Abstract
Osteoclasts, bone-resorptive multinucleated cells derived from hematopoietic stem cells, are associated with many bone-related diseases, such as osteoporosis. Osteoclast-targeting small-molecule inhibitors are valuable tools for studying osteoclast biology and for developing antiresorptive agents. Here, we have discovered that methyl-gerfelin (M-GFN), the methyl ester of the natural product gerfelin, suppresses osteoclastogenesis. By using M-GFN-immobilized beads, glyoxalase I (GLO1) was identified as an M-GFN-binding protein. GLO1 knockdown and treatment with an established GLO1 inhibitor in osteoclast progenitor cells interfered with osteoclast generation, suggesting that GLO1 activity is required for osteoclastogenesis. In cells, GLO1 plays a critical role in the detoxification of 2-oxoaldehydes, such as methylglyoxal. M-GFN inhibited the enzymatic activity of GLO1 in vitro and in situ. Furthermore, the cocrystal structure of the GLO1/M-GFN complex revealed the binding mode of M-GFN at the active site of GLO1. These results suggest that M-GFN targets GLO1, resulting in the inhibition of osteoclastogenesis.
Keywords: osteoclast, small molecule, crystal structure
Osteoclasts are bone-resorptive multinucleated cells derived from hematopoietic stem cells of the monocyte/macrophage lineage. Together with osteoblasts/stromal cells, osteoclasts are involved in remodeling bone matrix (1, 2). It is well known that two critical factors supplied by osteoblasts, the receptor activator of NF-κB ligand (RANKL) and macrophage colony stimulating factor (M-CSF), are essential for the differentiation and maturation of osteoclast precursors in bone (1, 2). During resorption, osteoclasts are highly polarized; form tightly sealed compartments on the bone surface; and secrete protons and proteases such as cathepsin K from the ruffled border; resulting in the degradation of bone matrix (1). Hence, the abnormal enhancement of osteoclasts is implicated in a variety of human diseases, including osteoporosis, rheumatoid arthritis, and cancer bone metastasis (3).
Osteoclast-targeting small-molecule inhibitors would be useful not only as tools for basic research on osteoclasts, but also as therapeutic drugs for these bone-related diseases. Bisphosphonates, synthetic analogs of pyrophosphate, are currently the most important and effective antiresorptive drugs available. Elucidation of the mechanisms underlying bisphosphonate activity, especially the identification of target proteins, has led to a deep understanding of osteoclast function (4). In addition, many naturally occurring small molecules have been reported to inhibit the differentiation and function of osteoclasts (5–12).
Cellular phenotype-based assays can be used to identify osteoclast-targeting small-molecule inhibitors (9). After cellular phenotype-based assays, the target identification of bioactive small molecules is one of the most important steps. To make biologically active tagged probes such as biotin conjugates, we devised a coupling method that enables the introduction of a variety of small molecules onto solid supports through a photoaffinity reaction (13). In this method, aryldiazirine groups covalently attached to solid supports are transformed upon UV irradiation into highly reactive carbenes, which are expected to bind to or insert irreversibly into proximal small molecules in a functional group-independent manner. We have applied this method to construct small-molecule microarrays (14, 15) and small-molecule affinity matrices (16).
The glyoxalase system, consisting of the enzymes GLO1 (EC 4.4.1.5) and glyoxalase II (GLO2, EC 3.1.2.6), is an integral component of cellular metabolism in mammalian cells (17). A major function of the glyoxalase pathway is believed to be detoxification of α-ketoaldehydes, especially a cytotoxic metabolite methylglyoxal (MG). The substrate for GLO1 is the hemithioacetal formed through the nonenzymatic conjugation of MG with GSH. The product of the GLO1-catalyzed reaction is S-d-lactoylglutathione, which is then hydrolyzed by GLO2 to d-lactate.
Here, we screened for new small-molecule inhibitors targeting osteoclasts from the chemical library of RIKEN Natural Products Depository (NPDepo) (18) and found that M-GFN, the methyl ester of gerfelin (GFN) isolated from a fungal strain, Beauveria felina QN22047 (19, 20), suppressed osteoclastogenesis. By using the photocross-linked M-GFN affinity matrix, we identified GLO1 as the molecular target of M-GFN that is involved in osteoclastogenesis inhibition. Furthermore, to clarify the binding mode of M-GFN, we determined the crystal structure of GLO1 complexed with M-GFN.
Results
M-GFN Inhibits Osteoclastogenesis.
