Abstract
TRIM RING finger proteins have been shown to play an important role in cancerogenesis, in the pathogenesis of some human hereditary disorders, and in the defense against viral infection, but the function of the majority of TRIM proteins remains unknown. Here, we show that TRIM RING finger protein TRIM2, highly expressed in the nervous system, is an UbcH5a-dependent ubiquitin ligase. We further demonstrate that TRIM2 binds to neurofilament light subunit (NF-L) and regulates NF-L ubiquitination. Additionally, we show that mice deficient in TRIM2 have increased NF-L level in axons and NF-L-filled axonal swellings in cerebellum, retina, spinal cord, and cerebral cortex. The axonopathy is followed by progressive neurodegeneration accompanied by juvenile-onset tremor and ataxia. Our results demonstrate that TRIM2 is an ubiquitin ligase and point to a mechanism regulating NF-L metabolism through an ubiquitination pathway that, if deregulated, triggers neurodegeneration.
Keywords: RING finger protein, axonopathy, ataxia, ubiquitination
Ubiquitination of a protein can influence its stability, interactions, activity, or intracellular localization. Three main enzyme families are involved in ubiquitination: ubiquitin activating enzymes, ubiquitin conjugating enzymes, and ubiquitin ligases. Correct localization, timing and specificity of the ubiquitination reaction are ensured mainly by ubiquitin ligases (E3s). RING finger E3s are the most abundant E3 class, characterized by the presence of the cysteine-rich RING finger domain (1). Tripartite (TRIM) RING finger proteins have been defined based on their conserved modular structure (RING finger, B-box, coiled-coil domains) as a subgroup of the RING finger proteins (2). Despite their well conserved modular structure, no common biological role has yet been discovered for TRIM proteins. Recently, some members of the TRIM family have been identified as ubiquitin ligases, involved in cancerogenesis and the defense against viral infection (3, 4). Several human diseases have been linked to mutant RING finger and TRIM E3s. Mutations in the RING finger protein parkin have been shown to trigger a juvenile form of Parkinson's disease (PD) (5). A mutant TRIM37 has been found to cause mulibrey nanism in human (6) and translocation of the TRIM gene pml has been identified in patients suffering from acute promyelocytic leukemia (3). The function of the most of TRIM proteins has not yet been discovered.
TRIM2, highly expressed in the nervous system, has been linked to neuronal activity because its expression in hippocampus correlates with the activity of NMDA receptor (7). In addition, it has been shown to interact with the unconventional motor protein myosin V (7). In the present study, we demonstrate that TRIM2 is an ubiquitin ligase with its activity confined to the RING finger domain. In addition, we show that TRIM2 interacts with the neurofilament light subunit (NF-L) and that ubiquitination of NF-L significantly increases after expression of the full-length TRIM2, but not TRIM2 ligase dead mutant. To examine the function of TRIM2 in vivo, we generate a mouse line carrying a gene trap vector insertion within the Trim2 gene (Trim2GT mice). We analyze TRIM2 expression in the developing and adult nervous system and demonstrate that mice deficient in TRIM2 have increased NF-L levels in axons and show juvenile-onset ataxia. Moreover, Trim2GT mice have swollen axons in several brain areas, including the cerebellum, retina, and spinal cord. This axonopathy is characterized by disorganized intermediate filaments and accumulation of NF-L in axons and is followed by a progressive neurodegeneration. Taken together, our results introduce TRIM2 as a ubiquitin ligase that binds to and regulates NF-L metabolism by ubiquitination.
Results
Generation and Characterization of the Trim2GT Mouse Gene Trap Line.
To characterize the function of TRIM2 in vivo, we took advantage of a large-scale mouse gene trap (GT) screen performed in our laboratory (8). In the screen, mutant embryonic stem (ES) cell lines were generated that carried a randomly integrated promoterless GT vector containing a 5′ splice acceptor site (allowing a vector to be spliced out as a novel exon) and 3′ polyadenylation signal (terminating transcription of the targeted allele) (Fig. 1A). Genes interrupted by the GT vector insertion were identified by a 5′ RACE PCR (supporting information (SI) Fig. S1), and selected ES cell lines were used for generation of mutant mouse lines.
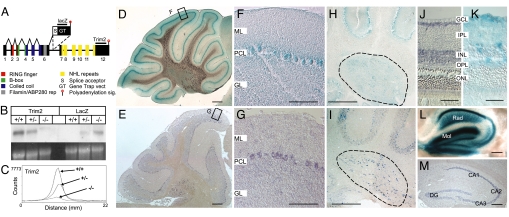
Fig. 1.
