Abstract
The molybdenum nitrogenase, present in a diverse group of bacteria and archea, is the major contributor to biological nitrogen fixation. The nitrogenase active site contains an iron–molybdenum cofactor (FeMo-co) composed of 7Fe, 9S, 1Mo, one unidentified light atom, and homocitrate. The nifQ gene was known to be involved in the incorporation of molybdenum into nitrogenase. Here we show direct biochemical evidence for the role of NifQ in FeMo-co biosynthesis. As-isolated NifQ was found to carry a molybdenum–iron–sulfur cluster that serves as a specific molybdenum donor for FeMo-co biosynthesis. Purified NifQ supported in vitro FeMo-co synthesis in the absence of an additional molybdenum source. The mobilization of molybdenum from NifQ required the simultaneous participation of NifH and NifEN in the in vitro FeMo-co synthesis assay, suggesting that NifQ would be the physiological molybdenum donor to a hypothetical NifEN/NifH complex.
Keywords: nif, iron-sulfur cluster, Azotobacter vinelandii
Biological nitrogen fixation performed by microorganisms that have nitrogenase(s) accounts for roughly two-thirds of the nitrogen fixed globally. Most nitrogen fixation is carried out by the activity of molybdenum nitrogenase (1), which is widely distributed in nature (2). The molybdenum-nitrogenase enzyme is composed of two proteins (3): a heterotetrameric NifDK-protein component (a2β2 dinitrogenase) and a homodimeric NifH-protein component (dinitrogenase reductase). NifDK contains one iron–molybdenum cofactor (FeMo-co) within the active site of each α-subunit (NifD), and one [8Fe–7S] P-cluster at the interface of the α- and β-subunits in each αβ pair (4). NifH contains a [4Fe–4S] cluster bridging the two subunits and one site for Mg-ATP binding and hydrolysis in each subunit (5).
FeMo-co is a unique cofactor‖ composed of a [Mo–3Fe–3S] partial cubane bridged to a [4Fe–3S] partial cubane by three sulfur ligands and one unidentified light atom in the center of the cofactor (6). The molybdenum atom is also coordinated to the organic acid homocitrate. The biosynthesis of FeMo-co is a complex process that involves the activities of several nitrogen fixation (nif) gene products that function as molecular scaffolds, enzymes, or escort proteins that carry FeMo-co precursors between assembly sites in the pathway (7, 8). Following assembly, FeMo-co is inserted into apo-NifDK, a P-cluster-containing but FeMo-co-deficient form of NifDK that is matured into functional NifDK simply by FeMo-co insertion, to generate the mature dinitrogenase enzyme that is competent for nitrogen fixation.
The NifEN scaffold protein is believed to function in the pathway as a central node to which an [Fe–S]-containing FeMo-co precursor, molybdenum, and homocitrate might converge to complete the assembly of FeMo-co (9–12). In this model, the [Fe–S]-containing FeMo-co precursor, NifB-co**, would be initially assembled by NifB and then transferred to NifEN for its conversion into the [Fe–S] VK-cluster (13, 14). Homocitrate would be provided by the homocitrate-synthase activity of NifV (15, 16). Although molybdate satisfies the molybdenum requirement for FeMo-co synthesis in vitro, the physiological source of molybdenum remains unknown.
Genetic evidence from Azotobacter vinelandii, Klebsiella pneumoniae, and other nitrogen-fixing bacteria has suggested the involvement of NifQ in molybdenum processing for nitrogenase. In all cases tested, nifQ has been shown to be essential for diazotrophic growth, although excess molybdate in the growth medium alleviated the nifQ phenotype (17–19). The role of NifQ is specifically related to the Mo-nitrogenase because nifQ mutants were not affected in either the capacity to grow by using the alternative vanadium or iron-only nitrogenase systems (20) or in the activity of molybdopterin-containing enzymes (21).
NifQ homologues are present in all diazotrophic species of the Proteobacteria phylum, with the exception of some Rhizobium species. NifQ proteins have a conserved Cx4Cx2Cx5C amino acid motif that has been suggested to bind a molybdenum-containing metal cluster or a Mo-S intermediate for FeMo-co synthesis (22). However, NifQ proteins are unique and their primary sequences are unrelated to other proteins shown to be involved in molybdenum trafficking. NifQ proteins do not carry molbindin-like domains that are involved in molybdate binding (23). Molbindin domains are found in the ModA component of molybdate-transport systems, in the molybdate-responsive ModE transcriptional regulator, and in the A. vinelandii ModG protein involved in molybdenum homeostasis. Additionally, NifQ does not exhibit significant sequence similarity to the molybdenum-storage-protein (Mosto) MosA and MosB polypeptide components, which are able to bind large amounts of molybdenum, presumably as heptamolybdate (24).
In this work, we show that NifQ carries a molybdenum–iron–sulfur cluster that serves as a specific molybdenum donor for the synthesis of FeMo-co in vitro. Mobilization of molybdenum from NifQ required the simultaneous presence of the NifH and NifEN proteins. These findings strongly support a role for NifQ as the physiological molybdenum donor to a putative NifEN/NifH complex during FeMo-co biosynthesis.
Results
NifQ Is an Iron-Sulfur Molybdoprotein.
Expression of a His-tagged version of NifQ in UW300 cells was 50-fold higher than NifQ expression in wild-type cells. His-NifQ overexpression did not alter in vitro nitrogenase activity [supporting information (SI) Table S1] or the regulation by ammonium of the expression of NifQ or any other essential nif gene analyzed in UW300 (data not shown). Hereafter, we refer to the A. vinelandii His-tagged NifQ variant as NifQ. The NifQ protein was purified under strict anaerobic conditions from UW300 cells (Fig. 1A and Experimental Procedures). The average purification yield was 280 mg NifQ/kg UW300 cell paste. The A. vinelandii NifQ was also purified from recombinant Escherichia coli cells overexpressing GST-NifQ fusion protein (Fig. 1B). NifQ protein purified from E. coli cells was denominated as rNifQ. The average purification yield was 4 g rNifQ/kg E. coli cell paste.
