Abstract
Mutation of the RB-1 and p53 tumor suppressors is associated with the development of human osteosarcoma. With the goal of generating a mouse model of this disease, we used conditional and transgenic mouse strains to inactivate Rb and/or p53 specifically in osteoblast precursors. The resulting Rb;p53 double mutant (DKO) animals are viable but develop early onset osteosarcomas with complete penetrance. These tumors display many of the characteristics of human osteosarcomas, including being highly metastatic. We established cell lines from the DKO osteosarcomas to further investigate their properties. These immortalized cell lines are highly proliferative and they retain their tumorigenic potential, as judged by their ability to form metastatic tumors in immunocompromised mice. Moreover, they can be induced to differentiate and, depending on the inductive signal, will adopt either the osteogenic or adipogenic fate. Consistent with this multipotency, a significant portion of these tumor cells express Sca-1, a marker that is typically associated with stem cells/uncommitted progenitors. By assaying sorted cells in transplant assays, we demonstrate that the tumorigenicity of the osteosarcoma cell lines correlates with the presence of the Sca-1 marker. Finally, we show that loss of Rb and p53 in Sca-1-positive mesenchymal stem/progenitor cells is sufficient to yield transformed cells that can initiate osteosarcoma formation in vivo.
Keywords: osx-cre, Sca-1, hibernoma mouse model
Osteosarcomas account for ≈30% of malignant bone tumors and 3–4% of all childhood malignancies (1, 2). They arise primarily around the knee joint, lower femur and upper tibia, which are all regions of active bone growth and repair. These tumors are predominantly osteoblastic in nature, although there is a correlation between loss of differentiation and poor prognosis. The generation of new therapeutic treatments for osteosarcoma has improved the 5-year survival rate of affected individuals. However, like other mesenchymal neoplasms, osteosarcomas are predisposed to metastasize via the hematogenous route, and thus, pulmonary metastasis is a major cause of death. Analyses of both sporadic and hereditary tumors show that inactivation of the p53 and RB-1 tumor suppressors plays a key role in the development of this tumor type (1, 2). Li-Fraumeni patients, who often carry germ-line mutations in p53, are predisposed to a variety of tumors, 12% of which are bone sarcomas (3, 4). p53 mutations are also observed in 20–60% of sporadic osteosarcomas (5–7). Similarly, patients carrying germ-line mutations in RB-1 have an ≈500-fold higher incidence of osteosarcoma than the general population (8). Moreover, RB-1 mutations are detected in 70% of all adolescent osteosarcomas (9). Finally, human osteosarcomas can carry mutations in both p53 and RB-1 (10).
Mouse models have provided considerable insight into the role of p53 in bone development and tumorigenesis. Experiments from three different settings suggest that p53 plays an important role in bone development by modulating the differentiation of osteoblasts. First, p53-deficient mice display both accelerated osteoblast differentiation and increased bone density (11). Second, hyperactivation of p53, via deletion of the p53-inhibitor Mdm2, suppresses osteoblast differentiation by inhibiting expression of the bone-specific transcription factor Runx2 (12). Finally, in vitro studies show that deletion of p53 from mesenchymal stem cells (MSCs) and osteoblast precursors in vitro promotes transcriptional changes associated with the early stages of osteogenesis but impairs end-stage differentiation to mature osteocytes (13). Together, these experiments suggest that p53-loss promotes commitment to the osteoblast lineage but blocks the terminal differentiation of these progenitors. Importantly, mice carrying tumor-associated alleles of p53 develop a variety of tumor types including osteosarcoma (14). The status of Rb in these tumors has not been investigated. However, sarcomas arising in Rb+/−;p53−/− mice do undergo loss of heterozygosity of Rb (15).
Analyses of cell lines and mouse models also provide intriguing links between Rb and osteogenesis. The retinoblastoma protein pRb has been shown to physically interact with Runx2, and the resulting complex transcriptionally activates the late osteoblast marker osteocalcin (16). Loss of pRb, but not the pRb-related pocket proteins p107 and p130, can suppress the terminal osteogenic differentiation of cultured cell lines (16). Moreover, we have recently shown that embryos conditionally deleted for Rb display defects in both endochondral and intramembranous ossification that result, at least in part, from a cell cycle exit defect (17). Unfortunately, these conditional Rb mutant animals die at birth, precluding analysis of adult bone phenotypes. Heterozygous Rb mutant mice and Rb−/−/wild type chimeras are viable, but they develop pituitary and thyroid tumors, never osteosarcomas (18). Thus, to date, there is no mouse model of Rb mutant osteosarcoma.
In this study, we have used conditional and transgenic mouse strains to inactivate Rb and/or p53, specifically in osteoblast precursors. The resulting compound mutant animals developed metastatic osteosarcomas that closely resemble human tumors. Analysis of these tumors shows that their tumorigenic potential correlates with their expression of the Sca-1 stem cell marker and other aspects of the stem cell gene expression program.
Results
Mutation of Rb and p53 in Osteoblast Precursors Results in Osteosarcomas.