Generation and characterization of TRIM2GT mice. (A) Integration site of the GT vector in the Trim2GT mouse line within the Trim2 locus, inside intron 6. (B) Expression of Trim2 in WT (+/+), Trim2GT heterozygous (+/−), and homozygous (−/−) mice analyzed by Northern blotting with Trim2 3′ UTR probe (Trim2) and a GT vector-specific probe (LacZ) (loading control, ethydium bromide-stained gel). (C) Quantification of Trim2 expression from B by instant imager. (D–M) Trim2 expression in cerebellar Purkinje cells (D–G), deep cerebellar nuclei (H and I, encircled), retina (J and K), and hippocampus (L and M), by β-gal staining of Trim2GT heterozygous mice (D, F, H, K, L) and by in situ hybridization using Trim2 probe (E, G, I, J, M). M, molecular layer; PCL, Purkinje cell layer; GL, granule cell layer; GCL, ganglionic cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; Rad, stratum radiatum; Mol, stratum moleculare; DG, dentate gyrus; CA1–3 hippocampal areas (Scale bars: 200 μm in D and E; 100 μm in F–I, L, and M; and 30 μm in J and K.)
For our analysis, we selected an ES cell line with GT vector insertion site within the Trim2 coding sequence as indicated by 5′ RACE PCR sequence (Fig. S1 A and B). By comparison of the 5′ RACE PCR product with the mouse Trim2 cDNA and with the genomic sequence, we determined that the GT vector integrated inside the Trim2 locus between exons 6 and 7 (Fig. 1A). The insertion site was verified by PCR amplification of the genomic DNA between exon 6 of Trim2 and the 5′ part of the GT vector (Fig. S1C) that exactly matched the sequence of intron 6 of Trim2 gene. The mutant locus generated a predicted 7.0-kb transcript containing the initial 1,719 bp of Trim2 fused to the RNA transcript of the gene trap vector (5.3 kb), which was terminated by the vector's polyA signal. Northern blot analysis using GT vector specific (lacZ) and WT 3′ UTR-specific probes (Trim2) and quantified by instant imager, revealed a residual of ≈5% of the WT transcript (≈7.2 kb) in the homozygous mutants (Fig. 1 B and C), probably due to partial elimination of the GT vector by alternative splicing. We designated this insertional mutation Trim2GT.
TRIM2 Expression in the Mouse Nervous System.
To characterize the expression pattern of TRIM2, we used β-galactosidase staining of the GT vector reporter gene (β-gal) driven by the promoter of Trim2, and nonradioactive in situ hybridization (ISH) using a Trim2-specific probe. We detected high Trim2 expression in the cerebellum, hippocampus, retina, and spinal cord. In the adult cerebellum, the strongest expression was in Purkinje cells and in the deep cerebellar nuclei (Fig. 1). In retina, we detected high expression of Trim2 in the ganglionic cell layer, inner nuclear layer and in the outer plexiform layer by β-gal staining (Fig. 1). We found particularly high expression level of Trim2 in the adult hippocampus: in pyramidal cells of CA1-CA3 hippocampal areas and in granule cells of the dentate gyrus (Fig. 1). Intense β-gal staining found in stratum radiatum of the hippocampus proper and in the molecular layer of the dentate gyrus corresponds to the dendritic field of pyramidal and granule neurons respectively (Fig. 1L). Using β-gal staining, we detected TRIM2 expression also in the developing nervous system, particularly in the spinal cord, dorsal root ganglia, hindbrain, and midbrain (Fig. S2).
Trim2GT Mice Develop an Early-Onset Neurodegeneration.
Until ≈1.5 months of age, the Trim2GT homozygotes were indistinguishable from the WT and heterozygous littermates. At this age, the homozygous mice started to show intention tremor, followed by gait ataxia (Movie S1). In later stages, the mutant mice suffered from episodes of spontaneous generalized seizures.
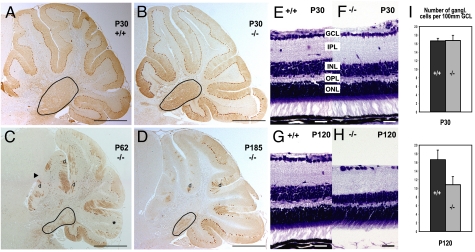
Because Trim2 is expressed in cerebellum (Fig. 1), tremor and ataxia were indicative of a cerebellar-related phenotype. We therefore analyzed cerebella of homozygous mice at several time intervals. In 1-month-old animals, we did not detect significant difference in cerebellar anatomy or number of Purkinje cells between homozygous mice and their WT littermates by calbindin D-28K (Purkinje cell marker) immunostaining (602.0 ± 63.1, n = 3; 607.8 ± 85.6, n = 3 respectively, midsagittal sections of the vermis) (Fig. 2 A and B). However, starting at approximately postnatal day 50 (P50), the homozygous mice developed a substantial (73%; 224.2 ± 25.7, n = 3) and progressive loss of Purkinje cells, particularly marked in the anterior and posterior lobes of the vermis (Fig. 2 C and D). In 5-month-old Trim2GT mice, we observed 85% (125.4 ± 20.6, n = 3) decrease in Purkinje cells as compared with WT mice (839.0 ± 31.1, n = 4). The degeneration of Purkinje cells manifested by the loss of calbindin D-28K immunoreactivity (Fig. 2 A–D) and inositol-3-phosphate receptor (data not shown), was further supported by hematoxylin/eosin staining showing a deficit in Purkinje cell somata (Fig. S3). Several caspase-3 immunoreactive Purkinje cells were detected in 2-month-old, but not 1-month-old, homozygotes (Fig. S4), indicating that the death of the Purkinje cells is apoptotic. The deep cerebellar nuclei showed a similar progressive neurodegeneration in the mutant animals (Fig. 2 B–D and Fig. S5).