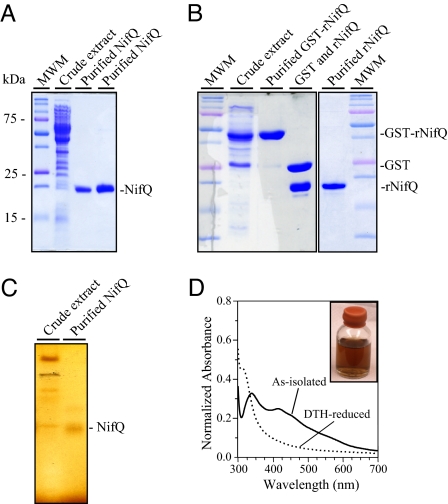
Fig. 1.
Purification and UV-visible spectra of native A. vinelandii NifQ and recombinant rNifQ. (A) SDS/PAGE analysis of A. vinelandii NifQ purification steps (cell-free extract lane contains 40 μg of protein; purified NifQ lanes contain 5 μg and 15 μg of protein, respectively). Molecular weight markers are indicated to the left. The position of NifQ is indicated to the right. (B) SDS/PAGE analysis of recombinant rNifQ purification steps (cell-free extract lane contains 25 μg of protein; purified protein lanes contain 8 μg of protein). Protein positions in the gel are indicated to the right. (C) Fe staining of NifQ purified from A. vinelandii cells after anoxic native PAGE (cell-free extract lane contains 100 μg of protein; purified NifQ lane contains 25 μg of protein). (D) UV-visible spectra of as-isolated NifQ (solid line) and DTH-reduced NifQ (dotted line). DTH-treated NifQ samples (33 μM NifQ) were gel-filtered on a Sephadex G-25 resin to remove excess DTH before recording the spectrum. Spectra were normalized by absorbance at 280 nm.
NifQ and rNifQ preparations were reddish brown and exhibited a positive signal in gels stained for Fe after anoxic native PAGE (Fig. 1C). Exposure of the purified proteins to air resulted in color bleaching indicative of sensitivity to oxygen (data not shown). The native molecular weight of as-isolated NifQ (Mr = 25.7 ± 0.4 kDa) is similar to the molecular weight deduced from the A. vinelandii nifQ primary sequence (Mr = 19.7 kDa) and thus consistent with a monomeric structure. As-isolated NifQ contained 3.1 ± 0.1 Fe atoms and 0.30 ± 0.04 Mo atoms per NifQ molecule as determined by inductively coupled plasma optical emission spectrometry (ICP-OES). Interestingly, ICP-OES analysis showed that rNifQ contained 4.6 ± 0.2 Fe atoms but no molybdenum. NifQ displayed a UV-visible spectrum typical of [Fe–S] proteins with a sharp peak at 340 nm, a broad peak at 410 nm, and an A410/A280 ratio of 0.2 (Fig. 1D). This ratio decreased after incubating NifQ with 5 mM sodium dithionite (DTH), which is characteristic of [Fe–S] cluster reduction by DTH.
The Molybdenum Within NifQ Is in a [Mo–3Fe–4S] Cluster Environment.
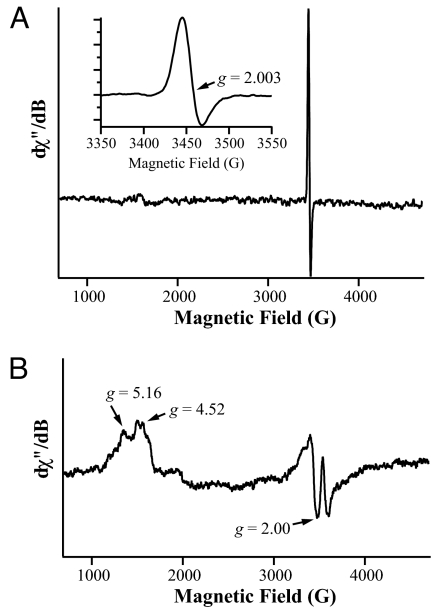
Fig. 2A shows the X-band continuous wave electron paramagnetic resonance (CW-EPR) spectrum of the as-isolated NifQ protein. The spectrum exhibits a fairly isotropic signal centered at g = 2.00 with a peak-to-peak line width of ≈27 G (Fig. 2A magnification). This EPR signal has an asymmetric shape that is characteristic of a S = 1/2 [3Fe–4S]+ cluster (25, 26); it exhibits line width similar to that of aconitase (25) and line shape similar to that of ferredoxin II from Desulfovibrio gigas (27). It has been proposed that the EPR signal of oxidized [3Fe–4S]+ clusters arises from the antiferromagnetic exchange coupling of three high-spin (S = 5/2) ferric ions to form an S = 1/2 ground state (26). Power- and temperature-dependence profiles of the EPR signal from NifQ are also consistent with the presence of [3Fe–4S]+ clusters in the as-isolated NifQ (Fig. S1).
Fig. 2.
CW-EPR analysis of NifQ from A. vinelandii. Samples contained ≈1 mM NifQ. (A) As-isolated NifQ. The magnification is an enlarged image in the g = 2 region. (B) NifQ protein (1 mM) incubated with 5 mM DTH, 1 mM Na2MoO4, and 1 mM Na2S. EPR spectra were recorded under the following experimental conditions: temperature, 12 K; microwave frequency, 9.70 GHz; microwave power, 0.03 mW for as-isolated NifQ and 5 mW for reduced NifQ; modulation amplitude, 8 G; modulation frequency, 100 kHz; time constant, 80.97 ms; conversion time, 10.24 ms; and resolution, 4096 points.
Treatment of NifQ with DTH gave rise to a different EPR-active species resembling spectra of chemically synthesized compounds comprising S = 3/2 [Mo–3Fe–4S]3+ clusters (28–30) (data not shown). This new EPR signal did not arise from reduction of the [3Fe–4S]+ cluster to the [3Fe–4S]0 state (S = 2), because such a species would be EPR silent in the perpendicular mode of CW-EPR. When NifQ treatment with DTH was carried out in the presence of molybdate and sulfide, the intensity of the [Mo–3Fe–4S]3+-like signal increased and the features at the g = 4 region of the spectra were sharpened (Fig. 2B).