To generate a mouse model of osteosarcoma, we used mice carrying three alleles: the conditional alleles of Rb (19) and p53 (20) and the Osx1-GFP::Cre transgene (21). In this Cre transgene (herein called Cre), expression of Cre recombinase is driven by promoter sequences of Osterix1 (Osx1), a master regulator of bone differentiation, and is therefore restricted to osteogenic precursors derived from skeletal progenitors (21). By crossing Rb+/c;Cre+, p53+/c;Cre+ or Rb+/c;p53+/c;Cre+ males with Rbc/c,p53c/c, or Rbc/c;p53c/c females, we generated animals carrying every possible combination of Rb and p53 alleles, with or without Cre. All genotypes arose at approximately the expected frequency [supporting information (SI) Table S1]. Mice carrying Cre were slightly smaller than their littermates at birth, but this did not affect their survival. By 2–3 months of age, mice of all genotypes were of similar size (data not shown). Consistent with previous reports (21), we confirmed that Cre was expressed specifically in osteoblasts and not other mesenchymal lineages using reporter mice (A.S.L. and J.A.L., unpublished data). We also showed that the Cre transgene catalyzed efficient recombination of the conditional Rb and p53 alleles in the bone, by using PCR-based genotyping assays (Fig. S1).
To screen for tumors, we established an aging colony of the various Rb;p53 mutant genotypes and monitored them carefully. Moribund animals were euthanized and all tissues were analyzed for tumor phenotypes by histopathology. Up to 1 year of age (Fig. 1A and Table 1) and beyond (data not shown), the vast majority of Rbc/c;Cre+ mice remained tumor-free. Two of these animals did develop tumors at 9 and 12 months of age. However, these were pituitary tumors, the typical tumor of Rb+/− germ-line mutant and Rb−/− chimeric mutant animals (18). This result suggests that the Cre transgene is expressed at low levels in neuroendocrine tissues/precursors. Because the Cre transgene is known to act in osteoblast precursors and histological analysis did not reveal tumorigenic lesions in the bones of adult Rbc/c;Cre+ animals (data not shown), we conclude that Rb loss is not sufficient to promote the transformation of murine osteoblast precursors.
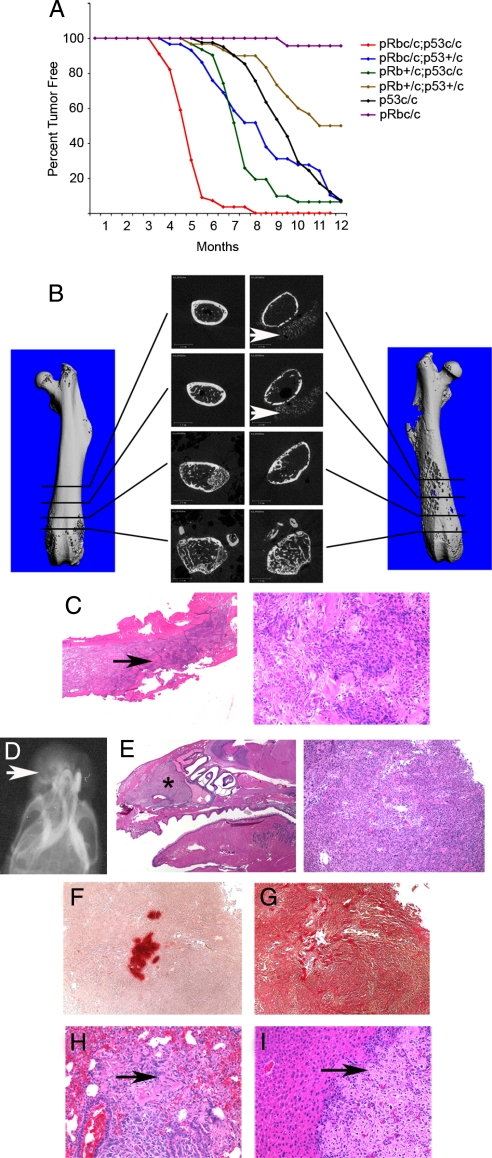
Fig. 1.
A mouse model of metastatic osteosarcoma. (A) Kaplan–Meier plot of the indicated genotypes carrying Osx1-GFP::Cre up to 12 months of age. (B–I) Analyses of osteosarcomas and associated metastases arising in DKO mice. (B) 3D reconstructed images from microComputerised Tomography are shown for a control femur (Left) versus a femur containing an osteosarcoma (Right). Central panels show 2D images at the indicated positions. Note the loss of bone cortex and the presence of bone spicules located in the tumor that has grown beyond the periosteum (arrows). (C) Histological analyses of an osteosarcoma in a femur show areas of bone cortex erosion (Left, arrow) and the presence of little mineralized bone within the tumor (Right). (D–G) Analysis of a representative snout tumor by soft x-ray image to show the typical sunburst pattern (arrow) (D), H&E staining and analysis of adjacent sections of undecalcified tumor (E) with Alizarin Red to detect calcified bone matrix (F) or Sirius Red to detect collagen (G). (H and I) Representative examples of osteosarcoma metastases (arrow), in lung (H) and liver (I) containing detectable bone matrix. (Magnification: C and E ×2; F–I ×40.)
Table 1.