Fig. 2.
Degeneration in Trim2GT homozygous mice. (A–D) Calbindin D-28K immunostaining of WT (A) and homozygous (B) cerebellar Purkinje cells at postnatal day P30. Degeneration of vermal Purkinje cells and the deep nuclei (encircled) detected in 2- (C) and 6-month-old (D) mutants (anterior lobes, arrowhead; posterior lobe, asterisk; Purkinje cell dendrites, d). (E–H) Nissl staining of retina in WT and Trim2GT homozygous mice at P30 (E and F) and P120 (G and H). Inner nuclear layer, INL; outer plexiform layer, OPL. (I) Quantification of the number of ganglionic cells per 100 μm of the ganglionic cell layer (GCL) in 1- and 4-month-old mice (n = 3). (Scale bars: 700 μm in A–D, 30 μm in E–H.)
Similar to the cerebellum, retinas of 1-month-old homozygous mice did not show any detectable histological alterations (Fig. 2 E and F). However, at 4 months of age, retinas in homozygous mice displayed decreased thickness of the inner nuclear layer and a reduced number of ganglionic cells (Fig. 2 G and H). The average number of ganglionic cells per 100 μm of the ganglionic cell layer was reduced by one-third, from 16.4 to 10.8 in 4-month-old homozygotes (Fig. 2I). Consistently, the outer plexiform layer (formed by axodendritic synapses between INL and photoreceptors) was also reduced (Fig. 2H), whereas the size of the photoreceptor layer was not altered in the mutants.
RING Finger Protein TRIM2 Is a Ubiquitin Ligase.
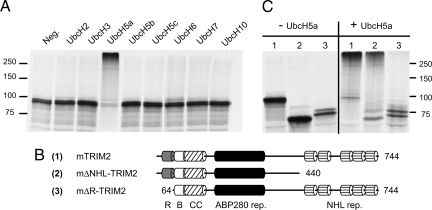
To get an insight into how TRIM2 deficiency causes neurodegeneration in our animal model, we first analyzed the function of TRIM2 in vitro. A common feature of many RING finger-type ubiquitin ligases is their ability to autoubiquitinate (9); thus, we decided to test ubiquitination activity of TRIM2. We synthesized TRIM2 by using the rabbit reticulocyte lysate TNT reaction in the presence of [35S]methionin and incubated the 35S-labeled TRIM2 with ubiquitin, proteasome inhibitors, rabbit E1, and a set of E2s (UbcH2, UbcH3, UbcH5a, UbcH5b, UbcH5c, UbcH6, UbcH7, UbcH10) in separate reactions. In the presence of UbcH5a, but no other tested E2s, TRIM2 became polyubiquitinated (Fig. 3A) indicating that it is able to function as an ubiquitin ligase in vitro by using UbcH5a as ubiquitin-conjugating enzyme (E2).
Fig. 3.
Ubiquitin ligase activity of TRIM2. (A) [35S]Met-labeled TRIM2 autoubiquitination in an in vitro ubiquitination assay in the presence of various ubiquitin-conjugating enzymes (E2). (B) Schematic representation of TRIM2 full-length and mutant constructs. (R, RING finger; B, B box; CC, coiled-coil). (C) In vitro autoubiquitination of full-length and mutant [35S]Met TRIM2 in the absence (−) or presence (+) of UbcH5a.
TRIM2 contains the N-terminal tripartite motif domain (RING finger and B-box domains, followed by an α-helical coiled-coil domain), filamin/ABP280 domain, and five NHL repeats at the C terminus (2, 7) (Fig. 3B). To test whether the RING finger domain is responsible for the ubiquitination activity of TRIM2, we generated TRIM2 mutants lacking the C-terminal NHL repeats or the N-terminal RING finger domain (Fig. 3B). We could detect the autoubiquitination activity of the full-length TRIM2 and in the TRIM2 with NHL deletion (Fig. 3C). In contrast, TRIM2 lacking the RING finger domain was not able to catalyze its autoubiquitination (Fig. 3C), which indicates that the RING finger domain is necessary for TRIM2 ubiquitination activity.
TRIM2 Interacts with Neurofilament Light Chain.
TRIM2 has been shown to interact with the molecular motor protein myosin V (7) responsible for transport of smooth endoplasmic reticulum (SR) into dendritic spines of Purkinje cells (10). By analyzing the ultrastructure of Purkinje cell dendritic spines of the Trim2GT mutants, we could clearly detect SR in the spines (Fig. S6), indicating that interaction between TRIM2 and myosin V is not necessary for transport of SR. Additionally, we did not see any changes in myosin V level in Neuro2a cell line after Trim2 transfection (data not shown) or any difference in the intensity or distribution of myosin V immunoreactivity in Trim2GT homozygous mice (Fig. S6), suggesting that myosin V is not a substrate for TRIM2-catalyzed ubiquitination.
