Ionizing radiation (IR) poses a severe threat to genome stability, inducing base changes, cross-links, and double-stranded breaks (DSBs). Improper repair of IR damage has severe consequences, frequently resulting in formation of chromosomal translocations leading to gene dysregulation at translocation breakpoints and/or oncogenic fusions. Moreover, cancer genomes are very severely rearranged. Thus, there is great interest in determining the origins of spontaneous genome rearrangement. Homologous recombination (HR) has long been implicated in the recovery from irradiation, as evidenced by the sensitivity of rad52 yeast strains to irradiation (1, 2). In this issue of PNAS, Argueso et al. (3) demonstrate not only that HR is the primary mechanism by which DSBs are repaired in diploid yeast cells recovering from IR, but also that chromosomal aberrations (CAs) are generated by recombination between nonallelic repeats and that this process profoundly reshapes the genome of surviving cells.
DSBs are so deleterious that even a single DSB, left unrepaired, can be lethal in yeast (4). DSB formation leads to a cell cycle delay (5, 6), allowing repair by either HR or nonhomologous end joining (NHEJ). In yeast, HR is the preferred mechanism, yet even the limited repetitive DNA content in yeast presents a formidable problem by presenting an opportunity for ectopic (nonallelic) HR. Repair by HR requires an extensive homology search to find a partner for repair; sister chromatids are preferential substrates in diploid G2 cells (7). Yeast cells contain diverse dispersed repeats, including subtelomeric X and Y′ repeats, dispersed tDNA loci, and numerous Ty retrotransposons and solo-LTR elements. Each of these is a potential site for ectopic HR, and tandemly oriented elements, inverted repeats, and dispersed copies can generate duplications, deletions, and translocations depending on their relative orientation and their genomic positions. The potential for Ty elements to participate in ectopic HR is well known (8, 9). The risk of HR among dispersed repeats is exacerbated by induction of DSBs in or near repeats; one DSB increases translocation frequency of 1,000-fold, and two DSBs can increase it by 107-fold (10). As with DSBs occurring in unique genomic regions, those generated in repeat elements are repaired efficiently by recombination with both allelic and ectopic elements (11).
Better understanding of the role of transposable elements in generating CAs and genomic mechanisms to restrain this deleterious process are of great interest. Most investigations into the mechanism of DSB repair in yeast have exploited inducible DSBs made by targeted endonuclease cutting. Argueso et al. (3) have taken a broader view by subjecting yeast cells to extensive irradiation, inducing random genome-wide DSBs. By exploiting diploid yeast arrested in G2, they generated a pure population of 4n cells, which were subjected to high doses of IR, inducing ≈250 DSBs per cell (Fig. 1). Not only were the yeast able to cope with this extraordinary level of damage, the bulk populations rapidly restored their genome to a native chromosomal configuration after just 3 h of recovery. Survival rates were astonishingly high: 7–28% of the cells formed colonies. This surprising result dramatically demonstrates the ability of a complex eukaryotic genome to undergo extensive repair and rebuilding from the most extreme genomic insults rapidly.
Fig. 1.
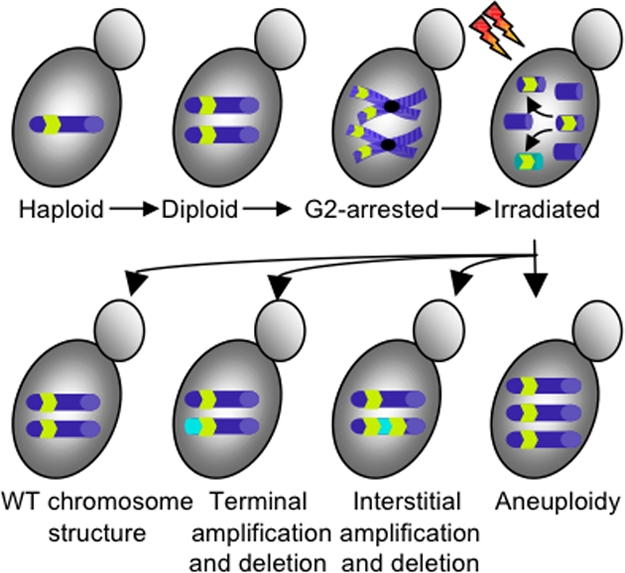
Rebuilding shattered yeast genomes. (Upper) Diploid yeast cells arrested in G2 were used to maximize the number of genome copies available as repair templates. After IR, the genome was shattered to ≈250 DSBs per cell. Repetitive DNA elements (yellow arrows) can mispair during strand invasion, leading to ectopic HR. (Lower) After 3 h of recovery, CAs are widespread, with formation of amplifications, deletions, translocations, and aneuploidy, often with multiple events per cell.