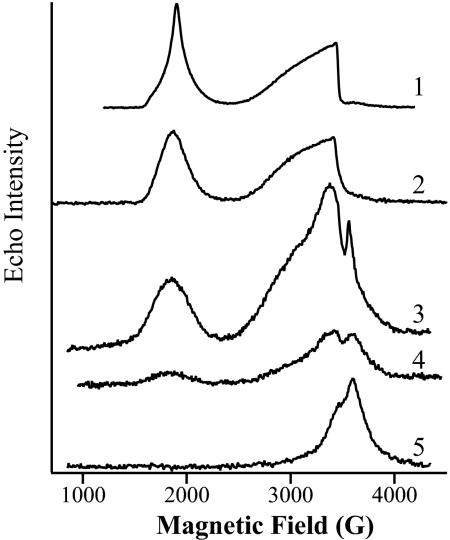
Electron spin echo (ESE) field-sweep EPR spectrum of DTH-reduced NifQ showed a line shape (Fig. 3 Line 4) with features similar to that of FeMo-co associated with the NifDK component of nitrogenase (Fig. 3 Line 1) or with the carrier protein nitrogenase accessory factor Y (NafY) (Fig. 3 Line 2). This line shape is consistent with the presence of an S = 3/2 [Mo–Fe–S] cluster in NifQ. Treatment of NifQ with DTH, molybdate, and sulfur markedly increased the [Mo–Fe–S] cluster signal (Fig. 3 Line 3). On the other hand, the molybdenum-free and DTH-reduced rNifQ lacked the [Mo–Fe–S] cluster signal in the ESE field-sweep EPR spectrum (Fig. 3 Line 5), as expected.
Fig. 3.
Two-pulse ESE-EPR field-sweep spectra recorded at 4.2 K of DTH-reduced NifDK protein (Line 1); DTH-reduced NafY:FeMo-co complex generated in vitro (44) (Line 2); NifQ protein (1 mM) incubated with 5 mM DTH, 1 mM Na2MoO4, and 1 mM Na2S (Line 3); DTH-reduced NifQ (1 mM NifQ and 5 mM DTH) (Line 4); and DTH-reduced rNifQ (1 mM rNifQ and 5 mM DTH) (Line 5). EPR spectra were recorded under the following experimental conditions. Trace 1: microwave frequency, 9.74 GHz; microwave power, 25 W; t = 140 ns; and repetition time, 8 ms. Trace 2: microwave frequency, 9.74 GHz; microwave power, 39 W; t = 140 ns; and repetition time, 8 ms. Trace 3: microwave frequency, 9.73 GHz; microwave power, 63 W; t = 136 ns; and repetition time, 10 ms. Trace 4: microwave frequency, 9.73 GHz; microwave power, 63 W; t = 136 ns; and repetition time, 10 ms. Trace 5: microwave frequency, 9.72 GHz; microwave power, 79 W; t = 144 ns; and repetition time, 20 ms.
In conclusion, CW-EPR and ESE field sweep EPR analysis, combined with Fe and Mo determinations, indicate that NifQ is able to coordinate a redox responsive [Mo–3Fe–4S] cluster.
NifQ Serves as a Molybdenum Donor for FeMo-co Synthesis in Vitro.
The assay for in vitro FeMo-co synthesis with purified components used in this work requires a FeMo-co precursor (NifB-co or VK-cluster††), NifH, NifEN, homocitrate, Mg-ATP, DTH, and a source of molybdenum (either Na2MoO4 or NifQ). Synthesized FeMo-co binds and activates FeMo-co-deficient apo-NifDK protein, generating active NifDK that is routinely assayed by its acetylene-reducing activity.
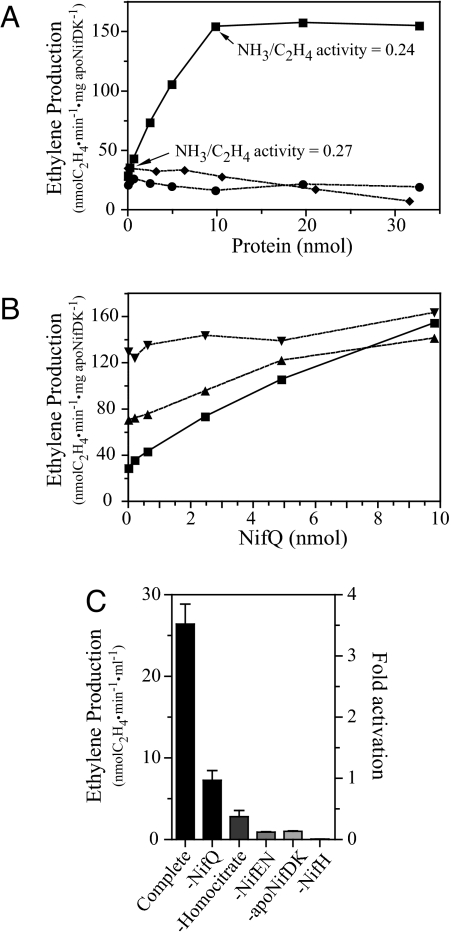
Fig. 4A shows the titration with NifQ of VK-cluster-dependent in vitro FeMo-co biosynthesis reactions. Because no molybdate was added to the reactions, this result shows that as-isolated NifQ substituted for Na2MoO4 in the assay. Similar results were obtained when NifB-co and precursor-free NifEN [purified from UW243 cells (13)] were substituted for VK-cluster-loaded NifEN in the assay (data not shown). We note that the background level of apo-NifDK activation observed in the reaction lacking NifQ was because of mobilization of the molybdenum present in purified NifEN preparations (11).
Fig. 4.
NifQ serves as a molybdenum donor for in vitro synthesis of FeMo-co with purified components. FeMo-co synthesis was determined by the acetylene reduction activity of matured NifDK. In addition, the dinitrogen reduction activity of matured NifDK was determined in selected samples. All vials were acid-washed and thoroughly rinsed with Milli-Q water to remove traces of molybdenum. FeMo-co synthesis reactions contained 0.43 nmol of apo-NifDK, 0.48 nmol of VK-cluster loaded NifEN, 2.4 nmol NifH, and the indicated amount of NifQ (0–30 nmol). (A) Titration of FeMo-co synthesis reactions with increasing amounts of purified NifQ (squares), purified rNifQ (circles), or purified cyanobacterial nitrate reductase (NarB) (diamonds). No molybdate was added to the reactions. (B) Titration of FeMo-co synthesis reactions with increasing amounts of purified NifQ in the presence of 18 μM molybdate (inverted triangles), 0.18 μM molybdate (triangles), or no molybdate (squares). (C) Requirement for different components in the FeMo-co synthesis reaction with purified proteins. No molybdate was added to the reactions. All reactions contained 4.9 nmol of NifQ except the reaction lacking NifQ. Fold activation with respect to the reaction lacking NifQ is indicated in the right y axis. In the experiments lacking NifH, FeMo-co synthesis was stopped by the addition of 0.3 mM (NH4)2MoS4 before assaying the activity of reconstituted NifDK. Data are the average of two to four independent determinations.