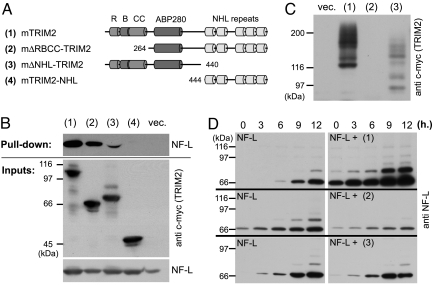
Interestingly, TRIM32, structurally related to TRIM2 (11), has been shown to bind skeletal myosin but to ubiquitinate actin (12). The binding partner of TRIM2, myosin V, has been shown to bind neurofilament light chain (NF-L) and through this interaction to modulate neurofilament organization in axons (13). Because changes in NF-L metabolism have been shown to trigger neurodegeneration in mice (14), we tested whether TRIM2 could interact with NF-L and affect its metabolism. Using TRIM2 expressed in neuro2a cell line as a bait, we were able to pull down endogenous NF-L from mouse brain lysate, suggesting that TRIM2 and NF-L can, indeed, interact (Fig. S7A). To analyze the interaction in more detail, we expressed NF-L together with full-length or truncated forms of TRIM2 in HeLa cells and tested their binding. In these experiments, full-length TRIM2 and TRIM2 with deletion of RBCC domain could pull down NF-L (Fig. 4B), but deletion of the NHL repeats reduced the binding, and deletion of both the central region and RBCC domains abolished the NF-L–TRIM2 interaction (Fig. 4 A and B).
Fig. 4.
TRIM2–NF-L interaction and ubiquitination. (A) Schematic representation of TRIM2 full-length and mutant constructs. (B) NF-L pull-down assay with the distinct TRIM2 constructs in HeLa cells. (C) Autoubiquitination of TRIM2 and its truncated forms in HeLa cells. (D) NF-L ubiquitination time course in HeLa cells—alone (NF-L) and after coexpression of the full-length TRIM2 (NF-L + 1), the ligase-dead mutant (NF-L + 2), or the C-terminally truncated TRIM2 (NF-L + 3).
Regulation of NF-L Ubiquitination by TRIM2.
Given that TRIM2 is an ubiquitin ligase and interacts with NF-L, we analyzed whether NF-L can be ubiquitinated in vitro and whether TRIM2 is involved in the process. By an in vitro ubiquitination assay, we determined that NF-L is ubiquitinated in the presence of UbcH5a E2 (Fig. S7B). To test the role of TRIM2 in the NF-L ubiquitination, we established an “in vivo” ubiquitination assay. We transfected HeLa cells with NF-L, His-tagged ubiquitin, and either full-length c-myc-tagged TRIM2 or TRIM2 lacking NHL repeats or RBCC domain. After 1 day in culture, we stopped protein degradation by adding MG132 proteasomal inhibitor for 12 h, lysed the cells, and pulled down all of the His-tag-ubiquitinated proteins with Ni+ beads. By subsequent NF-L or TRIM2 Western blot analysis, we were thus detecting only their ubiquitinated forms pulled down with Ni+-beads. TRIM2 showed the same self-ubiquitination activity in HeLa cells (Fig. 4C) as observed in the in vitro ubiquitination assay (Fig. 3C). Similar to the in vitro system (Fig. S7B), we detected a basal level of ubiquitinated NF-L after its transfection into the HeLa cells (Fig. 4D), however, coexpression of TRIM2 led to a significant increase of the NF-L ubiquitination (P(Mann–Whitney) ≤ 0.0078; n = 6; Median = 1.22) (Fig. 4D), whereas coexpression of the ligase-dead mutant did not have a significant effect on NF-L ubiquitination (P(Mann-Whitnay) ≤ 0.21, n = 6; Median = 0.82). Overall, the NF-L ubiquitination level was 2.48 times higher (SE = 0.58, n = 6) in the samples transfected with TRIM2 than in the samples transfected with TRIM2 ligase-dead mutant, which often led to reduction of NF-L ubiquitination (Fig. 4D), and is consistent with the function of a dominant-negative construct. We have seen similar reduction of NF-L ubiquitination using C-terminally truncated TRIM2 (Fig. 4D). Taken together these results demonstrate that manipulating the level of TRIM2 affects NF-L ubiquitination.
Neurodegeneration in Trim2GT Mutant Mice Is Accompanied by Axonal Swelling and Accumulation of NF-L.
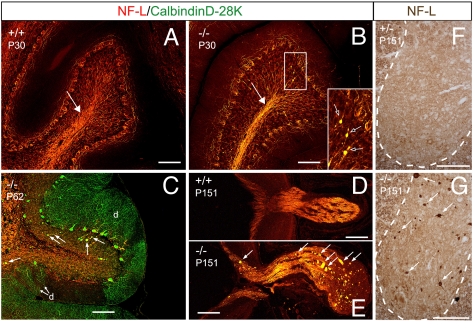
To test whether TRIM2 deficiency affects NF-L metabolism in Trim2GT mice, we analyzed the distribution of NF-L in the mutant brains before and after the onset of the neurodegeneration. We found increased NF-L immunoreactivity and the presence of sporadic NF-L aggregates in the cerebellar white matter and granule cell layer of 1-month-old homozygotes (Fig. 5B), i.e., before the onset of neurodegeneration. The NF-L aggregates colocalized with a Purkinje cell-specific marker, calbindin D-28K, indicating that the NF-L accumulated in swollen axons of Purkinje cells (Fig. 5C). In addition we observed NF-L-rich axonal swellings in the optic nerve (Fig. 5 D and E), brainstem, spinal cord (Fig. 5 F and G) and frontal cortex (Fig. S8) of Trim2GT mutant mice.