Are the reconstructed yeast chromosomes truly restored to a wild-type configuration? More often than not, the reassembly process generated misassembled genomes unique to each randomly chosen strain. There was extensive variation in chromosome structure, with 54 of 71 clones showing at least one CA. Despite the random distribution of DSBs throughout the genome, almost all (91%) translocation breakpoints were located within repetitive sequences, revealing the remarkable fidelity with which breaks in unique genomic sequences are repaired and heavily implicating repetitive DNA in the generation of CAs that reshape the genome through ectopic HR. Extensive genome analysis of individual survivor clones by electrophoretic karyotyping, array-CGH, band-CGH, PCR, and Southern blotting revealed dramatic variation in genome structure. Within individual survivors, genomic changes included interstitial duplications, loss of heterozygosity, and chromosomal translocations (Table 1). The large number of breakpoints, including a high frequency of complex CAs, suggests that any and all Ty elements of the same type can serve as HR partners. Complex CAs were remarkably common, with examples of tripartite recombination (Fig. 2) frequently detected. The stunning diversity of genome structures recovered suggests that yeast genome structure is remarkably pliable. After formation of the Rad51 filament during HR, an extensive search is undertaken to find homologous sequences (12, 13); although the chance of choosing the correct partner sequence is inversely proportional to its copy number in the genome (14), the frequent tripartite recombination observed in this study suggests that both ends of the DSB independently undergo extensive homology searches able to scan the genome quickly and efficiently.
Table 1.
Aberrations observed in survivors
| Aberration type | % of survivors* |
|---|---|
| Aneuploidy | |
| Monosomy | 31 |
| Trisomy | 70 |
| Chromosome rearrangements | |
| Deletions (terminal) | 36 |
| Deletions (interstitial) | 17 |
| Amplifications (terminal) | 35 |
| Amplifications (interstitial) | 13 |
| Translocations† | 68 |
*Percentage of survivors with this rearrangement type.
†Estimated.
Fig. 2.
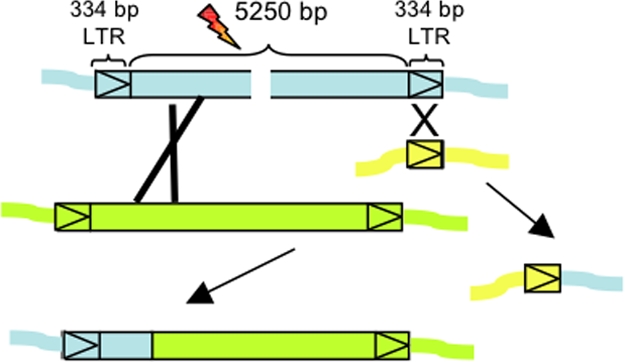
Ty-mediated recombination. (Top and Middle) Structure of a Ty retrotransposon, comprising two long terminal repeats (LTRs) that flank unique TyA and TyB protein-coding sequences. (Bottom) Tripartite recombination between dispersed Ty elements located either inter- or intrachromosomally and generating complex rearrangements.
The ability to recover cells after extensive IR is a powerful mechanism to investigate both the immediate repair of damage and the long-term consequences for cells coping with CAs. This ability of yeast to rebuild its genome after extensive fragmentation is reminiscent of the biology of Deinococcus radiodurans, a highly IR-resistant bacterium with an ≈8n genome. After IR that shatters the genome into 20- to 30-kb fragments, the Deinococcus genome is perfectly reconstructed in a two-step assembly process: “extended synthesis-dependent strand annealing” followed by RecA-dependent HR. The Deinococcus and Saccharomyces processes share two fundamental properties: the requirement for multiple genomes in the cell to be shattered and iterative rounds of chromosome reassembly by HR (15). Is this mechanism of genome reassembly common to prokaryotes and lower eukaryotes with a lower repetitive DNA content and therefore a stronger bias toward HR or is this a more universally conserved phenomenon?