Incidence of osteosarcoma, neuroendocrine, hibernoma, and other tumor types in Rb;p53;Osx1-GFP::Cre genotypes
| Genotype (all Cre+) | Fraction of mice with tumors by 1 year | Mice analyzed by histopathy | Tumor type* |
Mice with mets, % | Average age of euthanasia†, days ± SD | |||
|---|---|---|---|---|---|---|---|---|
| OS | NE | HIB | Other | |||||
| Rbc/c | 1/23 | 2 | 2 (2 pit) | 0 | ND | |||
| p53c/c | 36/41 | 25 | 25 | 32 | 281 ± 55 | |||
| Rb+/c;p53+/c | 15/30 | 16 | 16 | 19 | 299 ± 84 | |||
| Rbc/c;p53+/c | 26/29 | 18 | 17 | 4 (3 pit) | 22 | 251 ± 87 | ||
| Rb+/c;p53c/c | 29/31 | 21 | 21 | 1 | 43 | 207 ± 33 | ||
| Rbc/c;p53c/c | 56/56 | 43 | 28 | 24 | 19 | rhabdo | 37 | 147 ± 31 |
*Tumor types: OS, osteosarcoma; NE, neuroendocrine tumor; HIB, hibernoma; rhabdo, rhabdomyosarcoma.
†Age of euthanasia comparison t test: DKO vs. p53, P < 0.0001; Rb+/c;p53+/c vs. p53, P = 0.13; Rb+/c;p53c/c vs. p53, P < 0.0001; and Rbc/c;p53+/c vs. p53, P = 0.17.
Consistent with the presence of osteosarcoma in humans and mice with germ-line p53 mutations (1), a large fraction of the p53c/c;Cre+ mice developed osteosarcoma, but not other tumor types, by 1 year of age (Fig. 1A and Table 1). Although p53 loss is clearly sufficient to promote tumorigenesis, our data reveal strong synergy between Rb and p53 mutations in osteosarcoma development (Fig. 1A and Table 1). The Rb+/c;p53c/c;Cre+ and Rbc/c;p53+/c;Cre+ genotypes were highly predisposed to develop osteosarcoma, and their mean survival time was considerably shorter than that of the p53c/c;Cre+ animals (Fig. 1A and Table 1). In addition, osteosarcomas arose in a significant fraction of the Rb+/c;p53+/c;Cre+ animals, but rarely (p53+/c;Cre+) or never (Rb+/c;Cre+) in the single heterozygous mutants (Table 1 and data not shown). Importantly, with the exception of the occasional neuroendocrine tumor, osteosarcoma was the only tumor type arising in Rb+/c;p53c/c;Cre+, Rbc/c;p53+/c;Cre+, p53c/c;Cre+ and Rb+/c;p53+/c;Cre+ animals. This observation supports the view that the Cre transgene is highly tissue-specific and strongly suggests that these osteosarcomas arise through transformation of osteoblast precursors. Like human osteosarcomas, a significant fraction of these tumors were metastatic (Table 1). The metastases were most commonly seen in the lung and liver, but they also arose in the spleen, kidney, ovary, and adrenal glands (Fig. 1 and Table S2).
The synergy between Rb and p53 is underscored by the phenotype of the Rbc/c;p53c/c;Cre+ (herein called DKO) mice. These animals had a substantially shorter mean lifespan than the intermediate genotypes (Fig. 1A and Table 1) and developed osteosarcomas (75% of animals), neuroendocrine tumors (60% of animals), and hibernomas (44% of animals), tumors derived from brown adipose tissue (Fig. S2). Many DKOs presented with multiple tumor types, and in 40% of cases metastasis of at least one of the primary tumors was observed (Table 1 and Table S2). There was no obvious correlation between the time of death of the DKOs and their associated tumor types (data not shown). Lack of correlation suggests that the shortened lifespan of the DKOs, vs. other genotypes, is not due simply to the presence of additional tumor types but likely reflects the accelerated onset and/or aggressiveness of the tumors.
The osteosarcomas arose in a variety of locations, including the femur, a major site for human osteosarcoma, and the snout (the most common site in our model), spine, and skull. These tumors displayed characteristics typical of human osteosarcomas (Fig. 1 and data not shown). For example, microComputerized Tomography and H&E staining of femoral osteosarcomas showed destruction of the bone cortex and the presence of ossified spicules in the tumor mass located outside of the periosteum (Fig. 1 B and C). Similarly, x-ray analysis of a typical snout tumor revealed the classic sunburst pattern indicative of osteoid tissue (osseous tissue before calcification: Fig. 1D). Moreover, the osteosarcomas were largely composed of osteoblastic cells, as judged by H&E staining and Sirius Red staining for collagen (Fig. 1 C, E, and G). However, like many human osteosarcomas, these tumors were predominantly poorly differentiated or undifferentiated, as judged by low levels of Alizarin Red staining of calcified bone matrix (Fig. 1F). We also used quantitative real-time PCR (qRT-PCR) to analyze the expression of differentiation markers in primary osteosarcomas derived from DKO mice (Fig. S3). These tumors contained mRNAs associated with early to mid stages of bone differentiation, such as Runx2, Osx, Alkaline Phosphatase (Alp), and Collagen1 (Col1), at the same or higher levels than control bone tissue. In contrast, Osteocalcin (Oc) mRNA, associated with fully differentiated osteoblasts that have secreted bone matrix, was present at lower levels than in the control. Notably, mRNAs associated with adipose tissue were not expressed in the primary osteosarcomas, but were present in hibernomas (Fig. S3). Finally, as noted above, a significant fraction of the osteosarcomas metastasized to lung and liver (Fig. 1 H–I, Table 1, and Table S2). Thus, mutation of Rb and p53 using this Cre transgene induces formation of metastatic osteosarcomas that resemble the human disease.
