50) that result from substitution of the second shell residue at H47 illustrates the importance of structural data for rational protein engineering. Zahnd et al. (19) made a comparable observation when they investigated single chain antibody fragments subjected to directed evolution. Crystallographic analysis revealed that mutations of noncontact residues confer significant improvement in antigen affinity. Likewise, for antibody 26-10, it was shown that even conservative mutations of a noncontact residue significantly affected the affinity for ligand (20, 21). Dubreuil et al. (22) used homology modeling combined with in vitro scanning saturation mutagenesis and error-prone PCR to improve the specificity of anti-progesterone antibody P15G12C12G11 by engineering of first and second sphere residues. Thus, for protein engineering, although it may often be difficult to obtain such optimized binding affinities without some structural data in hand, directed evolution can sometimes achieve that goal (23). The 1E9/DB3 case again highlights the importance of second shell residues to both stabilize and enhance the properties and interactions of the first shell and directly determine the functionality of protein binding sites. Therefore, the second interaction shells are at least equally valuable targets as the first shell in the rational design and engineering of protein binding sites (24, 25).
Biochemical analysis revealed similar nanomolar steroid binding affinities for 1E9dm and DB3 (12). Therefore, it was tempting to assume that the mutations of 1E9 LeuH47 and ArgH100 to tryptophan, as found in DB3, would permit 1E9 to interact in the same way with steroids as DB3. However, structural investigation of the steroid interactions of 1E9dm reveals that, although TrpH47 and TrpH100 are absolutely essential for steroid binding, their interaction mechanisms are distinct from DB3. The differences are associated with the deeper ligand binding site of 1E9dm that results from a less bulky and more flexible methionine at position H100b at the base of the pocket, instead of the phenylalanine that is found in DB3. Deeper penetration of the steroids into the 1E9dm interior (Fig. 3B) obviates the need for the different syn and anti binding modes seen in DB3 for steroids with different configurations. Rather, progesterone and 5β-androstane-3,17-dione are both bound in the syn-mode and are located at approximately the same location in the 1E9dm combining site (Fig. 3 B and C).
Hydrogen bonds, salt bridges, and polar interactions are important for discrimination and specificity of molecular recognition processes (26–28). Directional interactions, such as those provided by the polar atoms of the highly conserved AsnH35, contribute significantly to catalysis and ligand recognition of 1E9 and DB3 (4, 8, 10). Antigen specificity of DB3 has been proposed to be focused on conserved interactions with the steroid D rings, such as hydrogen bond acceptor requirements at C17 and C20 of the steroids, and conserved van der Waals contacts with the buried steroid D ring (4, 6). DB3 derives from immunization with a progesterone conjugate (11-hemisuccinyl-progesterone-BSA) (2) that constrains the steroid orientation in the combining site because of the attachment of the linker to the exposed carrier protein (29). Although the steroid coupling position predetermines ligand binding with buried D rings in DB3, no such restrictions apply to 1E9dm. The inverted head-to-tail arrangement of progesterone with a buried A ring demonstrates that interactions that provide specific recognition of the steroid D ring in DB3 are not essential for steroid binding by 1E9dm. Similar Kd values for the 1E9dm and DB3 steroid complexes (12) further suggest that the increased number of hydrophobic interactions associated with the deeper penetration of the ligands into the 1E9dm interior energetically compensates for lack of specific directional and hydrogen bond interactions (16, 21). Steroid binding by 1E9dm is thus reminiscent of the anti-digoxin antibody 26–10 (21), which also exploits shape complementarity rather than hydrogen bonding or electrostatic interactions to achieve specificity and high-affinity ligand recognition.
In summary, the Diels–Alderase 1E9, the 1E9 LeuH47Trp/ArgH100Trp mutant 1E9dm, and the steroid-binding antibody DB3 use the same restricted hydrophobic scaffold for ligand binding, and only two residues are needed to control specificity. As demonstrated by 1E9dm and DB3, a common set of binding interactions can be exploited in very different ways to achieve the same functional result, namely nanomolar affinity binding to an entire range of structurally distinct steroids.
What do those findings imply for steroid-binding by proteins in particular and ligand recognition by immune receptors in general? Hydrophobic interactions are more adaptable than directional electrostatic interactions and correlate with a more plastic binding site. The high degree of conservation between two independently raised progesterone-binding antibodies, DB3 (2) and P15G12C12G11 (22), indicates that steroid binding requires a predefined set of residues that form a structurally optimal, hydrophobic scaffold to provide essential van der Waals interactions to hydrophobic ligands. As demonstrated by 1E9dm, small mutational changes can then generate different binding modes while retaining similar affinities for the same ligands. The plasticity of the hydrophobic scaffold initially determines the degree of cross-reactivity for distinct ligands, whereas directed interactions can (but may not necessarily) provide more selectivity and higher affinity. Very small changes in a binding site can completely change the balance between the energetic contributions of the hydrophobic scaffold and directional interactions that enable a variety of different structural solutions to be explored to achieve the same functional outcome. Although this represents an economic way to extend the pathogen recognition profile of immune receptors, it also brings along a potential drawback of increased receptor promiscuity.