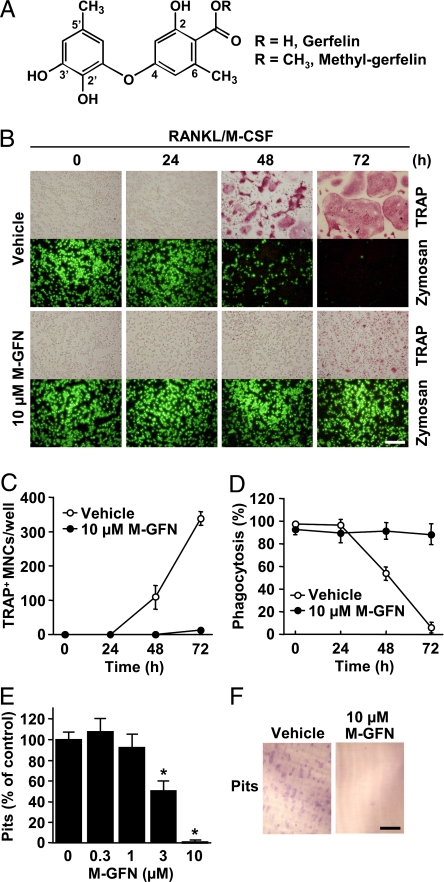
To identify small molecules that inhibit osteoclast function, we performed cellular phenotype-based screening from our natural product libraries. Mouse bone marrow-derived macrophages (BMMs) were differentiated mostly into tartrate-resistant acid phosphatase-positive (TRAP+) multinucleated osteoclasts after 72 h in the presence of RANKL and M-CSF (Fig. 1 B and C), after which TRAP+ multinucleated osteoclasts were no longer capable of phagocytosis, a characteristic of macrophages (Fig. 1 B and D). During this screening, we identified GFN and M-GFN as inhibitors of osteoclast differentiation (Fig. 1A). M-GFN (10 μM) strongly suppressed the formation of TRAP+ multinucleated osteoclasts induced by RANKL and M-CSF, after which M-GFN-treated cells retained their ability to phagocytose (Fig. 1 B–D). GFN and M-GFN suppressed osteoclastogenesis with 61 and 2.8 μM (IC50 value), respectively [supporting information (SI) Fig. S1], suggesting that the weak inhibitory effect of GFN is due to poor cell permeability, because it has a carboxylic group in its structure (Fig. 1A). When BMMs were cultured on dentine slices for 72 h in the presence of RANKL and M-CSF, resorption pits were formed (Fig. 1F). Treatment of BMMs with M-GFN reduced the number of pits in a dose-dependent manner (Fig. 1 E and F). Thus, M-GFN suppressed osteoclastogenesis with no influence on the phagocytic ability of BMMs. However, the effect of M-GFN on the survival and function of mature osteoclasts was negligible compared with the effective concentrations of M-GFN that inhibit osteoclastogenesis (Fig. S2).
Fig. 1.
M-GFN suppresses osteoclastogenesis with no influence on the phagocytic ability of BMMs. (A) Chemical structures of GFN and M-GFN. (B–D) BMMs were treated with M-GFN (10 μM) in the presence of RANKL and M-CSF for the indicated times. Then, cells were treated with fluorescein-conjugated zymosan A (FL-zymosan A), which is a model substrate to detect phagocytosis, and further stained for TRAP (B). TRAP+ multinucleated cells (MNCs) (C) and FL-zymosan A-incorporated phagocytic cells (D) were counted. Phagocytosis (%) means the ratio of the number of FL-zymosan A-incorporated cells to the total cell count. (E and F) Effect of M-GFN on the pit-forming activity of osteoclasts. BMMs were cultured on dentine slices with the indicated concentrations of M-GFN in the presence of RANKL and M-CSF for 72 h. Resorption pits on the slices, which are only formed by osteoclasts but not by BMMs, were stained with toluidine blue O (F), and the pits were counted (E). Data are shown as the mean ± SD (n = 4). *, P < 0.01 vs. Vehicle. (Scale bars, 200 μm.)
Identification of M-GFN-Binding Proteins.
GFN was originally discovered in the culture broth of a fungus as an inhibitor of human geranylgeranyl diphosphate (GGPP) synthase. GFN and M-GFN inhibited GGPP synthase in a noncompetitive manner against isopentenyl diphosphate (IPP) at 19.0 and 2.5 μM (Ki value), respectively (ref. 19, Fig. S3). In addition, nitrogen-containing bisphosphonates such as alendronate are known to inhibit farnesyl diphosphate (FPP) synthase and thereby prevent protein prenylation, resulting in the inhibition of osteoclastogenesis in a GGPP-sensitive manner (21, 22). Therefore, it seems likely that M-GFN and GFN inhibit GGPP synthase, thereby suppressing osteoclastogenesis. However, the inhibition of osteoclastogenesis by M-GFN and GFN, unlike that by alendronate, was not rescued by the addition of geranylgeraniol (GGOH), a cell-permeable analog of GGPP (Fig. S4 A and B). Moreover, treatment of BMMs with alendronate or mevastatin for 24 h caused the accumulation of unprenylated Rap1A, which is known to be geranylgeranylated in cells, whereas M-GFN (up to 66 μM) and GFN (up to 300 μM) had no effect (Fig. S4C and D). These results suggested that GGPP synthase inhibition by M-GFN and GFN has little or no involvement in the inhibition of osteoclastogenesis. Then, we attempted to determine whether M-GFN could interact with another protein to cause osteoclastogenesis inhibition in the cell. To pursue this question, we made an M-GFN affinity matrix by using our recently developed photocross-linking method.
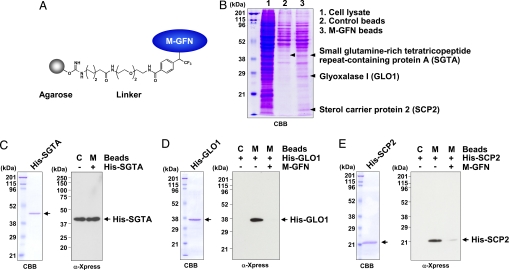
M-GFN was immobilized on agarose beads through a photoaffinity linker (Fig. 2A). Lysates of the mouse monocyte line RAW264 were precleared with control beads, then incubated with M-GFN-immobilized beads (M-GFN beads). The reacted beads were washed, and the coprecipitated proteins were analyzed by SDS/PAGE and visualized by Coomassie brilliant blue (CBB) staining. We observed three major protein bands that coprecipitated specifically with M-GFN beads, whereas the control beads had little or no such bands (Fig. 2B). We identified these proteins as mouse small glutamine-rich tetratricopeptide repeat-containing protein A (SGTA), GLO1, and sterol carrier protein 2 (SCP2). GLO1 and SCP2 were seen only in coprecipitants of M-GFN beads, whereas SGTA was detected slightly in coprecipitants of control beads (Fig. 2B).