Cell Lines Derived from Osteosarcomas Are Immortal and Form Osteogenic Tumors When Transplanted in Nude Mice.
To further characterize these tumors, we dissected primary osteosarcomas from three different DKO mice, mechanically disaggregated the cells, and placed them in culture. The tumors used for this experiment span the range of osteosarcoma phenotypes seen in our mice: two of the tumors (985 and 2674) were largely undifferentiated, whereas the third (2380) had a higher level of osteoid matrix (Fig. 2 A and B). All three tumors yielded rapidly growing cell populations, and PCR verified that the Rbc/c and p53c/c conditional alleles had undergone complete recombination (data not shown). The resulting cell lines (called DKO-OS-985, DKO-OS-2380, and DKO-OS-2674) were fully immortalized.
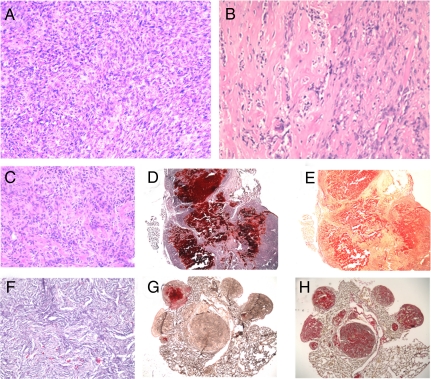
Fig. 2.
OS cell lines can form bone tumors in immuno-compromised mice. (A and B) H&E stained section of the primary osteosarcomas 985 and 2380, respectively. Tumors derived from s.c. (C–E) or i.v. (F–H) injection of DKO-OS-985. (C and F) H&E staining. Adjacent sections were stained with either Alizarin Red (D, G) or Sirius Red (E, H) to stain calcified bone matrix and collagen, respectively. (Magnification: A–C and F ×40; D, E, G, and H ×2.)
To investigate their tumorigenic potential, we injected the osteosarcoma (OS) cell lines into immuno-compromised mice, both s.c. and i.v. DKO-OS-985, DKO-OS-2380, and DKO-OS-2674 all yielded ≥1 cm3 masses (s.c.) or bone nodules in the lungs (i.v.) between 50 and 100 days (Fig. 2 and Table S3). The resulting tumors closely resembled the parental osteosarcomas. They were osteoblastic in nature, as determined by H&E, Sirius Red, and Alizarin red staining (Fig. 2 C–H). However, they were poorly differentiated or undifferentiated, as only small regions of the tumor produced calcified bone (Fig. 2 C–H). Moreover, the s.c. tumors were highly invasive and in some (DKO-OS-2380 and DKO-OS-2674) or all (DKO-OS-985) instances, they metastasized to the liver and other organs (data not shown). Thus, the OS cell lines retained their ability to form metastatic osteosarcomas in vivo.
Osteosarcoma Cell Lines Demonstrate Properties of Mesenchymal Stem/Progenitor Cells in Vitro.
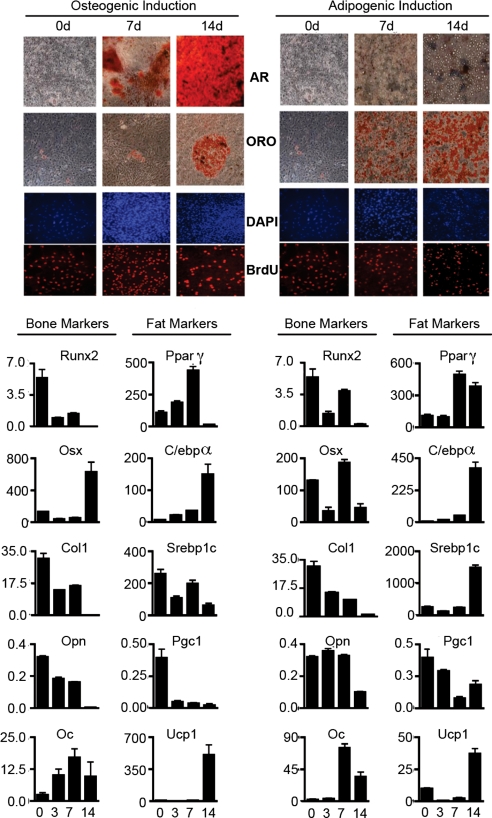
The specificity of the Cre transgene, characteristics of the primary osteosarcomas, and osteoblastic properties of the transplanted tumor cell lines all suggest that the tumors result from transformation of cells committed to the bone lineage. Thus, we asked whether the cultured tumor cells retained their ability to differentiate into bone in vitro. For these experiments, we allowed the tumor cells to reach confluence and then cultured them in osteogenic induction media. DKO-OS-985 (Fig. 3), DKO-OS-2380, and DKO-OS-2674 (data not shown) all gave similar results: The bone differentiation program was rapidly activated as judged by the detection of bone matrix by Alizarin Red staining and by the expression of key bone differentiation markers. Notably, the OS cell lines all retained a large number of proliferating cells throughout the differentiation time course, as assessed by BrdU incorporation (Fig. 3 and data not shown). In contrast, wild-type osteoblast and MSC preparations consistently stopped proliferating before they produced bone matrix (data not shown). The OS cells lines displayed one other unexpected phenotype: Some of the cells in bone differentiation media adopted the adipogenic fate, as judged by Oil Red O staining for lipid droplets (Fig. 3). Consistent with this