The level of NifDK-specific activity reconstituted in vitro was limited by several factors. First, at saturating levels of NifQ (10 nmol), the amount of NifEN present in the assay (0.48 nmol) became limiting. When 2.4 nmol of NifEN were used in the assay, the level of reconstituted NifDK activity and the amount of NifQ required to saturate the assay increased accordingly (data not shown). Second, molybdenum donation by NifQ was incomplete under the in vitro FeMo-co biosynthesis assay conditions. In vitro FeMo-co synthesis reactions in which denatured NifQ protein served as the molybdenum source yielded 2-fold higher NifDK activity levels than reactions in which the same amount of native NifQ served as the molybdenum donor (data not shown). Third, saturating amounts of apo-NifDK were added to the assay to obtain higher absolute levels of apo-NifDK activation at the expense of obtaining lower specific activity per mg of NifDK, as described in ref. 14.
NifQ supported the formation of fully functional NifDK protein. In vitro reconstituted NifDK reduced dinitrogen to ammonia with a ratio of specific activity of ammonia-to-ethylene production similar to native NifDK (Fig. 4A). This ratio was different from those of the alternative nitrogenases (especially the iron-only nitrogenase), demonstrating that NifQ mostly supported the synthesis of FeMo-co and not that of the analogous cofactor FeFe-co.
Neither purified rNifQ, determined to be molybdenum-free by ICP-OES, nor the molybdoprotein nitrate reductase (NarB) were able to activate the apo-NifDK in parallel assays (Fig. 4A). Purified NarB contained 1 mol of Mo per mol of protein and was fully active in a methyl-viologen-dependent nitrate reductase activity assay (31) (data not shown). The fact that the molybdenum-free NifQ and the molybdoprotein NarB could not activate apo-NifDK suggested that (i) the molybdenum-loaded form of NifQ was specifically required for FeMo-co synthesis and (ii) NarB could not act as a source of molybdenum for FeMo-co synthesis.
The effect of NifQ in apo-NifDK activation was less pronounced when a subsaturating amount of molybdate was additionally included in the reaction mixture and was very small in a reaction additionally containing a saturating amount of molybdate (Fig. 4B). Thus, NifQ could serve as a molybdenum donor as well as molybdate in the in vitro FeMo-co synthesis assay.
Fig. 4C shows the effect of omitting specific protein components from the apo-NifDK activation mixture in reactions lacking molybdate. The reaction containing NifQ showed maximal NifDK activity, whereas the reaction lacking NifQ exhibited 20% of such a maximum. The NifDK activity observed in the reconstituting reaction lacking NifQ has been reported (11) and attributed to the presence of substoichiometric amounts of molybdenum in purified NifEN that are incorporated into FeMo-co. Reactions carried out in the absence of either NifEN, NifH, apo-NifDK, or homocitrate showed very low NifDK activities, indicating their requirement for apo-NifDK activation using NifQ as the molybdenum donor.
Mobilization of Molybdenum from NifQ Requires the Simultaneous Presence of NifH and NifEN Proteins.
The features of the stimulatory effect of NifQ on apo-NifDK activation shown in Fig. 4A demonstrate that the molybdenum carried by NifQ was used for FeMo-co synthesis in vitro. However, the identity of the molybdenum acceptor protein(s) in the reaction remained unclear. To investigate this aspect, as-isolated NifQ samples were incubated under FeMo-co synthesis conditions in the presence of either (i) NifH, (ii) NifEN, (iii) NifH and NifEN, or (iv) NifH, NifEN, and apo-NifDK. After incubation, the protein components of each reaction mixture were reisolated by chromatography (Fig. S2) and their molybdenum contents were determined by ICP-OES. The result of this analysis is shown in Table 1.
Table 1.
Mobilization of molybdenum from NifQ under conditions of FeMo-co synthesis
| Protein | Molybdenum, mol |
|---|---|
| NifQ | |
| Before incubation (as isolated) | 0.36 ± 0.03 |
| After incubation with NifEN | 0.28 ± 0.02 |
| After incubation with NifH | 0.29 ± 0.01 |
| After incubation with NifEN and NifH | 0.11 ± 0.01 |
| After incubation with NifEN, NifH and apo-NifDK | 0.15 ± 0.02 |
| NifEN | |
| Before incubation (as isolated) | 0.24 ± 0.03 |
| After incubation with NifQ | 0.19 ± 0.02 |
| After incubation with NifQ and NifH | 0.45 ± 0.06 |
| NifH | |
| Before incubation (as isolated) | 0.009 ± 0.002 |
| After incubation with NifQ | 0.013 ± 0.001 |
| After incubation with NifQ and NifEN | 0.060 ± 0.010 |
Purified as-isolated NifQ was incubated with purified (i) NifH, (ii) NifEN, (iii) NifH and NifEN, or (iv) NifH, NifEN and apo-NifDK in reaction mixtures that also contained Mg-ATP, DTH, and homocitrate. No external source of molybdenum was added to the mixtures. NifQ, NifH, and NifEN proteins were reisolated by chromatography after incubation. Molybdenum content was quantified by ICP-OES and is shown in mol of molybdenum per mol of protein (i.e. NifQ monomer, NifH dimer, or NifEN tetramer). Values are the average of at least two independent determinations ± SD.
The molybdenum content of NifQ was not significantly affected after incubation with NifH or NifEN. Likewise, the molybdenum content of either NifEN or NifH was unaltered by incubation with NifQ. These results suggest a lack of molybdenum exchange between NifQ and NifH or between NifQ and NifEN.
On the other hand, a major alteration in the molybdenum distribution among NifQ, NifH, and NifEN occurred when all three proteins were incubated together under FeMo-co synthesis conditions and then reisolated (Table 1). Upon reisolation, the molybdenum content of NifQ was >3-fold lower, with a concomitant significant increase in the molybdenum content of both NifEN and NifH. The addition of apo-NifDK as a “sink” of in vitro synthesized FeMo-co did not seem to further stimulate mobilization of molybdenum from NifQ toward NifEN/NifH.