Fig. 2.
Identification of M-GFN-binding proteins. (A) Model structure of M-GFN beads. (B) Detection of the coprecipitated proteins for M-GFN beads from RAW264 cell lysates. RAW264 cell lysates were precleared with control beads and incubated with M-GFN beads. The reacted beads were washed, and the eluted proteins were subjected to SDS/PAGE and visualized by CBB staining. The coprecipitated proteins for M-GFN beads were identified as described in Materials and Methods. (C–E) Purified His-tagged SGTA (C), GLO1 (D), or SCP2 (E) protein was incubated with control beads and M-GFN beads in the presence or absence of M-GFN as a competitor. The reacted beads were washed, and the eluted proteins were immunoblotted with anti-Xpress Ab, which allows detection of recombinant His-tagged proteins containing the N-terminal leader peptide (Xpress epitope). C, Control beads; M, M-GFN beads.
To determine whether M-GFN could bind directly to SGTA, GLO1, or SCP2, we performed pull-down experiments using purified (His)6-tagged (His-) proteins (Fig. 2 C–E). His-SGTA coprecipitated at nearly the same levels in both control and M-GFN beads (Fig. 2C), suggesting that the binding of SGTA is a nonspecific interaction. However, His-GLO1 and His-SCP2 coprecipitated only with M-GFN beads, and competition was clearly observed in the presence of M-GFN (Fig. 2 D and E). These results suggest that M-GFN binds directly to both GLO1 and SCP2, but not to SGTA.
GLO1 Activity Is Required for Osteoclastogenesis.
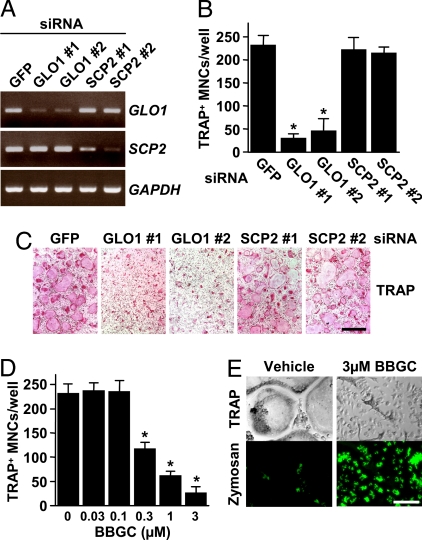
To investigate whether the loss of GLO1 or SCP2 activity could suppress osteoclastogenesis, we performed RNAi experiments in RANKL-induced osteoclastogenesis. We generated five siRNAs: one that was specific for GFP as a control, two that were specific for GLO1 (termed GLO1 nos. 1 and 2), and two that were specific for SCP2 (termed SCP2 nos. 1 and 2). RT-PCR analysis showed that GLO1 and SCP2 mRNA levels were reduced by 85%, 74%, 70%, and 85% in GLO1 no. 1, GLO1 no. 2, SCP2 no. 1, and SCP2 no. 2 siRNA-treated cells, respectively, as determined by densitometric analysis (Fig. 3A). Similarly, GLO1 enzymatic activities were significantly reduced by 79% and 85% in GLO1 nos. 1 and 2 siRNA-treated cells, respectively (data not shown). After 72 h of culture, TRAP staining revealed that treatment of BMMs with GLO1 siRNAs reduced the number of TRAP+ multinucleated osteoclasts induced by RANKL and M-CSF to a level significantly lower than the control (GFP siRNA), whereas treatment with SCP2 siRNAs had almost no effect (Fig. 3 B and C), suggesting that GLO1, but not SCP2, is involved in osteoclastogenesis. As expected, a known GLO1 inhibitor, S-p-bromobenzylglutathione cyclopentyl diester (BBGC) (23), dose-dependently inhibited osteoclastogenesis, and BBGC-treated cells retained the ability of phagocytosis (Fig. 3 D and E).
Fig. 3.
GLO1 activity is required for osteoclastogenesis. (A–C) The knockdown of GLO1, but not of SCP2, fails to generate osteoclasts. (A) BMMs were transfected with siRNAs derived from mouse GLO1 and SCP2, or GFP, and then cultured in the presence of RANKL and M-CSF for 48 h. To confirm the depletion of these mRNAs, RT-PCR was performed. (B and C) BMMs were transfected with siRNAs derived from mouse GLO1 and SCP2, or GFP, and then cultured in the presence of RANKL and M-CSF for 72 h. Cells were fixed and stained for TRAP (C), and TRAP+ MNCs were counted (B). Data are shown as the mean ± SD (n = 5). *, P < 0.01 vs. GFP siRNA. (Scale bar, 500 μm.) (D and E) A known GLO1 inhibitor suppresses osteoclastogenesis. BMMs were treated with the indicated concentrations of BBGC in the presence of RANKL and M-CSF for 72 h. Then, cells were treated with FL-zymosan A and further stained for TRAP (E). TRAP+ MNCs were counted (D). Data are shown as the mean ± SD (n = 4). *, P < 0.01 vs. vehicle. (Scale bar, 150 μm.)