Fig. 5.
Increased NF-L axonal density and NF-L axonal swellings in Trim2GT homozygous mice. (A) WT littermate of B. (B) Trim2GT homozygotes show increased NF-L level at P30 in cerebellar white matter (arrow) and pathological accumulation of NF-L (arrows in the Inset). (C) NF-L-filled axonal swellings colocalize with calbindin D-28K staining of Purkinje cell axons (arrows); d, remaining Purkinje cell dendrites. (E–G) NF-L-filled axonal spheroids (arrows) are also present in the optic nerve (E) and the gray matter of spinal cord (G) of the Trim2GT homozygotes, but not in their WT or heterozygous (D and F) littermates. (Scale bars: 100 μm.)
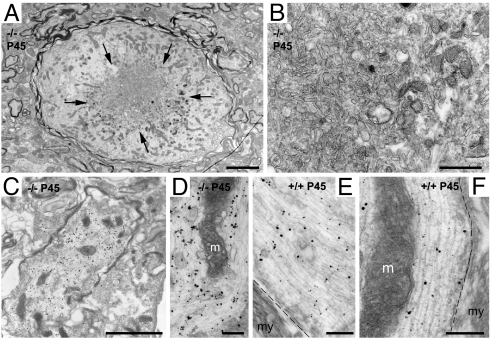
Transmission electron microscopy performed in the cerebellum of 1.5-month-old mice confirmed the presence of axonal swellings, which consisted of the accumulation of disorganized neurofilaments and microtubules, mitochondria, and vesicles (Fig. 6). Some swollen axons showed signs of degeneration, which varied from few degenerating mitochondria to numerous dense and multivesicular bodies accumulated in the center of the swelling (Fig. 6). Preembedding immunogold/silver labeling confirmed the presence of accumulated NF-L in the axonal dilation (Fig. 6).
Fig. 6.
Ultrastructure and NF-L immunoelectronmicroscopy of dilated axons in the deep cerebellar nuclei of Trim2GT mice. (A–D) Swollen axons dilated by the accumulation of neurofilaments, microtubuli, mitochondria, and vesicles. (A) Dilated axon with clear signs of degeneration, with the accumulation of dense and multivesicular bodies (arrows) tightly packed (B). In A, numerous regular-size axons can be observed nearby. (C–F) Preembedding immunoelectronmicroscopy using NF-L antibody. Immunometal particles are associated to the disorganized neurofilaments in the dilated axons (C and D) or to the properly oriented parallel neurofilaments in normal axons (E and F). m, mitochondria; my, myelin; dashed line, separation between the axoplasm and the ensheathing myelin. (Scale bars: 2.5 μm in A; μm in C; 500 nm in B; 250 nm in D and E; and 200 nm in F.)
Discussion
In the present study, we demonstrate that TRIM2 is a UbcH5a-dependent ubiquitin ligase that interacts with NF-L through its central region. These results suggest that TRIM2 might be responsible for NF-L ubiquitination. In agreement with this hypothesis, we demonstrate that expression of full-length TRIM2 (but not of the ligase-dead mutant) significantly increases NF-L ubiquitination. Moreover, expression of the ligase-dead mutant TRIM2 shows dominant-negative effect on NF-L ubiquitination. We further demonstrate that TRIM2-deficient mice exhibit juvenile-onset tremor and ataxia and abnormal accumulation of NF-L in the axons of some of the neurons most prominently expressing TRIM2. These pathological changes are followed by progressive neurodegeneration. Together, our results indicate that TRIM2 is an ubiquitin ligase that binds to and ubiquitinates NF-L and that TRIM2 deficiency leads to neurodegeneration in mice likely by altering NF-L metabolism with consequent NF-L accumulation in axons and impairment of axonal transport.
NF-L as a Substrate of TRIM2.
TRIM proteins are often found as parts of multiprotein complexes ubiquitinating one or several of their binding partners (12, 15, 16). TRIM32, a member of the same TRIM subfamily as TRIM2 (11), has been shown to bind skeletal myosin but to ubiquitinate actin. Similarly, TRIM2 has been shown to bind myosin V, but we could not detect its effect on myosin V. In contrast, expression of TRIM2 (but not its ligase-dead mutant) was in our ubiquitination assays consistently increasing NF-L ubiquitination. Moreover, even though the effect of the mutant construct was more variable, transfection of the ligase-dead mutant repeatedly showed a dominant-negative effect on NF-L ubiquitination. Additional evidence corroborating the view that NF-L is a substrate for TRIM2-driven ubiquitination comes from our in vivo data. We have demonstrated that TRIM2 deficiency leads in 1-month-old homozygotes to higher level of NF-L in axons and NF-L-filled axonal swellings (Fig. 5), which are largely devoid of ubiquitin immunoreactivity (Fig. S9), indicating that NF-L in the swollen axons is not ubiquitinated. Ubiquitin immunoreactivity found in a small number of swollen axons can be attributed to the hypomorphic phenotype of TRIM2GT mice in which 5% of TRIM2 is still present in the homozygous mice.