In conclusion, significant mobilization of molybdenum from NifQ required simultaneous incubation with NifH and NifEN under FeMo-co synthesis conditions. Donation of molybdenum from NifQ contributed to FeMo-co synthesis, as demonstrated by its ability to activate apo-NifDK when included in the reaction mixtures (Fig. 4 and Table 1).
Discussion
Molybdenum is normally present in the environment as trace amounts of molybdate (<50 nM) or as highly insoluble minerals such as molybdenite (MoS2) or wulfenite (PbMoO4) (32). Consequently, microorganisms have developed different strategies to cope with molybdenum limitation. A model microorganism with which to study molybdenum uptake and metabolism is A. vinelandii, which carries Mo-co-containing enzymes and nitrogenase. A. vinelandii expresses several high-affinity molybdate-transport systems, a Mosto, and additional proteins involved in molybdenum trafficking for the biosynthesis of FeMo-co and Mo-co. One such protein is NifQ, for which genetic evidence has suggested a role in molybdenum processing for nitrogenase.
Alignment of NifQ amino acid sequences from a variety of bacteria shows a conserved C-terminal region including a Cx4Cx2Cx5C motif suggested to be a metal cluster binding site (Fig. S3). Herein, we provide definitive evidence for the presence of metal cluster(s) on NifQ.
The CW-EPR spectrum of as-isolated NifQ resembles a [3Fe–4S]+-containing protein (25), whereas the EPR spectrum of DTH-reduced NifQ resembles model compounds having a [Mo–3Fe–4S]3+ cluster unit (28–30). Two interpretations can be considered to rationalize these observations. First, each NifQ molecule carries two types of [Fe–S] clusters: a [3Fe–4S] and a [Mo–3Fe–4S] cluster. Second, a percentage of the NifQ molecules in a purified preparation contains a [3Fe–4S] cluster (observable in the as-isolated state of NifQ), whereas the rest of the NifQ molecules contain a [Mo–3Fe–4S] cluster (observable in the DTH-reduced state of NifQ). We favor the second interpretation because of the average contents of iron (3.1 mol Fe/mol NifQ) and molybdenum (0.3 mol Mo/mol NifQ) in NifQ preparations and because the [Mo–3Fe–4S] signal in NifQ could be enhanced by incubation with sulfur and molybdenum under reducing conditions, indicating that in vitro cluster reconstitution was taking place. Coordination of a heterometal by a reduced [3Fe–4S]0 cluster to complete a [M–3Fe–4S] cubane has been reported before (33).
In any case, the results presented herein indicate that NifQ is able to coordinate a redox responsive [Mo-3Fe-4S] cluster. Consistently, extended X-ray absorption fine structure (EXAFS) analysis of NifQ, to be reported elsewhere, indicates the presence of a [Mo-3Fe-4S] cluster in this protein (J.A.H. et al., unpublished results). Although a significant percentage (≈70%) of NifQ molecules in a purified preparation carry [3Fe-4S] clusters, incubation of NifQ with molybdate and sulfide under reducing conditions appears to increase [Mo-3Fe-4S] cluster occupancy.
Why is the as-isolated NifQ protein loaded with substoichiometric amounts of molybdenum? There are three nonexclusive possible explanations. First, as-isolated NifQ might represent a snapshot of molybdenum trafficking through NifQ during active FeMo-co synthesis. Second, molybdenum binding to NifQ might be labile under purification conditions. Third, because NifQ is present at very low levels in nitrogenase-derepressed A. vinelandii wild-type cells (data not shown), strain UW300, which overexpresses a his-tagged variant of NifQ, was used in this study. NifQ overexpression in UW300 cells might have overwhelmed the capacity of the cells to load NifQ with molybdenum.
Interestingly, NifQ expressed and purified from E. coli cells (rNifQ) was found to be a molybdenum-free [4Fe–4S]-cluster-containing protein. The CW-EPR spectrum of DTH-reduced rNifQ, recorded at 12 K, showed an axial EPR signal (g1 = g2 = 2.05 and g3 = 1.92) with a line shape typical of [4Fe–4S]+ clusters (Fig. S4). In addition, temperature- and power-dependence profiles of this EPR signal were indicative of the presence of a [4Fe–4S]+ cluster in rNifQ (power dependence shown in Fig. S5). These observations suggest that the loading of molybdenum into NifQ is carried out by a specific mechanism in A. vinelandii (a mechanism that is absent in E. coli) rather than by a process in which NifQ captures free molybdate from the cytoplasm.
The present study shows a clear linkage between NifQ and molybdenum trafficking for nitrogen fixation. NifQ served as direct source of molybdenum for FeMo-co biosynthesis in vitro. The role of NifQ appears to be in substituting sulfur by oxygen ligands to the molybdenum atom by incorporating it into an [Fe–S] cluster frame. This role is consistent with previous genetic evidence showing suppression of a nifQ mutant phenotype by cystine in the growth medium (34). It is not clear why molybdate can substitute for NifQ in the in vitro FeMo-co biosynthesis assay, but it is possible that molybdate reacts in vitro with excess DTH (35), thus generating Mo–S compounds suitable for FeMo-co synthesis.
The requirement for the simultaneous presence of NifH and NifEN in the reaction suggests that a NifEN/NifH complex could be required to unload the molybdenum from NifQ. Neither NifH nor NifEN alone were capable of mobilizing molybdenum from NifQ. These results also suggest specificity in the process of molybdenum transfer rather than leakage of molybdenum from NifQ into the solvent followed by capture of molybdenum by another protein. The ability of NifEN to associate with wild-type NifH can be inferred from the observation that a NifH mutant variant (L127Δ) forms a stable complex with a form of NifEN that is loaded with a FeMo-co precursor (36). Such a FeMo-co precursor, denominated as the VK-cluster, appears to be a molybdenum-free complex [Fe–S] cluster (13, 37).