M-GFN Inhibits GLO1 Activity in a Competitive Manner.
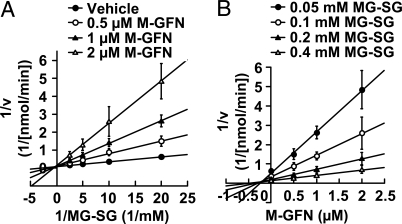
To test whether M-GFN could inhibit GLO1 activity, we performed an in vitro GLO1 assay using a spectrophotometric method that monitored the increase in absorbance at 240 nm because of the formation of S-d-lactoylglutathione. Kinetic analysis showed that M-GFN exhibited a competitive-type inhibition pattern, and the Ki value was 0.23 ± 0.10 μM (Fig. 4). GFN also showed the same inhibitory activity with a Ki value of 0.15 ± 0.07 μM (Fig. S5). In addition, intracellular MG levels were significantly increased in M-GFN-treated BMMs (Fig. S6A), and MG treatment suppressed osteoclastogenesis (Fig. S6 B and C). Thus, we concluded that M-GFN and GFN acted as competitive inhibitors of GLO1.
Fig. 4.
M-GFN inhibits GLO1 activity in a competitive manner. (A and B) Kinetic analysis of M-GFN against mouse His-GLO1. (A) Lineweaver–Burk plot of initial velocity vs. varying MG-SG concentrations. (B) Dixon plot of initial velocity vs. varying M-GFN concentrations. Data are shown as the mean ± SD (n = 3).
Crystal Structure of GLO1 Complexed with M-GFN.
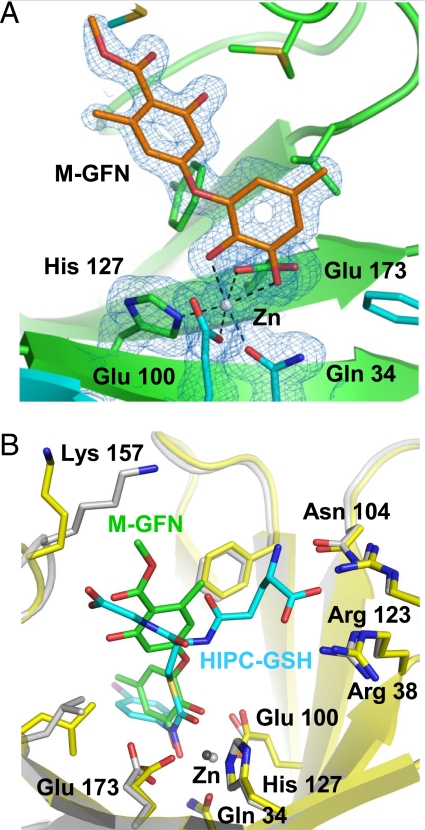
To clarify the binding mode of M-GFN, we determined the crystal structure of mouse GLO1 complexed with M-GFN at 1.7-Å resolution. This structural model contains one protein homodimer, two zinc ions, and two M-GFN molecules in an asymmetric unit. The two active sites of the GLO1 homodimer are located at the dimer interface and are characterized by a zinc ion-binding site, a glutathione-binding site, and a hydrophobic pocket. In the active site, the zinc ion is in octahedral coordination and is bound by Gln 34, Glu 100, His 127, Glu 173, and two hydroxyl groups of M-GFN directly (Fig. 5A). The crystal structures of human GLO1 in a complex with some glutathione derivative inhibitors have been reported (24, 25). The binding mode of human GLO1/S-(N-hydroxy-N-p-iodophenylcarbamoyl)glutathione (HIPC-GSH) complex is thought to mimic the enediolate reaction intermediate in catalysis (25). The binding mode of M-GFN/zinc ion resembles that of the HIPC-GSH (Fig. 5B) and is suspected to mimic one intermediate form. In the glutathione binding region, the opposite side of the zinc coordination moiety of M-GFN is located near the glycyl moiety of glutathione of superimposed HIPC-GSH. However, no hydrogen-bonding interaction between M-GFN molecule and the water molecules/amino acid residues is observed in this region. In the hydrophobic pocket, the 5′-methyl group of M-GFN points to the region and is surrounded by hydrophobic residues. These arrangements of M-GFN confirm the competitive inhibition data of M-GFN and suggest that the coordination between the two hydroxyl groups of M-GFN and the zinc ion contributes primarily to the binding between GLO1 and M-GFN.
Fig. 5.
Cocrystal structure of GLO1 complexed with M-GFN. (A) Structure of M-GFN binding to the active site zinc ion of mouse GLO1. 2Fobs−Fcalc electron density map contored at 1.0 sigma and overlaid on its structural model. Carbon atoms in one molecule of GLO1 dimer, the other molecule, and M-GFN are drawn in green, cyan, and orange, respectively. Gray sphere represents zinc ion. (B) Comparison of M-GFN/GLO1 complex and superimposed HIPC-GSH/GLO1 complex. M-GFN and HIPC-GSH molecules are shown as stick models in green and cyan, respectively. Carbon atoms of the protein (stick and ribbon model) and zinc ion in M-GFN/GLO1 model are drawn in yellow and dark gray, respectively, and those of HIPC-GSH/GLO1 are in gray. Oxygen, nitrogen, and sulfur atoms are drawn in red, blue, and gold, respectively, in both models. The half side of M-GFN partially overlaps the glycyl moiety of superimposed HIPC-GSH.