finding, adipocyte differentiation markers were induced in these cells (Fig. 3). To explore adipocyte differentiation further, we cultured the tumor cells in adipogenic differentiation media (Fig. 3). Under these conditions, a significant fraction of the cells differentiated into adipocytes, as confirmed by both Oil Red O staining and gene expression analysis of adipocyte differentiation markers (Fig. 3). Notably, these cells also expressed bone differentiation markers. They did not stain with Alizarin Red, but this is likely because of the absence of inorganic phosphate (a component of osteogenic but not adipogenic differentiation media), which is essential for formation of the mineralized bone matrix. Contrary to normal adipogenesis, proliferating cells persisted throughout the differentiation time course. Thus, for both bone and fat differentiation, the normal link between differentiation stimuli and cell cycle exit is disrupted in these OS cell lines. Finally, preliminary studies suggest that the OS cell lines can also be induced to differentiate into cartilage-producing chondrocytes when cultured in chondrogenic media (data not shown). Taken together, these data suggest that the DKO-OS cell lines possess characteristics reminiscent of MSCs/mesenchymal progenitor cells (MPCs).
Fig. 3.
Osteosarcoma cells lines are multipotent in vitro. DKO-OS-985 cells were induced to differentiate into the bone (Left) and fat (Right) lineages and assayed at the indicated time points (days). Mineral deposits were stained with Alizarin Red (AR) as a marker for osteogenic differentiation. Oil-Red O (ORO) was used to stain lipid droplet accumulation during adipogenic induction. Cells were pulsed with BrdU to determine the proliferative status during differentiation. Expression of differentiation markers for bone and fat was determined by qRT-PCR.
Osteosarcoma Cell Lines Express Sca-1, a Marker of Early Mesenchymal Progenitors, and This Correlates with Their Tumorigenic Potential.
Given the multipotency of the OS cell lines, we tested them for the expression of a known MSC/MPC marker, Sca-1. We found that a significant fraction of the DKO-OS-985, DKO-OS-2380, and DKO-OS-2674 cells expressed Sca-1 (Fig. 4A and data not shown). We then asked whether the presence or absence of Sca-1 influenced the tumorigenicity of the OS cell lines. To answer this question, we used FACS to isolate populations of DKO-OS-985 that had either high or low/no Sca-1 expression and were all CD45− (to eliminate any hematopoietic stem cells) and assayed their tumorigenicity by s.c. injection in immunocompromised mice. In one experiment, tumors arose only from the Sca-1high population (Table S3). In another experiment, the Sca-1high cells produced a much larger tumor than the Sca-1low/− cells (Fig. 4B). Therefore, the tumorigenicity of the OS cell lines correlates with the presence of the Sca-1 marker.
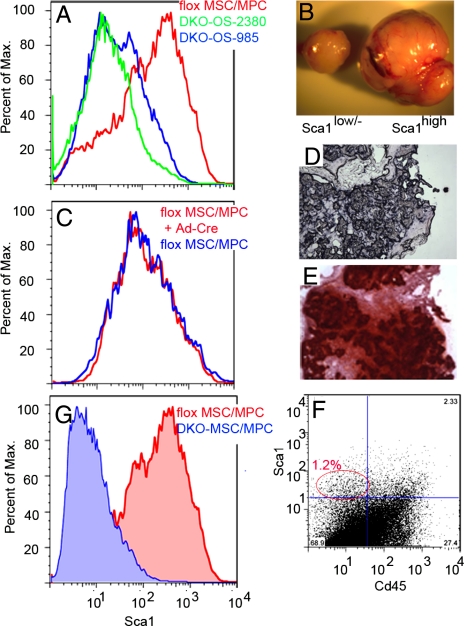
Fig. 4.
Sca-1 expression and Rb- and p53-loss are both required for efficient tumorigenesis in vivo. Sca-1 expression in DKO-OS-985 and DKO-OS-2380 cell lines versus flox MSC/MPCs (A) or flox MSC/MPC+Ad-Cre cells, in which Rb and p53 have been inactivated, versus flox MSC/MPCs (C). (B) Tumors arising in immunocompromised mice injected s.c. with 105 DKO-OS-985 cells sorted for either ScaIlow/− or ScaIhigh. (D and E) Tumors arising in immunocompromised mice injected s.c. with 106 flox MSC/MPC+Ad-cre stained for Alp expression (D) or Alizarin Red (E). Sca-1 expression in primary DKO osteosarcomas (F) and DKO MSC/MPCs versus flox MSC/MPCs (G).