Determination of the entry point of molybdenum into the pathway for FeMo-co biosynthesis has been elusive. In vitro FeMo-co synthesis reactions using Na2MoO4 as source of molybdenum showed mutual NifH and NifEN dependence in the incorporation of molybdenum into FeMo-co precursors (38, 39). Based on these results, NifH was suggested to be an entry point for molybdenum (38) or even asserted to be a molybdenum/homocitrate insertase (39). On the other hand, the existence of a NifH-independent molybdenum binding site within NifEN was proposed based on the presence of substoichiometric amounts of molybdenum in NifEN purified from a ΔnifH strain (11). Molybdenum within NifEN was shown to be part of a [Mo–Fe–S] cluster different from the previously characterized FeMo-co precursor VK-cluster (12) and thought to represent a transient site for intramolecular molybdenum donation toward the VK-cluster. However, these observations could not rule out a NifH's possible role in enhancing molybdenum binding into this transient site in NifEN.
This study provides strong evidence indicating that molybdenum trafficking for FeMo-co synthesis involves the specific donation of molybdenum from NifQ to NifEN/NifH. Although genetic and biochemical evidence shows that excess molybdate can substitute for NifQ, this protein has an essential role when the levels of molybdenum available for FeMo-co synthesis are low. This result accredits NifQ as a physiological molybdenum donor and suggests that a putative NifEN/NifH complex might be the molybdenum acceptor during FeMo-co biosynthesis. The chemistry underlying this process and specifically whether or not NifQ transfers the entire [Mo–3Fe–4S] cluster remains to be elucidated.
Experimental Procedures
Expression and Purification of NifQ from A. vinelandii.
NifQ was purified from cell-free extracts of A. vinelandii strain UW300 (PnifH::his9-nifQ) by affinity chromatography to a Co2+ resin (Talon resin, Clontech) under anaerobic conditions inside a glove box. Buffers for protein purification and analysis were sparged with purified N2 for 20–30 min. The A. vinelandii cell-free extracts were prepared by osmotic shock according to ref. 40, followed by centrifugation at 30,000 × g for 1 h to remove cell debris. Cell-free extracts were loaded onto a 20-ml Co2+-affinity column equilibrated in buffer A [10 mM sodium phosphate, 1.8 mM potassium phosphate buffer (pH 7.3), 140 mM NaCl, 2.7 mM KCl, 10% glycerol]. The column was washed with 200 ml of buffer B [50 mM Tris-HCl buffer (pH 7.9), 500 mM NaCl, 25 mM imidazole], and the NifQ protein was eluted from the column by applying 40 ml of buffer C [50 mM Tris-HCl buffer (pH 7.9), 150 mM NaCl, 300 mM imidazole]. Eluted NifQ was concentrated by ultrafiltration through a YM10 membrane in an Amicon cell under a N2 atmosphere and then subjected to a cycle of dilution-concentration in buffer D [50 mM Tris-HCl buffer (pH 7.9), 150 mM NaCl] to remove the residual imidazole. A typical purification procedure yielded 100 mg of NifQ from 340 g of UW300 cell paste. NifQ preparations were estimated to be >95% pure based on SDS/PAGE analysis. The preparations of purified NifQ were frozen as droplets into liquid nitrogen until use.
In Vitro FeMo-co Synthesis Assays with Purified Components.
The in vitro FeMo-co synthesis reactions were performed in 9-ml serum vials sealed with rubber stoppers. All of the vials were acid-washed overnight in 4 M HCl and thoroughly rinsed with Milli-Q water (Millipore) before use. The vials were repeatedly evacuated and flushed with argon gas and finally rinsed with 0.3 ml of anaerobic 25 mM Tris-HCl (pH 7.5), 1 mM DTH. The complete reactions contained 25 mM Tris-HCl (pH 7.5), 0.182 mM homocitrate, 2.92 mM DTH, 3.3% glycerol, an ATP-regenerating mixture (1.32 mM ATP, 18.7 mM phosphocreatine, 2.27 mM MgCl2, 45.4 μg/ml creatine phosphokinase), 150 μg of NifH (2.4 nmol), 100 μg of apo-NifDK (0.43 nmol), and 100 μg of NifEN (0.48 nmol) as a source of FeMo-co precursor (purified NifEN protein carries the VK-cluster). When indicated, Na2MoO4, purified NifQ, rNifQ, or NarB were used as a source of molybdenum in the reaction. The reactions (total volume of 350 μl) were incubated at 30°C for 35 min to allow for the FeMo-co synthesis and insertion reactions. The resulting activation of apo-NifDK was analyzed by the acetylene reduction assay after addition of 0.2 mg of NifH and 0.8 ml of ATP-regenerating mixture (41). Ammonia production by activated apo-NifDK was determined as described (14).
Analysis of Molybdenum Transfer Among NifQ, NifH, and NifEN.
In vitro FeMo-co synthesis mixtures were scaled up ≈20-fold to a volume of 7 ml, keeping the concentration of all chemicals constant. Na2MoO4 was omitted from the reaction mixtures, and all vials were acid-washed and thoroughly rinsed with Milli-Q water to remove molybdenum traces. Two hundred nanomoles of NifQ were mixed with either (i) 112 nmol of NifH, (ii) 9.6 nmol of NifEN, (iii) 112 nmol of NifH plus 9.6 nmol of NifEN, or (iv) 112 nmol of NifH, 9.6 nmol of NifEN, and 8.6 nmol of apo-NifDK. Reactions mixtures were incubated for 35 min at 30°C under conditions of in vitro FeMo-co synthesis, and then the proteins in each mixture were separated by chromatography as follows. NifQ and NifH mixtures were separated by chromatography in Co2+ affinity resin, which binds NifQ but not NifH. NifH was then desalted by gel filtration on a PD-10 column (GE Healthcare). NifQ and NifEN were separated by preparative gel filtration on a HiLoad 16/60 Sephadex 200 PG column (GE Healthcare). NifQ, NifH, and NifEN were separated by sequential chromatography in Co2+ affinity resin and preparative gel filtration on a HiLoad 16/60 Sephadex 200 PG column. Finally, when the mixture contained NifQ, NifH, NifEN, and apo-NifDK, NifQ was separated by affinity chromatography on a Co2+ resin followed by preparative gel filtration on a HiLoad 16/60 Sephadex 200 PG column. In all cases, reisolated proteins were concentrated in Centricon devices (Amicon) and stored as droplets in liquid nitrogen.
To determine the molybdenum content associated with each isolated component, protein samples were heat-denatured and centrifuged, and the supernatant was analyzed by ICP-OES as described in SI Experimental Procedures.
EPR Spectroscopy.