Discussion
The present study has shown that M-GFN targets GLO1, thereby inhibiting osteoclastogenesis. This conclusion is supported by the following findings: (i) GLO1 coprecipitated with M-GFN beads from whole-cell lysates (Fig. 2B), (ii) M-GFN bound directly to GLO1 (Fig. 2D) in the active site by coordination bonding via the zinc ion (Fig. 5A), (iii) M-GFN inhibited the enzymatic activity of GLO1 in a competitive manner (Fig. 4), and (iv) GLO1 knockdown by siRNA and GLO1 inhibitor treatment in BMMs inhibited osteoclastogenesis (Fig. 3). Furthermore, M-GFN increased the intracellular MG levels in BMMs (Fig. S6A), and MG treatment suppressed osteoclastogenesis (Fig. S6 B and C), suggesting that M-GFN inhibits GLO1 activity, thereby allowing the accumulation of MG, resulting in the inhibition of osteoclastogenesis.
MG is a highly reactive α-oxoaldehyde and can react with and modify both proteins and nucleic acids, leading to protein cross-linking, gene transcription, and cellular apoptosis (17). We have confirmed that high concentrations of M-GFN (>66 μM) and MG (>3 mM) induced cell death in BMMs (data not shown). The effective concentrations of M-GFN and MG that inhibit osteoclastogenesis might modify protein(s) or nucleic acid(s) that play critical roles in it. In fact, we confirmed that M-GFN and MG strongly inhibited RANKL-induced nuclear factor of activated T cells, calcineurin-dependent 1 (NFATc1) expression (Fig. S7), an essential transcription factor for osteoclastogenesis (26). In addition, to assess the selectivity of osteoclastogenesis inhibition by M-GFN, we examined the effect of M-GFN on osteoblast differentiation. Although 10 μM M-GFN inhibited osteoblast mineralization by 40%, considering the IC50 values and treatment times, M-GFN had a greater effect on osteoclastogenesis (IC50 = 3.1 μM, 72-h treatment) than on osteoblast differentiation (IC50 > 10 μM, 12-day treatment), as shown in Fig. 1E and Fig. S8.
GFN was originally identified as an inhibitor of GGPP synthase (19, 20). However, (i) GGOH did not reverse M-GFN and GFN inhibition of osteoclastogenesis (Fig. S4 A and B) (ii) M-GFN and GFN did not prevent prenylation of Rap1A (Fig. S4 C and D), and (iii) GGPP synthase did not coprecipitate with M-GFN beads in whole-cell lysates (Fig. 2B), suggesting that GGPP synthase was not involved in osteoclastogenesis inhibition by M-GFN and GFN. Moreover, the inhibitory activity of M-GFN against GLO1 (Ki = 0.23 μM) is ≈10 times that of GGPP synthase (Ki = 2.5 μM). GLO1 inhibition must be the principal cause of M-GFN inhibition in osteoclastogenesis. According to the pull-down experiments using M-GFN beads, SCP2, like GLO1, is an M-GFN-binding protein (Fig. 2 B and E). SCP2 is one of several lipid-binding proteins thought to be involved in the trafficking and metabolism of cholesterol and other lipids in mammalian cells (27). Because SCP2 knockdown by siRNA in BMMs had little effect on osteoclastogenesis (Fig. 3B), M-GFN-SCP2 interaction is not involved in the osteoclastogenesis inhibition by M-GFN, even if M-GFN could inhibit SCP2 activity.
So far, a series of GSH-conjugates have been mainly developed as GLO1 inhibitors (28). S-p-bromobenzyl-glutathione is a potent inhibitor (Ki = 0.16 μM), and di-ester prodrugs of this compound, such as BBGC, which have been developed to improve cellular uptake, have antitumor effects in vitro and in vivo (23, 29). In addition, the S-(N-aryl/alkyl-N-hydroxycarbamoyl)-glutathione derivatives are transition state analogs that represent a novel class of very potent GLO1 inhibitors (28, 30–32). However, nonsteroidal anti-inflammatory drugs (NSAIDs) have been reported to inhibit not only cyclooxygenases but also GLO1; the Ki value of indomethacin against GLO1 is 18.1 μM (33). Our present study shows that M-GFN and GFN are previously uncharacterized non-GSH-based GLO1 inhibitors. In the cocrystal structure of mouse GLO1 complexed with M-GFN, two oxygen atoms of the hydroxy groups of M-GFN coordinated directly with the zinc ion (Fig. 5). The methyl group of the methyl ester of M-GFN is directed toward the flexible loop region (residues 153–160); however, the interaction between the M-GFN molecule and the residues on the loop is not observed. In the case of GFN, the GFN molecule has a carboxylic group, and this moiety can interact with the side chain of Lys 157 of the flexible loop region. This probability supports the result that the affinity of GFN for GLO1 is slightly stronger than that of M-GFN (Fig. 4 and Fig. S5).