Given this finding, we wished to establish whether the inactivation of Rb and p53 in Sca-1high MSC/MPC preparations is sufficient to confer tumorigenicity. For this experiment, we isolated stromal cells from the bone marrow of Rbc/c;p53c/c mice and placed the cells in culture to establish flox MSC/MPCs. After two passages, the flox MSC/MPCs were infected with a Cre-expressing adenovirus and recombination of the conditional alleles was confirmed by PCR genotyping (data not shown). Untreated and recombined (flox MSC/MPC+Ad-Cre) MSC/MPCs were briefly expanded to yield sufficient cells for s.c. injection into immunocompromised mice. At this time point, the two populations were similarly composed of predominantly Sca-1high/CD45− cells (Fig. 4C and data not shown). However, whereas the wild-type flox MSC/MPCs did not form tumors, the flox MSC/MPC+Ad-Cre yielded tumors that stained positive for both the bone marker Alp and Alizarin Red (Fig. 4 D and E and Table S3). Thus, we conclude that the loss of Rb and p53 in Sca-1high MSC/MPCs is sufficient to create osteosarcoma-initiating cells. Long-term passaging of the flox MSC/MPC+Ad-Cre cultures confirmed that these cells are fully immortalized in vitro. Furthermore, the composition of the cell population shifted over time to give a mixture of Sca-1high and Sca-1− cells (data not shown), indicating that division of the Sca-1+ tumor-initiating cells can yield Sca-1− progeny.
The presence of Sca-1+ cells within the OS cell lines was somewhat unexpected because Cre expression, and therefore p53 and Rb inactivation, occurs in committed osteoblast precursors (i.e., cells that are presumed to be Sca-1−). To determine whether these Sca-1+ cells exist in the endogenous tumors, we dissociated primary osteosarcomas from DKOs and analyzed them directly by FACS. Importantly, Sca-1+/CD45− cells consistently constituted a relatively small percentage (≈1%) of the tumor, with the bulk consisting of Sca-1−/CD45− cells (Fig. 4F). To further explore this finding, we isolated bone marrow stromal cells from 6- to 10-week-old DKO mice before the presence of gross osteosarcomas. We placed these cells in culture and assayed the passage 1 DKO MSC/MPC population by FACS. Remarkably, the majority of the DKO MSC/MPCs were Sca-1low/− (Fig. 4G). Notably, this cellular composition represents a clear departure from the properties of wild-type flox MSC/MPCs (which are predominantly Sca-1high) (Fig. 4G), and it more closely resembles that of the primary osteosarcoma. Thus, inactivation of Rb and p53 had greatly altered the properties of the bone marrow mesenchymal cells by 6–10 weeks of age. Given the short culture time of the DKO MSC/MPC preparations, we conclude that the Sca-1low/− osteoprogenitors must exist in the DKO bone marrow, and their predominance within the culture suggests that their levels are significantly elevated compared with wild-type bone marrow. Additionally, the absence of Rb and p53 may help enable these cells to be established in culture. We believe there are two potential sources for the Sca-1low/− osteoprogenitors in vivo. First, they could result from the accumulation and expansion of Sca-1low/−-committed osteoblast precursors that were the target of Rb and p53 loss. Second, they could be the progeny of the DKO Sca-1+ osteoprogenitors that arose after the loss of Rb and p53 in the committed osteoblast. Taken together, our findings provide insight into the cell lineages that contribute to osteosarcoma in our model. First, loss of Rb and p53 occurs in committed osteoblast precursors. Second, DKO Sca-1+ cells arise at low frequency in vivo and Sca-1 expression correlates with tumor-initiating capacity. Finally, the DKO Sca-1+ cells can give rise to Sca-1− progeny, and such Sca-1− cells constitute the bulk of the endogenous osteosarcomas.
Discussion
Mutation of Rb and p53 is associated with development of human osteosarcoma. We have used an Osx1-Cre transgene (21) to induce inactivation of these tumor suppressors in murine osteoblast precursors. Loss of Rb alone is insufficient to establish osteosarcoma in these animals. However, because other Rb/p53 genotypes are tumor prone, the lack of osteosarcomas is not because of an inability of the Cre-expressing precursors to become tumor-initiating cells. Instead, we presume that the tumorigenic consequences of Rb-loss are suppressed in these cells. It seems likely that other pocket proteins contribute to this suppression, becausee chimeras generated with Rb;p107, but not Rb, mutant ES cells develop osteosarcomas at low frequency (22). In addition, our data underscore the key role of p53 in osteosarcoma development. First, p53-loss in osteoblast precursors is sufficient to allow osteosarcoma formation. Second, we see robust synergy between p53 and Rb in tumorigenesis. The rapidity with which these mice die from osteosarcoma correlates with the dosage of p53 and Rb mutant alleles. Moreover, the DKO mice show a broadened tumor spectrum that includes hibernomas and neuroendocrine tumors and osteosarcomas. Indeed, these mice can develop multiple tumor types and die as early as 4 months of age. Importantly, irrespective of the starting genotype, the osteosarcomas display many of the characteristics of human osteosarcomas, including a shared predisposition to develop tumors within the femur, a similar cellular composition, and a high incidence of metastases.