Material and Methods
Summary.
For details on protein preparation, complex formation, crystallization, and structure determination, see SI Materials and Methods. In brief, the 1E9 LeuH47Trp/ArgH100Trp Fab (1E9dm) was produced as described in ref. 12. Steroid complexes were prepared by incubating 1E9dm with excess ligand and crystallized by vapor diffusion like apo 1E9dm in space group P3121. Diffraction data were collected at synchrotron sources and the apo 1E9dm structure was determined by molecular replacement (MR), using the coordinates of WT 1E9 Fab (PDB entry 1C1E). 1E9dm complex structures were determined by rigid body and restrained refinement with simulated annealing of the 1E9dm apo structure. Pronounced Fo − Fc difference electron density at the 3σ level was clearly defined for each steroid ligand. Data processing and final refinement statistics are shown in Table 1.
Supplementary Material
Acknowledgments.
We thank the staff of the Advanced Light Source Beam Lines 8.2.1, 8.2.2, and 8.3.1 and the Stanford Synchrotron Radiation Laboratory Beam Line 11-1 for support with data collection; E. W. Debler, R. L. Stanfield, J. G. Luz, A. Schiefner, A. L. Corper, and M. Elsliger for helpful discussions; and X. Dai for assistance on synchrotron trips. This work was supported by Erwin-Schrödinger Fellowship J2313 of the Austrian Science Fund (to P.V.), National Institutes of Health Grants AI042266 and CA58896 (to I.A.W.), and Eidgenössische Technische Hochschule Zürich (D.H.). This is publication 18621-MB from The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition note: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2O5X, 2O5Y, and 2O5Z).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801783105/DCSupplemental.
Despite their structural differences, the functional mimicry of 1E9dm and DB3 extends to low affinity ligands like testosterone. Fluorescence titration experiments (primary data not shown) gave a dissociation constant of 0.11 ± 0.03 M for the complex of this steroid with 1E9dm, which is within a factor of four or five of the Kd for DB3 (Kd = 0.5 ± 0.1 M) (3). Thus, the two antibodies achieve the same level of discrimination for testosterone as for steroids like progesterone and 5β-androstane-3,17-dione that bind two orders of magnitude more tightly (3,12).
References
- 1.Alt FW, Blackwell TK, Yancopoulos GD. Development of the primary antibody repertoire. Science. 1987;238:1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- 2.Wright LJ, Feinstein A, Heap RB, Saunders JC, Bennett RC, Wang MY. Progesterone monoclonal antibody blocks pregnancy in mice. Nature. 1982;295:415–417. doi: 10.1038/295415a0. [DOI] [PubMed] [Google Scholar]
- 3.Ellis ST, Heap RB, Butchart AR, Rider V, Richardson NE, Wang MW, Taussig MJ. Efficacy and specificity of monoclonal antibodies to progesterone in preventing the establishment of pregnancy in the mouse. J Endocrinol. 1988;118:69–80. doi: 10.1677/joe.0.1180069. [DOI] [PubMed] [Google Scholar]
- 4.Arevalo JH, Taussig MJ, Wilson IA. Molecular basis of crossreactivity and the limits of antibody–antigen complementarity. Nature. 1993;365:859–863. doi: 10.1038/365859a0. [DOI] [PubMed] [Google Scholar]
- 5.Arevalo JH, Stura EA, Taussig MJ, Wilson IA. Three-dimensional structure of an anti-steroid Fab′ and progesterone-Fab′ complex. J Mol Biol. 1993;231:103–118. doi: 10.1006/jmbi.1993.1260. [DOI] [PubMed] [Google Scholar]
- 6.Arevalo JH, et al. Structural analysis of antibody specificity. Detailed comparison of five Fab′-steroid complexes. J Mol Biol. 1994;241:663–690. doi: 10.1006/jmbi.1994.1543. [DOI] [PubMed] [Google Scholar]
- 7.Hilvert D, Hill KW, Nared KD, Auditor M-TM. Antibody catalysis of the Diels–Alder reaction. J Am Chem Soc. 1989;111:9261–9262. [Google Scholar]