In summary, we have demonstrated that M-GFN and GFN act as GLO1 competitive inhibitors, thereby suppressing osteoclastogenesis. Moreover, our study has shown that our photocross-linking method is quite useful for identifying target proteins for bioactive small molecules in future chemical genetic studies. Although the validation of GLO1 as a target of antiresorptive agents remains to be elucidated, our findings can facilitate the study of osteoclast biology and the development of therapeutic agents.
Materials and Methods
Reagents and Proteins.
M-GFN was synthesized by methylation of GFN (see SI Methods). The following were used: human soluble RANKL (Peprotech), human M-CSF (Leucoprol, Kyowa Hakko Kogyo), human TGF-β1 (R & D Systems); FL-zymosan A BioParticles (Molecular Probes), BBGC (kindly provided by Taiho Pharmaceutical), MG (Sigma-Aldrich), and Lipofectamine RNAiMAX and anti-Xpress Ab (Invitrogen). The mouse SGTA, GLO1, or SCP2 gene was cloned from a UAMS-32 (stromal/osteoblastic cell line) cDNA library and subcloned into the pRSET C vector (Invitrogen). These recombinant His-tagged proteins were expressed in the Escherichia coli BL21(DE3)pLysS strain and purified on a HisTrap HP (GE Healthcare) by using FPLC (Amersham Pharmacia Biotech).
Osteoclast Formation.
Five-week-old male ddY mice were from Japan SLC. Bone marrow cells were collected from their tibiae and femora and were cultured with M-CSF (50 ng/ml) and TGF-β1 (1 ng/ml) for 72 h in α-MEM (Sigma-Aldrich) supplemented with 10% FCS (Gibco) in type-I collagen-coated culture plates (Iwaki). After 72 h of culture, floating cells were removed by rinsing with PBS, and attached cells were used as BMMs. To induce osteoclast differentiation, BMMs were further cultured with RANKL (50 ng/ml) and M-CSF (50 ng/ml) for 72 h. Cells were then fixed in 3.7% formalin and stained for TRAP. TRAP+ multinucleated cells containing more than three nuclei were counted as osteoclasts. The experimental procedures and housing conditions for the animals were approved by the Animal Experiment Committee of RIKEN, and all of the animals were cared for and treated humanely in accordance with the Guidelines for Experiments Using Animals. The methods for TRAP staining, drug screening, phagocytosis assay, and pit formation assay are described in SI Methods.
Detection of Binding Proteins for M-GFN Beads.
Preparation of M-GFN beads is described in SI Methods. Mouse monocyte line RAW264 (RIKEN Cell Bank) was harvested and washed twice with PBS, then resuspended in binding buffer [10 mM Tris·HCl pH 7.6, 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, and protease inhibitor mixture (Roche)]. After cell lysis by homogenization with a syringe and sonication, the insoluble material was removed by centrifugation, and the supernatant was collected as a cell lysate. After the cell lysate (14 mg of protein) was precleared twice by incubation with control beads (150 μl) for 1 h at 4°C, the cleared cell lysate was incubated with M-GFN beads (200 μl) for 12 h at 4°C. The reacted beads were washed with binding buffer, and the bounded proteins were eluted with SDS/PAGE sample buffer, separated by SDS-polyacrylamide gel, and visualized by CBB staining. To identify the protein, the protein-containing region in the SDS-polyacrylamide gel was excised, washed, and dried in vacuo. After reduction and alkylation of the samples, in-gel digestion was performed with trypsin or Achromobacter protease I. The resulting peptides were extracted and analyzed by MALDI-TOF MS (16). The protein was identified from peptide mass fingerprinting by using the Mascot search program and the Swiss-Prot database. His-tagged SGTA, GLO1, or SCP2 protein (100 ng) was incubated with control or M-GFN beads (10 μl) in the presence or absence of M-GFN (200 μg) in binding buffer containing 1% BSA (1.6 ml) for 5 h at 4°C. The reacted beads were washed with binding buffer, and the bounded proteins were eluted with SDS/PAGE sample buffer. Proteins were resolved by SDS/PAGE and detected by Western blotting with anti-Xpress Ab.
RNAi and RT-PCR.
The methods for RNAi and RT-PCR are described in SI Methods.
In Vitro GLO1 Assay.
Kinetic measurements were carried out by using a thermostated spectrophotometer (Beckman Coulter, DU640) to monitor the increase in absorbance at 240 nm because of the formation of S-d-lactoylglutathione (ε = 3.37 mM−1·cm−1) at 30°C (34). The reaction buffer contained 0.1 M sodium phosphate (pH 7.0) and 14.6 mM MgSO4, and hemithioacetal (MG-SG) was preincubated for 5 min at 30°C. For the desired concentration of MG-SG, the concentrations of MG and GSH were calculated and varied by using the equation Kd = 3 mM = ([MG][GSH])/[MG-SG], and excess free GSH in the assay was 0.1 mM. The reaction was initiated by the addition of recombinant mouse His-GLO1 (3 nM) to the reaction buffer with or without compound.
Crystal Structure Analysis.