Our in vitro ubiquitination assays demonstrated that UbcH5a was functioning as E2 for both NF-L ubiquitination and TRIM2 autoubiquitination, and that particularly TRIM2 autoubiquitination was very efficient in the presence of UbcH5a (Fig. 3). These results imply that both lowering and increasing the UbcH5a level can have an adverse affect on NF-L ubiquitination—high UbcH5a level can lead to effective autoubiquitination of TRIM2 (therefore lowering NF-L ubiquitination), and low UbcH5a level can directly interfere with NF-L ubiquitination (Fig. S7B). To test the effect of different UbcH5a levels on NF-L ubiquitination, we have performed NF-L ubiquitination assays in HeLa cells with either endogenous level of UbcH5a or after overexpression of its WT or dominant-negative form (Fig. S7C). Overexpression of both constructs led to a slight reduction of NF-L ubiquitination, supporting our previous data and indicating that the level of UbcH5a is yet another factor regulating NF-L ubiquitination. Nevertheless, our data do not rule out the possibility that other E2s can substitute UbcH5a in NF-L ubiquitination in vivo.
NF-L Inclusions and TRIM2 Expression Pattern.
In TRIM2GT mutant mice, NF-L inclusions were found in neurons that normally express high levels of TRIM2. This applies to Purkinje cells, deep cerebellar nuclei neurons, and retinal ganglionic cells. In addition, we have detected NF-L accumulation in the cortical neurons, which normally express TRIM2 at a lower level. NF-L inclusions found in the gray matter of the spinal cord can be linked to the high TRIM2 expression we found there during embryonic development (Fig. S2). The spinal neuropathy can also partially contribute to the tremor and ataxia of the TRIM2GT homozygotes, because ataxia and tremor have been described also in some motor and peripheral neuropathies affecting spinal cord (17). Despite the high expression level of Trim2 in principal hippocampal neurons, we could neither detect axonal NF-L inclusions nor neurodegeneration in hippocampus. Although the actual cause of such a different outcome of TRIM2 deletion in hippocampus eludes us at present, it is possible that NF-L ubiquitination is differentially regulated in a cell-specific manner. Therefore, the hippocampus may contain compensatory pathways or molecules involved in the metabolism of neurofilaments that are not present in Purkinje or ganglionic cells. Future studies will have to address this divergence in neurofilament processing between different neural cells.
NF-L Axonal Inclusions as a Cause of Neurodegeneration.
Defects in the ubiquitin proteasome system have long been implicated in the pathogenesis of neurodegenerative disorders, but even though toxic protein accumulations have been found in numerous neuropathies, relatively few ubiquitin ligases have so far been directly linked to neurodegeneration. Probably the best analyzed is the RING finger protein parkin. Mutations compromising its ubiquitin ligase activity have been found to cause a juvenile form of PD (5). The exact mechanism how defective parkin triggers dopaminergic neurodegeneration has not been described yet, but because multiple parkin substrates have been shown to accumulate in the brains of parkin-null mice and PD patients carrying parkin mutations (18), it has been speculated that accumulation of the toxic parkin substrates initiates the neurodegeneration.
NF aggregates have been found in several neurodegenerative diseases (19). Although perikaryal NF inclusions seem not to affect neuronal survival (14, 20, 21), axonal NF inclusions, similar to those found in Trim2GT mutants, are reportedly toxic to neurons, likely through strangulation of the normal axonal transport (22, 23). Changes in NF-L metabolism seem to be particularly harmful to neurons because its overexpression leads to appearance of axonal neurofilament inclusions, neurodegeneration, and motor dysfunction (14, 23). Mutations in the NF-L gene have been reported in patients suffering from Charcot–Marie–Tooth disease type 2 (24), further indicating that alteration in NF-L metabolism is the etiopathogenic cause rather than the consequence of neurodegeneration. In our Trim2GT mutants the appearance of NF-L aggregates preceded the wave of neurodegeneration. Moreover, at the ultrastructural level, the accumulation of neurofilaments and organelles in the axoplasm of the Trim2GT mutants was suggestive of an impaired axonal transport and a functional axotomy. Thus, our data support the causal link between NF-L accumulation and neurodegeneration.
Despite the efforts devoted to the study of neurodegenerative disorders in recent decades, the cascades of events underlying the formation and neurotoxicity of NF accumulations are not fully understood. Our analysis of TRIM2 protein and Trim2GT mutant mice uncover an important player in NF-related neurodegeneration and points at the ubiquitination cascade as an important regulator of NF-L metabolism. Further dissection of the mechanisms that control neurofilament accumulation will lead to better understanding of processes underlying neurodegeneration.
Materials and Methods
ES Cells and Mice.