- 8.Xu J, et al. Evolution of shape complementarity and catalytic efficiency from a primordial antibody template. Science. 1999;286:2345–2348. doi: 10.1126/science.286.5448.2345. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Deng Q, Wang R, Houk KN, Hilvert D. Shape complementarity, binding-site dynamics, and transition state stabilization: A theoretical study of Diels–Alder catalysis by antibody 1E9. Chembiochem. 2000;1:255–261. doi: 10.1002/1439-7633(20001117)1:4<255::AID-CBIC255>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 10.Piatesi A, Hilvert D. Immunological optimization of a generic hydrophobic pocket for high affinity hapten binding and Diels–Alder activity. Chembiochem. 2004;5:460–466. doi: 10.1002/cbic.200300806. [DOI] [PubMed] [Google Scholar]
- 11.Haynes MR, Lenz M, Taussig MJ, Wilson IA, Hilvert D. Sequence similarity and cross-reactivity of a Diels–Alder catalyst and an anti-progesterone antibody. Isr J Chem. 1996;36:151–159. [Google Scholar]
- 12.Piatesi A, Aldag C, Hilvert D. Switching antibody specificity through minimal mutation. J Mol Biol. 2008;377:993–1001. doi: 10.1016/j.jmb.2008.01.069. [DOI] [PubMed] [Google Scholar]
- 13.Duax WL, Norton DA. Atlas of Steroid Structures. Vol 1. New York: Plenum; 1975. [Google Scholar]
- 14.Griffin JF, Duax WL, Weeks CM. Atlas of Steroid Structures. Vol 2. New York: Plenum; 1984. [Google Scholar]
- 15.Amzel LM, Poljak RJ. Three-dimensional structure of immunoglobulins. Annu Rev Biochem. 1979;48:961–997. doi: 10.1146/annurev.bi.48.070179.004525. [DOI] [PubMed] [Google Scholar]
- 16.Sekharudu C, et al. Crystal structure of the Y52F/Y73F double mutant of phospholipase A2: Increased hydrophobic interactions of the phenyl groups compensate for the disrupted hydrogen bonds of the tyrosines. Protein Sci. 1992;1:1585–1594. doi: 10.1002/pro.5560011206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burks EA, Chen G, Georgiou G, Iverson BL. In vitro scanning saturation mutagenesis of an antibody binding pocket. Proc Natl Acad Sci USA. 1997;94:412–417. doi: 10.1073/pnas.94.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He M, Hamon M, Liu H, Corper AL, Taussig MJ. Effects of mutation at the D–JH junction on affinity, specificity, and idiotypy of anti-progesterone antibody DB3. Protein Sci. 2006;15:2141–2148. doi: 10.1110/ps.062236806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zahnd C, et al. Directed in vitro evolution and crystallographic analysis of a peptide-binding single chain antibody fragment (scFv) with low picomolar affinity. J Biol Chem. 2004;279:18870–18877. doi: 10.1074/jbc.M309169200. [DOI] [PubMed] [Google Scholar]
- 20.Schildbach JF, et al. Modulation of antibody affinity by a non-contact residue. Protein Sci. 1993;2:206–214. doi: 10.1002/pro.5560020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffrey PD, et al. 26–10 Fab-digoxin complex: Affinity and specificity due to surface complementarity. Proc Natl Acad Sci USA. 1993;90:10310–10314. doi: 10.1073/pnas.90.21.10310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubreuil O, et al. Fine tuning of the specificity of an anti-progesterone antibody by first and second sphere residue engineering. J Biol Chem. 2005;280:24880–24887. doi: 10.1074/jbc.M500048200. [DOI] [PubMed] [Google Scholar]
- 23.Yuan L, Kurek I, English J, Keenan R. Laboratory-directed protein evolution. Microbiol Mol Biol Rev. 2005;69:373–392. doi: 10.1128/MMBR.69.3.373-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein–protein interface. J Mol Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Xiao G, et al. First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase. Biochemistry. 1996;35:4753–4765. doi: 10.1021/bi960189k. [DOI] [PubMed] [Google Scholar]
- 26.Koh JT. Engineering selectivity and discrimination into ligand-receptor interfaces. Chem Biol. 2002;9:17–23. doi: 10.1016/s1074-5521(02)00087-x. [DOI] [PubMed] [Google Scholar]
- 27.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 28.Schneider H.-J. Mechanisms of molecular recognition : Investigations of organic host-guest complexes. Angew Chem. 1991;30:1417–1436. [Google Scholar]
- 29.Gani M, et al. Monoclonal antibodies against progesterone: Effect of steroid-carrier coupling position on antibody specificity. J Steroid Biochem Mol Biol. 1994;48:277–282. doi: 10.1016/0960-0760(94)90156-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.