Nontagged recombinant mouse GLO1 was expressed in the E. coli BL21(DE3)pLysS strain and purified by using an anion exchange column and gel-filtration chromatography. Crystals of GLO1 containing M-GFN were obtained as described (24), but with the inclusion of M-GFN to a final concentration 1 mM in the drop. The crystal belongs to the space group P21, with unit cell dimensions of a = 42.0 Å, b = 65.3 Å, c = 66.2 Å, α = 90°, β = 101.18°, and γ = 90°. x-ray diffraction measurements were performed at the beamline SPring-8 BL26B2 at 100 K. Diffraction images of the dataset were indexed, integrated, and scaled by using the HKL2000 program suite (35). The data collection and refinement statistics are summarized in Table S1. Phase calculation was carried out by molecular replacement with the program, MOLREP (36), in the CCP4 program suite (37). Model building of the electron density map was performed automatically with the RESOLVE program (38) and LAFIRE program (39), and manually with the XtalView program (40). Crystallographic refinement was performed by using CNS (41).
Statistical Analyses.
Statistical analyses were performed by using one-way ANOVA followed by Dunnet's posthoc test. A value of P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments.
We thank M. Yamamoto and H. Miyatake for help with x-ray diffraction data collection; T. Usui, S. Kazami, and A. Mochizuki for support with the experiments; Y. Ichikawa and R. Nakagawa for DNA sequencing; R. Kamijo for support; and Taiho Pharmaceutical Co., Ltd., for providing BBGC. The synchrotron radiation experiments were performed at the BL26B2 in SPring-8 with mail-in data collection system. This work was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; by the Bioarchitect and Chemical Biology projects (RIKEN); and by funding for a special postdoctoral program of RIKEN (to M.K. and H.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.P.N. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinate has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2ZA0).
This article contains supporting information online at www.pnas.org/cgi/content/full/0712239105/DCSupplemental.
References
- 1.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 2.Suda T, et al. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 3.Hofbauer LC, Neubauer A, Heufelder AE. Receptor activator of nuclear factor-κB ligand and osteoprotegerin: Potential implications for the pathogenesis and treatment of malignant bone diseases. Cancer. 2001;92:460–470. doi: 10.1002/1097-0142(20010801)92:3<460::aid-cncr1344>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 4.Rogers MJ. New insights into the molecular mechanisms of action of bisphosphonates. Curr Pharmacol Des. 2003;9:2643–2658. doi: 10.2174/1381612033453640. [DOI] [PubMed] [Google Scholar]
- 5.Orcel P, Denne MA, de Vernejoul MC. Cyclosporin-A in vitro decreases bone resorption, osteoclast formation, and the fusion of cells of the monocyte-macrophage lineage. Endocrinology. 1991;128:1638–1646. doi: 10.1210/endo-128-3-1638. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura I, et al. Wortmannin, a specific inhibitor of phosphatidylinositol-3 kinase, blocks osteoclastic bone resorption. FEBS Lett. 1995;361:79–84. doi: 10.1016/0014-5793(95)00153-z. [DOI] [PubMed] [Google Scholar]
- 7.Blair HC, Jordan SE, Peterson TG, Barnes S. Variable effects of tyrosine kinase inhibitors on avian osteoclastic activity and reduction of bone loss in ovariectomized rats. J Cell Biochem. 1996;61:629–637. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C629::AID-JCB17%3E3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 8.Okahashi N, et al. Specific inhibitors of vacuolar H(+)-ATPase trigger apoptotic cell death of osteoclasts. J Bone Miner Res. 1997;12:1116–1123. doi: 10.1359/jbmr.1997.12.7.1116. [DOI] [PubMed] [Google Scholar]
- 9.Kazami S, et al. Iejimalides show anti-osteoclast activity via V-ATPase inhibition. Biosci Biotechnol Biochem. 2006;70:1364–1370. doi: 10.1271/bbb.50644. [DOI] [PubMed] [Google Scholar]
- 10.Woo J-T, Kasai S, Stern PH, Nagai K. Compactin suppresses bone resorption by inhibiting the fusion of prefusion osteoclasts and disrupting the actin ring in osteoclasts. J Bone Miner Res. 2000;15:650–662. doi: 10.1359/jbmr.2000.15.4.650. [DOI] [PubMed] [Google Scholar]
- 11.Woo J-T, et al. Reveromycin A, an agent for osteoporosis, inhibits bone resorption by inducing apoptosis specifically in osteoclasts. Proc Natl Acad Sci USA. 2006;103:4729–4734. doi: 10.1073/pnas.0505663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muguruma H, et al. Reveromycin A inhibits osteolytic bone metastasis of small-cell lung cancer cells, SBC-5, through an antiosteoclastic activity. Clin Cancer Res. 2005;11:8822–8828. doi: 10.1158/1078-0432.CCR-05-1335. [DOI] [PubMed] [Google Scholar]
- 13.Kanoh N, et al. Immobilization of natural products on glass slides by using a photoaffinity reaction and the detection of protein-small-molecule interactions. Angew Chem Int Ed. 2003;42:5584–5587. doi: 10.1002/anie.200352164. [DOI] [PubMed] [Google Scholar]
- 14.Kanoh N, et al. Photo-cross-linked small-molecule microarrays as chemical genomic tools for dissecting protein-ligand interactions. Chem Asian J. 2006;1:789–797. doi: 10.1002/asia.200600208. [DOI] [PubMed] [Google Scholar]