Our study also has important implications for questions regarding the osteosarcoma cell-of-origin. To date, much of our understanding of tumor stem cells has come from the study of hematological malignancies. For example, it has been shown that acute myeloid leukemia can arise from a committed progenitor cell (23). In these studies, although normal progenitor cells lost the expression of self-renewal pathways, transformed progenitor cells “acquired” the aberrant activation of self-renewal pathways. The resultant tumor-initiating cells thus contained a hybrid gene expression program, with some elements of progenitor cells and some elements of more primitive stem cells. In contrast to hematopoietic tumors, very little is known about tumor-initiating cells in osteosarcomas. The analysis of gene expression programs in Ewing's sarcoma, a tumor of bone and soft tissue, revealed an expression program that resembles MSCs (24). Notably, silencing or inhibiting the EWS/ETS fusion gene product in sarcoma cell lines released them from their undifferentiated state and permitted both adipocytic and osteoblastic differentiation, implying that Ewing's sarcomas retain a population of undifferentiated cells that resembles MSCs. However, whether these MSC-like cells could reinitiate tumors (and thus represent a putative tumor stem-cell population), or conversely, whether differentiated cells lost their tumor initiating potential, was not established.
Here, we show that cell-lines derived from DKO osteosarcomas can differentiate into at least two lineages in vitro and retain gene expression programs of multiple lineages even after commitment to one lineage. Thus, although these cells necessarily arise from a cell that expresses Osx1 (and has thus committed to the osteoblast pathway), they display a capacity for multipotent differentiation. Furthermore, these cell lines are also capable of reinitiating secondary tumors, and this capacity correlates with their expression of Sca-1, an antigen that is widely recognized as a marker of stem cells/uncommitted progenitors. Importantly, we confirm that these Sca-1+/CD45− cells exist in the endogenous osteosarcomas. How do these cells arise? One possibility (Model 1) is that Sca-1 and Osx1 are actually coexpressed in a small fraction of cells in vivo, presumably during the transition from uncommitted progenitor to early osteoblast precursor. These Sca-1+/Osx1+ cells would represent the key target for transformation by Rb and p53. Alternatively (Model 2), expression of Sca-1 and Osx1 is mutually exclusive, but loss of Rb and p53 in the Sca-1−/Osx+ committed bone precursor changes the property of these cells to allow, at low frequency, reactivation of a stem-cell-like phenotype that includes Sca-1 expression. Notably, by 6–10 weeks of age, the loss of Rb and p53 has altered the properties of the bone marrow mesenchymal cells such that MSC/MPC preparations shift from being predominately Sca-1high/CD45− (wild type) to predominantly Sca-1low/−/CD45− (DKO). We speculate that this shift reflects the expansion of the DKO Sca-1−/Osx+ osteoblast precursors in vivo. Presumably, this population either already contains rare DKO Sca-1+/Osx1+ recombinants (Model 1) or is a fertile ground for the rare dedifferentiation event that creates the DKO Sca-1+/Osx1+ (Model 2) cells.
Irrespective of the mechanism by which the DKO Sca-1+/Osx1+ cells arise, they clearly have hybrid properties. First, they have elements of more primitive stem cells that allow multilineage differentiation, expression of a stem cell antigen, and tumor reinitiating capacity. Second, they have elements of osteoblast precursor cells, as evidenced by their strong commitment to form osteosarcomas in vivo. Further experiments are required to understand the nature of this Sca-1+ cell population and, because Sca-1 is a murine marker, to translate these findings to human tumors. However, we hypothesize that these Sca-1+ cells represent, or at least include, the tumor-initiating cell for the osteosarcomas arising in this mouse model.
Materials and Methods
Animal Maintenance and Histological Analyses.
All animal procedures followed protocols approved by the Institute's Committee on Animal Care. The Rbc/c (19), p53c/c (20), and Osx1-GFP::Cre (21) mice were maintained on a mixed genetic background. The criteria for euthanizing aging animals and the preparation and staining of sections are described in SI Experimental Procedures. Analysis of 3D bone structure was performed by using high-resolution microtomographic imaging, as described in ref. 25.
Isolation and Analysis of OS Cell Lines and MSC/MPCs.
Osteosarcomas were dissected, minced, filtered through a 70-μm filter, and plated in normal growth medium (10% FBS in DMEM, 1% P/S, l-glutamine) to generate the OS cell lines. Cells were passaged as they reached confluence. For differentiation into bone and fat, cells were plated, allowed to reach confluence, and induced to differentiate as described in ref. 26. For RNA purification, cells were rinsed two times with PBS, and RNA extraction was performed by using the RNeasy kit (Qiagen). Gene expression was performed by SYBR-Green quantitative RT-PCR, using Ubiquitin mRNA to normalize RNA inputs. Primers used for qRT-PCR and mouse genotyping are shown in SI Experimental Procedures and Table S4.