The Trim2GT ES cell line, with IRESβgeo containing the GT vector inserted within Trim2 gene, was generated as described (25). The 5′RACE (GIBCO–BRL) on ES cells RNA amplified a 370-bp fragment (upstream of the insertional splice acceptor site) that exactly matched the mouse Trim2 sequence (7). The animals were genotyped by Southern blot analysis and PCR (details in SI Text). The Southern blot analysis was performed by using LacZ and Trim2 probes. The LacZ probe consisted of an AvaI fragment of pCH110 plasmid (Amersham Pharmacia). The Trim2 probe was made of a 1.1-kb EcoRI-HindIII fragment of Trim2 3′ untranslated region.
Ubiquitination Assays.
The in vitro ubiquitination was performed as described (26). Briefly, TRIM2 or NF-L was synthesized in vitro in the TNT T7 reticulocyte lysate (Promega) in the presence of [35S]Met. [35S]TRIM2 or [35S]NFL (3 μl) was incubated with 500 ng/μl E1 (Boston Biochem) and 500 ng/μl of different E2s (Boston Biochem) in the reaction buffer [50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 0.5 mM DTT, 100 mM ATP, 25 mg/ml ubiquitin (Boston Biochem), 5 mM MG-132 (Boston Biochem), 20 μM ubiquitin aldehyde (Boston Biochem)] in a total volume of 10 μl at 30°C for 20 min (TRIM2) or 90 min (NF-L).
The in vivo ubiquitination assay was performed in HeLa-tTA cells. The cells were transfected with His-tagged ubiquitin, NF-L, mTRIM2 (or mΔRBCC-TRIM2), and after 1 day in culture, 20 μM MG-132 was added to the media. The cells were lysed in lysis buffer [6 M urea; 40 mM Tris·HCl (pH 7.4), 20 mM imidazole, 0.5% Triton X-100] and subjected to the pull-down analysis with nickel–agarose beads to isolate His-tagged ubiquitin conjugates, followed by Western blot analysis with anti-NF-L (NR4; Sigma) and anti-c-myc (9E10; Santa Cruz Biotechnology) antibodies.
Histology and Immunocytochemistry.
Mice were intracardially perfused with ice-cold fixative solution [0.1 M phosphate buffer (pH 7.4), 4% paraformaldehyde (PFA), 0.2% picric acid), embedded in paraplast or cryomatrix, and cut sagittally 10 μm or 15 μm. β-Galactosidase staining was performed as described (27), on vibratome-cut brain sections (100 μm) for 24 h at 30°C. The sections were reembedded in paraplast and cut with microtome (10 μm). For immunostaining, the antibodies were used in the following dilutions: anti-calbindin D-28K (CB-955; Sigma) 1:50, anti-NF-L (NR4; Sigma) 1:300, anti-calbindin D-28K polyclonal (Sigma) 1:300, anti-cleaved caspase3 (Cell Signaling Technology) 1:200, anti-myosin V polyclonal [provided by P. Bridgman, Washington University in St. Louis, St. Louis, MO (28)] 1:500. The antigens were visualized with ABC universal kit (Vector) or with secondary fluorescent Alexa Fluor 488/Alexa Fluor 594-conjugated antibodies (Molecular Probes) 1:300. Fluorescently labeled tissues were counterstained with Hoechst 33258 (0.2 mg/ml).
Nonradioactive in situ hybridization was performed on 15-μm-thick cryostat sections as described (29). (For details see SI Text).
Electron Microscopy.
Male mice (1.5 months of age) were transcardially perfused with saline, followed by 4% paraformaldehyde, ≈0.2% picric acid in 0.1 M phosphate-buffer (PB) (pH 7.2–7.4) with 2% glutaraldehyde for structural electron microscopy (EM) or 0.05% glutaraldehyde for preembedding immunocytochemistry. Brains were removed, rinsed in PB, and sectioned on a vibratome at 70-μm thickness. Sections for structural EM were treated with 2% osmium, contrasted with uranyl acetate (1%), dehydrated, and embedded in Durcupan ACM (Fluka). The preembedding immunocytochemical procedures were identical to those described earlier (30). (For details see SI Text).
Pull-Down Assays.
The cells were transfected with NF-L and c-myc-tagged trim2 (or its truncated forms) and harvested at 24 h after transfection, homogenized in buffer A [50 mM Tris·HCl (pH 7.4), 150 mM NaCl; 0.9% SDS, EDTA-free protease inhibitors, Roche), and diluted 6× with buffer B [50 mM Tris·HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, EDTA-free protease inhibitors, Roche). The lysate was sonicated 2 times for 20 sec. and spun at 16,000 × g for 10 min at 4°C. The anti-c-myc agarose beads (Santa Cruz Biotechnology) were incubated overnight with the lysates, washed six times with buffer B and analyzed by Western blotting. The pulldown of endogenous NF-L was performed in a similar way (see SI Text).
Supplementary Material
Acknowledgments.
We thank Prof. M. Osborn and Drs. M. Kessel, X. Miro, P. Petrou, and M. Alberich Jorda for comments and suggestions; Prof. M. Osborn (Max Planck Institute of Biophysical Chemistry, Goettingen, Germany) for the generous gift of NF-L (NR4) antibodies; and Prof. P. Bridgman for myosin V antibodies. This work was supported by National Institutes of Health Grant AG022082 (to K.P.L.), Amgen, Inc., and the Max Planck-Society (P.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802261105/DCSupplemental.