- 15.Kanoh N, et al. SPR imaging of photo-cross-linked small-molecule arrays on gold. Anal Chem. 2006;78:2226–2230. doi: 10.1021/ac051777j. [DOI] [PubMed] [Google Scholar]
- 16.Kanoh N, Honda K, Simizu S, Muroi M, Osada H. Photo-cross-linked small-molecule affinity matrix for facilitating forward and reverse chemical genetics. Angew Chem Int Ed. 2005;44:3559–3562. doi: 10.1002/anie.200462370. [DOI] [PubMed] [Google Scholar]
- 17.Thornalley PJ. Glutathione-dependent detoxification of α-oxoaldehydes by the glyoxalase system: Involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem Biol Interact. 1998;111–112:137–151. doi: 10.1016/s0009-2797(97)00157-9. [DOI] [PubMed] [Google Scholar]
- 18.Tomiki T, et al. RIKEN natural products encyclopedia (RIKEN NPEdia), a chemical database of RIKEN natural products depository (RIKEN NPDepo) J Comput Aid Chem. 2006;7:157–162. [Google Scholar]
- 19.Zenitani S, et al. Gerfelin, a novel inhibitor of geranylgeranyl diphosphate synthase from Beauveria felina QN22047 I Taxonomy, fermentation, isolation, and biological activities. J Antibiot. 2003;56:617–621. doi: 10.7164/antibiotics.56.617. [DOI] [PubMed] [Google Scholar]
- 20.Zenitani S, et al. Gerfelin, a novel inhibitor of geranylgeranyl diphosphate synthase from Beauveria felina QN22047 II Structural elucidation. J Antibiot. 2003;56:658–660. doi: 10.7164/antibiotics.56.658. [DOI] [PubMed] [Google Scholar]
- 21.Fisher JE, et al. Alendronate mechanism of action: Geranylgeraniol, an intermediate in the mevalonate pathway, prevents inhibition of osteoclast formation, bone resorption, and kinase activation in vitro. Proc Natl Acad Sci USA. 1999;96:133–138. doi: 10.1073/pnas.96.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kavanagh KL, et al. The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs. Proc Natl Acad Sci USA. 2006;103:7829–7834. doi: 10.1073/pnas.0601643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornalley PJ, et al. Antitumour activity of S-p-bromobenzylglutathione cyclopentyl diester in vitro and in vivo. Biochem Pharmacol. 1996;51:1365–1372. doi: 10.1016/0006-2952(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 24.Cameron AD, Olin B, Ridderström M, Mannervik B, Jones TA. Crystal structure of human glyoxalase I–evidence for gene duplication and 3D domain swapping. EMBO J. 1997;16:3386–3395. doi: 10.1093/emboj/16.12.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron AD, et al. Reaction mechanism of glyoxalase I explored by an X-ray crystallographic analysis of the human enzyme in complex with a transition state analogue. Biochemistry. 1999;38:13480–13490. doi: 10.1021/bi990696c. [DOI] [PubMed] [Google Scholar]
- 26.Takayanagi H, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 27.Gallegos AM, et al. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog Lipid Res. 2001;40:498–563. doi: 10.1016/s0163-7827(01)00015-7. [DOI] [PubMed] [Google Scholar]
- 28.Creighton DJ, Zheng Z-B, Holewinski R, Hamilton DS, Eiseman JL. Glyoxalase I inhibitors in cancer chemotherapy. Biochem Soc Trans. 2003;31:1378–1382. doi: 10.1042/bst0311378. [DOI] [PubMed] [Google Scholar]
- 29.Sakamoto H, et al. Selective activation of apoptosis program by S-p-bromobenzylglutathione cyclopentyl diester in glyoxalase I-overexpressing human lung cancer cells. Clin Cancer Res. 2001;7:2513–2518. [PubMed] [Google Scholar]
- 30.Zheng Z-B, Creighton DJ. Bivalent transition-state analogue inhibitors of human glyoxalase I. Org Lett. 2003;5:4855–4858. doi: 10.1021/ol035917s. [DOI] [PubMed] [Google Scholar]
- 31.Kavarana MJ, Kovaleva EG, Creighton DJ, Wollman MB, Eiseman JL. Mechanism-based competitive inhibitors of glyoxalase I: intracellular delivery, in vitro antitumor activities, and stabilities in human serum and mouse serum. J Med Chem. 1999;42:221–228. doi: 10.1021/jm9708036. [DOI] [PubMed] [Google Scholar]
- 32.Sharkey EM, et al. Pharmacokinetics and antitumor properties in tumor-bearing mice of an enediol analogue inhibitor of glyoxalase I. Cancer Chemother Pharmacol. 2000;46:156–166. doi: 10.1007/s002800000130. [DOI] [PubMed] [Google Scholar]
- 33.Sato S, et al. Polyproline-rod approach to isolating protein targets of bioactive small molecules: Isolation of a new target of indomethacin. J Am Chem Soc. 2007;129:873–880. doi: 10.1021/ja0655643. [DOI] [PubMed] [Google Scholar]
- 34.Akoachere M, et al. Characterization of the glyoxalases of the malarial parasite Plasmodium falciparum and comparison with their human counterparts. Biol Chem. 2005;386:41–52. doi: 10.1515/BC.2005.006. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 37.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.Terwilliger TC. Map-likelihood phasing. Acta Crystallogr D. 2001;57:1763–1775. doi: 10.1107/S0907444901013749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao M, Zhou Y, Tanaka I. LAFIRE: Software for automating the refinement process of protein-structure analysis. Acta Crystallogr D. 2006;62:189–196. doi: 10.1107/S0907444905038965. [DOI] [PubMed] [Google Scholar]
- 40.McRee DE. XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 41.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.