X-band CW-EPR spectra were obtained by using a Bruker ECS106 spectrometer equipped with an ER4116DM dual-mode X-band cavity. Cryogenic temperature was obtained by using an Oxford Instrument ESR900 liquid helium flow cryostat, and temperature was controlled by using an ITC503 temperature controller (Oxford Instruments). ESE-detected EPR spectra were recorded on the Bruker Elexsys using the EN4118x-MD4 probe. ESE-EPR was collected by using the two-pulse sequence π/2–τ–π–τ–echo. The π/2 pulse was set to 16 ns, and τ ranged from 136–144 ns.
Additional methods, including the strains used in this work (Table S2), are detailed in SI Experimental Procedures.
Supplementary Material
Acknowledgments.
We thank Paul Ludden for discussions and support; Dennis Dean for providing strains DJ and DJ1041; Paul Brooks for assistance with ICP-OES analysis; Marcus Strawn for assistance with the high-performance liquid chromatography; and Basem Soboh, Robert Igarashi, Dehua Zhao, and Marie Demuez for helpful discussions. This work was supported by the National Institute of General Medical Sciences and National Institutes of Health Grants GM35332 and GM48242.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11589.
In addition to its presence in the FeMo-cofactor of nitrogenase, biologically active molybdenum can be found constituting molybdopterin cofactors (Mo-co). Molybdoenzymes containing Mo-co are widely distributed in nature and participate in essential redox reactions of C, N, and S metabolisms (42).
NifB-co, the metabolic product of NifB activity, is an isolatable [Fe–S] cluster of unknown structure that serves as precursor to FeMo-co. NifB-co has also been proposed to be a precursor of the cofactors for the alternative nitrogenases, FeV-co and FeFe-co (43).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803576105/DCSupplemental.
The VK-cluster is the [Fe–S] cluster precursor to FeMo-co that accumulates on NifEN in a ΔnifH mutant strain and is thought to represent an intermediate after NifB-co in FeMo-co synthesis (13).
References
- 1.Igarashi RY, Seefeldt LC. Nitrogen fixation: The mechanism of the Mo-dependent nitrogenase. Crit Rev Biochem Mol Biol. 2003;38:351–384. doi: 10.1080/10409230391036766. [DOI] [PubMed] [Google Scholar]
- 2.Raymond J, Siefert JL, Staples CR, Blankenship RE. The natural history of nitrogen fixation. Mol Biol Evol. 2004;21:541–554. doi: 10.1093/molbev/msh047. [DOI] [PubMed] [Google Scholar]
- 3.Bulen WA, LeComte JR. The nitrogenase system from Azotobacter: Two enzyme requirements for N2 reduction, ATP dependent H2 evolution and ATP hydrolysis. Proc Natl Acad Sci USA. 1966;56:979–986. doi: 10.1073/pnas.56.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Rees DC. Crystallographic structure and functional implications of the nitrogenase molybdenum-iron protein from Azotobacter vinelandii. Nature. 1992;360:553–560. doi: 10.1038/360553a0. [DOI] [PubMed] [Google Scholar]
- 5.Georgiadis MM, et al. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 6.Einsle O, et al. Nitrogenase MoFe-protein at 1.16 A resolution: A central ligand in the FeMo-cofactor. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 7.Rubio LM, Ludden PW. Maturation of nitrogenase: A biochemical puzzle. J Bacteriol. 2005;187:405–414. doi: 10.1128/JB.187.2.405-414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dos Santos PC, Dean DR, Hu Y, Ribbe MW. Formation and insertion of the nitrogenase iron-molybdenum cofactor. Chem Rev. 2004;104:1159–1174. doi: 10.1021/cr020608l. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin PJ, et al. The Azotobacter vinelandii NifEN complex contains two identical [4Fe-4S] clusters. Biochemistry. 1998;37:10420–10428. doi: 10.1021/bi980435n. [DOI] [PubMed] [Google Scholar]
- 10.Corbett MC, et al. Structural insights into a protein-bound iron-molybdenum cofactor precursor. Proc Natl Acad Sci USA. 2006;103:1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soboh B, Igarashi RY, Hernandez JA, Rubio LM. Purification of a NifEN protein complex that contains bound Mo and a FeMo-co precursor from an Azotobacter vinelandii ΔnifHDK strain. J Biol Chem. 2006;281:36701–36709. doi: 10.1074/jbc.M606820200. [DOI] [PubMed] [Google Scholar]
- 12.George SJ, et al. Identification of a Mo–Fe–S cluster on NifEN by Mo K-edge extended X-ray absorption fine structure. J Am Chem Soc. 2007;129:3060–3061. doi: 10.1021/ja0663428. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez JA, et al. NifX and NifEN exchange NifB cofactor and the VK-cluster, a newly isolated intermediate of the iron-molybdenum cofactor biosynthetic pathway. Mol Microbiol. 2007;63:177–192. doi: 10.1111/j.1365-2958.2006.05514.x. [DOI] [PubMed] [Google Scholar]
- 14.Curatti L, et al. In vitro synthesis of the iron-molybdenum cofactor of nitrogenase from iron, sulfur, molybdenum and homocitrate using purified proteins. Proc Natl Acad Sci USA. 2007;104:17626–17631. doi: 10.1073/pnas.0703050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoover TR, et al. Identification of the V factor needed for synthesis of the iron-molybdenum cofactor of nitrogenase as homocitrate. Nature. 1987;329:855–857. doi: 10.1038/329855a0. [DOI] [PubMed] [Google Scholar]