Is the use of nonallelic HR between Ty elements to generate CAs unique to recovery from high doses of IR? Although targeting a DSB to a Ty element induces Ty recombination, many forms of DNA damage that ought to create DSBs do not increase the level of recombination between specific pairs of Ty elements (11). Will spontaneous chromosome evolution (16) and CAs, such as those generated at collapsed replication forks, proceed similarly, despite the need for only a single-strand invasion step to restore the fork? Under conditions of spontaneous damage, where there is a lower incidence of simultaneous DSBs, is there a greater role for chromosome structure, chromosome looping, and intranuclear positioning of Ty elements that could mediate pairing of homologous sequences?
In mammalian cells, DSBs arise frequently from endogenously generated free oxygen radicals, collapsed replication forks, somatic recombination during lymphoid differentiation, topoisomerase-mediated DNA cleavage, viral transduction, and from exogenous damage. Perhaps because the repetitive DNA content of human cells is considerably higher than in yeast and Deinococcus (45% compared with 3%; Table 2), HR seems to be less dominant in mammals than in yeast, with more DSBs repaired by NHEJ. Yet the frequent association of CAs with mammalian repeats (for reviews, see refs. 17 and 18) suggests that the mechanism described by Argueso et al. (3) may operate in higher eukaryotic CA formation. It will be exciting to see what fraction of those CAs characteristic of oncogenic transformation are repeat-associated and whether the exchanges are predominantly HR-mediated. In this age of deep sequencing, the answer cannot be far away.
Table 2.
Genomes compared
| Parameter | Deinococcus | Saccharomyces | Homo |
|---|---|---|---|
| Type | Eubacteria | Simple eukaryote | Complex eukaryote |
| Genome size | 3.3 Mb | 15 Mb | 3000 Mb |
| Dominant ploidy | 8n | 2n; 4n in ref. 3 | 2n |
| Dominant repair type | HR | HR | NHEJ |
| Dispersed repeats, % | 3.8 | 3.3 | 45 |
| Dispersed repeat type(s) (not including tRNAs) | ∼10 families, ∼150 bp long | ∼10 families, 0.3–6 kb long | ∼35 families ranging from 0.3 to 10 kb |
| Repeat homogeneity | High | High | Low |
Footnotes
The authors declare no conflict of interest.
See companion article on page 11845.
References
- 1.Game JC, Mortimer RK. A genetic study of x-ray-sensitive mutants in yeast. Mutat Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 2.Saeki T, Machida I, Nakai S. Genetic control of diploid recovery after gamma-irradiation in the yeast Saccharomyces cerevisiae. Mutat Res. 1980;73:251–265. doi: 10.1016/0027-5107(80)90192-x. [DOI] [PubMed] [Google Scholar]
- 3.Argueso JL, et al. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci USA. 2008;105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resnick MA, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 5.Lee SE, et al. Saccharomyces Ku70, mre11/rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 6.Toczyski DP, Galgoczy DJ, Hartwell LH. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 7.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roeder GS, Fink GR. Movement of yeast transposable elements by gene conversion. Proc Natl Acad Sci USA. 1982;79:5621–5625. doi: 10.1073/pnas.79.18.5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surosky RT, Tye BK. Resolution of dicentric chromosomes by Ty-mediated recombination in yeast. Genetics. 1985;110:397–419. doi: 10.1093/genetics/110.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pannunzio NR, Manthey GM, Bailis AM. RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair. 2008;7:788–800. doi: 10.1016/j.dnarep.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parket A, Inbar O, Kupiec M. Recombination of Ty elements in yeast can be induced by a double-strand break. Genetics. 1995;140:67–77. doi: 10.1093/genetics/140.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman AS, Lichten M. The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics. 1996;144:43–55. doi: 10.1093/genetics/144.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lichten M, Haber JE. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics. 1989;123:261–268. doi: 10.1093/genetics/123.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson JH, Leung WY, Bosco G, Dieu D, Haber JE. The frequency of gene targeting in yeast depends on the number of target copies. Proc Natl Acad Sci USA. 1994;91:177–181. doi: 10.1073/pnas.91.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahradka K, et al. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature. 2006;443:569–573. doi: 10.1038/nature05160. [DOI] [PubMed] [Google Scholar]
- 16.Dunham MJ, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinstock DM, Richardson CA, Elliott B, Jasin M. Modeling oncogenic translocations: Distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair. 2006;5:1065–1074. doi: 10.1016/j.dnarep.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 18.Griffin CS, Thacker J. The role of homologous recombination repair in the formation of chromosome aberrations. Cytogenet Genome Res. 2004;104:21–27. doi: 10.1159/000077462. [DOI] [PubMed] [Google Scholar]