MSC/MPCs were generated as described in ref. 26. Conditional MSC/MPCs were infected with Ad5CMVCre-eGFP at ≈100 pfu per cell (University of Iowa Gene Transfer Vector Core). FACS analysis of OS and MSC/MPCs was performed on a FACSCalibur HTS (Becton-Dickinson) using ScaI and Cd45 antibodies (BD Pharmigen). For transplant assays, 105–106 unsegregated or sorted cells were injected either s.c. or i.v. into NOD/SCID mice. Moribund animals were euthanized, and tumors were collected for further experiments.
Supplementary Material
Acknowledgments.
We thank Stephen Rodda and Andrew McMahon for the Osx1-GFP::Cre transgenics, Tyler Jacks and Anton Berns for the Rb and p53 conditional strains, University of Iowa Gene Transfer Vector Core for Ad5CMVCre-eGFP, and Lees laboratoy members for helpful discussions. This project was supported by National Cancer Institute Grant CA121921 (to J.A.L.), a David H. Koch Graduate Fellowship (to S.D.B.), and a Ludwig Graduate Fellowship (to A.S.L.). J.A.L. is a Ludwig Scholar at Massachusetts Institute of Technology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805462105/DCSupplemental.
References
- 1.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- 2.Clark JC, Dass CR, Choong PF. A review of clinical and molecular prognostic factors in osteosarcoma. J Cancer Res Clin Oncol. 2008;134:281–297. doi: 10.1007/s00432-007-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malkin D. p53 and the Li-Fraumeni syndrome. Cancer Genet Cytogenet. 1993;66:83–92. doi: 10.1016/0165-4608(93)90233-c. [DOI] [PubMed] [Google Scholar]
- 4.Bell DW, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 5.Tsuchiya T, et al. Analysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prognostic implications in osteosarcoma and Ewing sarcoma. Cancer Genet Cytogenet. 2000;120:91–98. doi: 10.1016/s0165-4608(99)00255-1. [DOI] [PubMed] [Google Scholar]
- 6.Gokgoz N, et al. Comparison of p53 mutations in patients with localized osteosarcoma and metastatic osteosarcoma. Cancer. 2001;92:2181–2189. doi: 10.1002/1097-0142(20011015)92:8<2181::aid-cncr1561>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 7.Wunder JS, et al. TP53 mutations and outcome in osteosarcoma: A prospective, multicenter study. J Clin Oncol. 2005;23:1483–1490. doi: 10.1200/JCO.2005.04.074. [DOI] [PubMed] [Google Scholar]
- 8.Gurney JG, Severson RK, Davis S, Robison LL. Incidence of cancer in children in the United States. Sex-, race-, and 1-year age-specific rates by histologic type. Cancer. 1995;75:2186–2195. doi: 10.1002/1097-0142(19950415)75:8<2186::aid-cncr2820750825>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 9.Feugeas O, et al. Loss of heterozygosity of the RB gene is a poor prognostic factor in patients with osteosarcoma. J Clin Oncol. 1996;14:467–472. doi: 10.1200/JCO.1996.14.2.467. [DOI] [PubMed] [Google Scholar]
- 10.Toguchida J, et al. Chromosomal reorganization for the expression of recessive mutation of retinoblastoma susceptibility gene in the development of osteosarcoma. Cancer Res. 1988;48:3939–3943. [PubMed] [Google Scholar]
- 11.Wang X, et al. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol. 2006;172:115–125. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lengner CJ, et al. Osteoblast differentiation and skeletal development are regulated by Mdm2–p53 signaling. J Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tataria M, Quarto N, Longaker MT, Sylvester KG. Absence of the p53 tumor suppressor gene promotes osteogenesis in mesenchymal stem cells. J Pediatr Surg. 2006;41:624–632. doi: 10.1016/j.jpedsurg.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Iwakuma T, Lozano G, Flores ER. Li-Fraumeni syndrome: A p53 family affair. Cell Cycle. 2005;4:865–867. doi: 10.4161/cc.4.7.1800. [DOI] [PubMed] [Google Scholar]
- 15.Williams BO, et al. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Genet. 1994;7:480–484. doi: 10.1038/ng0894-480. [DOI] [PubMed] [Google Scholar]
- 16.Thomas DM, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 17.Berman SB, et al. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. doi: 10.1158/1541-7786.MCR-08-0176. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vooijs M, Berns A. Developmental defects and tumor predisposition in Rb mutant mice. Oncogene. 1999;18:5293–5303. doi: 10.1038/sj.onc.1202999. [DOI] [PubMed] [Google Scholar]
- 19.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 20.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 21.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 22.Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 2004;18:2952–2962. doi: 10.1101/gad.322004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 24.Tirode F, et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 25.Glatt V, Canalis E, Stadmeyer L, Bouxsein ML. Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J Bone Miner Res. 2007;22:1197–1207. doi: 10.1359/jbmr.070507. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee S, et al. Pharmacologic targeting of a stem/progenitor population in vivo is associated with enhanced bone regeneration in mice. J Clin Invest. 2008;118:491–504. doi: 10.1172/JCI33102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.