- 16.Zheng L, White RH, Dean DR. Purification of the Azotobacter vinelandii nifV-encoded homocitrate synthase. J Bacteriol. 1997;179:5963–5966. doi: 10.1128/jb.179.18.5963-5966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imperial J, Ugalde RA, Shah VK, Brill WJ. Role of the nifQ gene product in the incorporation of molybdenum into nitrogenase in Klebsiella pneumoniae. J Bacteriol. 1984;158:187–194. doi: 10.1128/jb.158.1.187-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno-Vivián C, Hennecke S, Pühler A, Klipp W. Open reading frame 5 (ORF5), encoding a ferredoxin-like protein, and nifQ are cotranscribed with nifE, nifN, nifX, and ORF4 in Rhodobacter capsulatus. J Bacteriol. 1989;171:2591–2598. doi: 10.1128/jb.171.5.2591-2598.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodríguez-Quiñones F, Bosch R, Imperial J. Expression of the nifBfdxNnifOQ region of Azotobacter vinelandii and its role in nitrogenase activity. J Bacteriol. 1993;175:2926–2935. doi: 10.1128/jb.175.10.2926-2935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joerger RD, Bishop PE. Nucleotide sequence and genetic analysis of the nifB-nifQ region from Azotobacter vinelandii. J Bacteriol. 1988;170:1475–1487. doi: 10.1128/jb.170.4.1475-1487.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pienkos PT, Shah VK, Brill WJ. Molybdenum cofactors from molybdoenzymes and in vitro reconstitution of nitrogenase and nitrate reductase. Proc Natl Acad Sci USA. 1977;74:5468–5471. doi: 10.1073/pnas.74.12.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubio LM, Ludden PW. In: Nitrogen Fixation at the Millenium. Leigh GJ, editor. Amsterdam: Elsevier; 2002. pp. 101–136. [Google Scholar]
- 23.Grunden AM, Shanmugam KT. Molybdate transport and regulation in bacteria. Arch Microbiol. 1997;168:345–354. doi: 10.1007/s002030050508. [DOI] [PubMed] [Google Scholar]
- 24.Fenske D, et al. A new type of metalloprotein: The Mo storage protein from Azotobacter vinelandii contains a polynuclear molybdenum-oxide cluster. Chembiochem. 2005;6:405–413. doi: 10.1002/cbic.200400263. [DOI] [PubMed] [Google Scholar]
- 25.Beinert H, Kennedy MC, Stout CD. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem Rev. 1996;96:2335–2374. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 26.Sanakis Y, et al. Evidence for antisymmetric exchange in cuboidal [3Fe–4S]+ clusters. J Am Chem Soc. 2000;122:11855–11863. [Google Scholar]
- 27.Cammack R, et al. Spectroscopic studies of the oxidation-reduction properties of three forms of ferredoxin from Desulphovibrio gigas. Biochim Biophys Acta. 1977;490:311–321. doi: 10.1016/0005-2795(77)90006-x. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong WH, Holm RH. Synthesis and structure of a new type of Mo-Fe–S double-cubane cluster and evidence for formation of magnetically uncoupled S = 3/2 MoFe3S4 sub-clusters. J Am Chem Soc. 1981;103:6246–6248. [Google Scholar]
- 29.Lee SC, Holm RH. The clusters of nitrogenase: Synthetic methodology in the construction of weak-field clusters. Chem Rev. 2004;104:1135–1158. doi: 10.1021/cr0206216. [DOI] [PubMed] [Google Scholar]
- 30.Mascharak PK, et al. Electronic properties of single-MoFe3S4 and double-MoFe3S4 cubane-type clusters. Inorg Chem. 1983;22:2851–2858. [Google Scholar]
- 31.Herrero A, Flores E, Guerrero MG. Regulation of nitrate reductase cellular levels in the cyanobacteria Anabaena variabilis and Synechocystis sp. FEMS Microbiol Lett. 1985;26:21–25. [Google Scholar]
- 32.Shah VK, Ugalde RA, Imperial J, Brill WJ. Molybdenum in nitrogenase. Annu Rev Biochem. 1984;53:231–257. doi: 10.1146/annurev.bi.53.070184.001311. [DOI] [PubMed] [Google Scholar]
- 33.Butt JN, et al. Investigation of metal-ion uptake reactivities of [3Fe–4S] clusters in proteins — Voltammetry of coadsorbed ferredoxin aminocyclitol films at graphite-electrodes and spectroscopic identification of transformed clusters. J Am Chem Soc. 1991;113:6663–6670. [Google Scholar]
- 34.Ugalde RA, Imperial J, Shah VK, Brill WJ. Biosynthesis of the iron-molybdenum cofactor and the molybdenum cofactor in Klebsiella pneumoniae: Effect of sulfur source. J Bacteriol. 1985;164:1081–1087. doi: 10.1128/jb.164.3.1081-1087.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pramanik P, Bhattacharya S. Deposition of molybdenum chalcogenide thin-films by the chemical-deposition technique and the effect of bath parameters on these thin-films. Mat Res Bull. 1990;25:15–23. [Google Scholar]
- 36.Rangaraj P, et al. Inhibition of iron-molybdenum cofactor biosynthesis by L127Delta NifH and evidence for a complex formation between L127Delta NifH and NifNE. J Biol Chem. 1999;274:29413–29419. doi: 10.1074/jbc.274.41.29413. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Fay AW, Ribbe MW. Identification of a nitrogenase FeMo cofactor precursor on NifEN complex. Proc Natl Acad Sci USA. 2005;102:3236–3241. doi: 10.1073/pnas.0409201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangaraj P, Ludden PW. Accumulation of 99Mo-containing iron-molybdenum cofactor precursors of nitrogenase on NifNE, NifH, and NifX of Azotobacter vinelandii. J Biol Chem. 2002;277:40106–40111. doi: 10.1074/jbc.M204581200. [DOI] [PubMed] [Google Scholar]
- 39.Hu YL, et al. Nitrogenase Fe protein: A molybdate/homocitrate insertase. Proc Natl Acad Sci USA. 2006;103:17125–17130. doi: 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah VK, Davis LC, Brill WJ. Nitrogenase. I. Repression and derepression of the iron-molybdenum and iron proteins of nitrogenase in Azotobacter vinelandii. Biochim Biophys Acta. 1972;256:498–511. doi: 10.1016/0005-2728(72)90078-3. [DOI] [PubMed] [Google Scholar]
- 41.Shah VK, Brill WJ. Nitrogenase. IV. Simple method of purification to homogeneity of nitrogenase components from Azotobacter vinelandii. Biochim Biophys Acta. 1973;305:445–454. doi: 10.1016/0005-2728(73)90190-4. [DOI] [PubMed] [Google Scholar]
- 42.Mendel RR, Bittner F. Cell biology of molybdenum. Biochim Biophys Acta. 2006;1763:621–635. doi: 10.1016/j.bbamcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Bishop PE, Joerger RD. Genetics and molecular biology of alternative nitrogen fixation systems. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:109–125. [Google Scholar]
- 44.Rubio LM, Singer SW, Ludden PW. Purification and characterization of NafY (apodinitrogenase gamma subunit) from Azotobacter vinelandii. J Biol Chem. 2004;279:19739–19746. doi: 10.1074/jbc.M400965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.