References
- 1.Freemont PS. RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 2.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de The H, et al. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 4.Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 5.Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 6.Avela K, et al. Gene encoding a new RING-B-box-Coiled-coil protein is mutated in mulibrey nanism. Nat Genet. 2000;25:298–301. doi: 10.1038/77053. [DOI] [PubMed] [Google Scholar]
- 7.Ohkawa N, et al. Molecular cloning and characterization of neural activity-related RING finger protein (NARF): A new member of the RBCC family is a candidate for the partner of myosin V. J Neurochem. 2001;78:75–87. doi: 10.1046/j.1471-4159.2001.00373.x. [DOI] [PubMed] [Google Scholar]
- 8.Stoykova A, et al. Gene trap expression and mutational analysis for genes involved in the development of the mammalian nervous system. Dev Dyn. 1998;212:198–213. doi: 10.1002/(SICI)1097-0177(199806)212:2<198::AID-AJA5>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takagishi Y, et al. The dilute-lethal (dl) gene attacks a Ca2+ store in the dendritic spine of Purkinje cells in mice. Neurosci Lett. 1996;215:169–172. doi: 10.1016/0304-3940(96)12967-0. [DOI] [PubMed] [Google Scholar]
- 11.Short KM, Cox TC. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem. 2006;281:8970–8980. doi: 10.1074/jbc.M512755200. [DOI] [PubMed] [Google Scholar]
- 12.Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354:413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 13.Rao MV, et al. Myosin Va binding to neurofilaments is essential for correct myosin Va distribution and transport and neurofilament density. J Cell Biol. 2002;159:279–290. doi: 10.1083/jcb.200205062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, Cork LC, Griffin JW, Cleveland DW. Increased expression of neurofilament subunit NF-L produces morphological alterations that resemble the pathology of human motor neuron disease. Cell. 1993;73:23–33. doi: 10.1016/0092-8674(93)90157-l. [DOI] [PubMed] [Google Scholar]
- 15.Witt SH, Granzier H, Witt CC, Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: Towards understanding MURF-dependent muscle ubiquitination. J Mol Biol. 2005;350:713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Yan Q, et al. CART: An Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol Biol Cell. 2005;16:2470–2482. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koeppen AH. The pathogenesis of spinocerebellar ataxia. Cerebellum. 2005;4:62–73. doi: 10.1080/14734220510007950. [DOI] [PubMed] [Google Scholar]
- 18.Ko HS, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Chalabi A, Miller CC. Neurofilaments and neurological disease. BioEssays. 2003;25:346–355. doi: 10.1002/bies.10251. [DOI] [PubMed] [Google Scholar]
- 20.Cote F, Collard JF, Julien JP. Progressive neuronopathy in transgenic mice expressing the human neurofilament heavy gene: A mouse model of amyotrophic lateral sclerosis. Cell. 1993;73:35–46. doi: 10.1016/0092-8674(93)90158-m. [DOI] [PubMed] [Google Scholar]
- 21.Wong PC, et al. Increasing neurofilament subunit NF-M expression reduces axonal NF-H, inhibits radial growth, and results in neurofilamentous accumulation in motor neurons. J Cell Biol. 1995;130:1413–1422. doi: 10.1083/jcb.130.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julien JP. Amyotrophic lateral sclerosis. Unfolding the toxicity of the misfolded. Cell. 2001;104:581–591. doi: 10.1016/s0092-8674(01)00244-6. [DOI] [PubMed] [Google Scholar]
- 23.Lee MK, Marszalek JR, Cleveland DW. A mutant neurofilament subunit causes massive, selective motor neuron death: Implications for the pathogenesis of human motor neuron disease. Neuron. 1994;13:975–988. doi: 10.1016/0896-6273(94)90263-1. [DOI] [PubMed] [Google Scholar]
- 24.Mersiyanova IV, et al. A new variant of Charcot–Marie–Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am J Hum Genet. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowdhury K, et al. Evidence for the stochastic integration of gene trap vectors into the mouse germline. Nucleic Acids Res. 1997;25:1531–1536. doi: 10.1093/nar/25.8.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee TH, et al. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J Biol Chem. 2006;281:759–768. doi: 10.1074/jbc.M509855200. [DOI] [PubMed] [Google Scholar]
- 27.Gossler A, Joyner AL, Rossant J, Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- 28.Bridgman PC. Myosin Va movements in normal and dilute-lethal axons provide support for a dual filament motor complex. J Cell Biol. 1999;146:1045–1060. doi: 10.1083/jcb.146.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moorman AF, Houweling AC, de Boer PA, Christoffels VM. Sensitive nonradioactive detection of mRNA in tissue sections: Novel application of the whole-mount in situ hybridization protocol. J Histochem Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- 30.Corti C, Aldegheri L, Somogyi P, Ferraguti F. Distribution and synaptic localisation of the metabotropic glutamate receptor 4 (mGluR4) in the rodent CNS. Neuroscience. 2002;110:403–420. doi: 10.1016/s0306-4522(01)00591-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.