Abstract
The Arp2/3 complex is an essential component of the yeast actin cytoskeleton that localizes to cortical actin patches. We have isolated and characterized a temperature-sensitive mutant of Schizosaccharomyces pombe arp2 that displays a defect in cortical actin patch distribution. The arp2+ gene encodes an essential actin-related protein that colocalizes with actin at the cortical actin patch. Sucrose gradient analysis of the Arp2/3 complex in the arp2-1 mutant indicated that the Arp2p and Arc18p subunits are specifically lost from the complex at restrictive temperature. These results are consistent with immunolocalization studies of the mutant that show that Arp2-1p is diffusely localized in the cytoplasm at restrictive temperature. Interestingly, Arp3p remains localized to the cortical actin patch under the same restrictive conditions, leading to the hypothesis that loss of Arp2p from the actin patch affects patch motility but does not severely compromise its architecture. Analysis of the mutant Arp2 protein demonstrated defects in ATP and Arp3p binding, suggesting a possible model for disruption of the complex.
INTRODUCTION
The involvement of the actin cytoskeleton in a variety of basic cellular processes such as polarized growth, organelle trafficking, cell motility, and cellular division is well known. All of these processes require reorganization and assembly of actin structures in response to cellular and environmental signals during the life of the cell. The numerous and diverse functions of the actin cytoskeleton appear to be accomplished by its interactions with a large number of regulatory and accessory proteins (Ayscough, 1998).
In recent years, a number of actin-related proteins have been identified that appear to share a common ancestor with conventional actin but are divergent from actin in their cellular functions (for reviews, see Frankel and Mooseker, 1996; Mullins et al., 1996; Machesky, 1997). Arp1p has been well characterized and is a component of the dynactin complex required for dynein-based vesicle motility (reviewed by Schroer et al., 1996). Arp2p and Arp3p were first isolated in Saccharomyces cerevisiae and Schizosaccharomyces pombe, respectively (Lees-Miller et al., 1992; Schwob and Martin, 1992), and recognized as components of a larger complex in Acanthamoeba by affinity chromatography on profilin-agarose (Machesky et al., 1994). This complex, known as Arp2/3, is composed of seven subunits and has since been found in humans (Machesky et al., 1997; Welch et al., 1997a), yeast (Balasubramanian et al., 1996; McCollum et al., 1996; Winter et al., 1997), and Dictyostelium (Murgia et al., 1995), suggesting a conserved molecular function for the complex (Machesky and Way, 1998; for reviews, see Machesky and Gould, 1999). In addition to Arp2p and Arp3p, the complex contains five subunits, known in humans as ARPC1B (p41-Arc), ARPC2 (p34-Arc), ARPC3 (p21-Arc), ARPC4 (p20-Arc), and ARPC5 (p16-Arc), the sequences of which offer no clues to their functions (Machesky et al., 1997; Welch et al., 1997a). The only exception is the ARPC1B (p41-Arc) subunit, which contains WD-40 repeats, motifs thought to play a role in protein-protein interactions.
In vitro studies with purified Arp2/3 from Acanthamoeba demonstrate roles for the complex in cross-linking actin filaments, enhancing actin filament nucleation, and pointed end capping of filaments (Mullins et al., 1997, 1998a,b). In mammalian cells, the complex localizes to regions of lamellipodial protrusions, where it is thought to play a role in promoting actin assembly (Machesky et al., 1997; Welch et al., 1997a; Bailly et al., 1999). High-resolution electron microscopy of lamellipodia has shown a highly branched network of actin filaments with the Arp2/3 complex localized to filament junctions, indicating a role for the complex in branch formation (Svitkina et al., 1997; Svitkina and Borisy, 1999). The Arp2/3 complex also localizes to the actin-rich tails of motile Listeria monocytogenes, and, along with the bacterial protein ActA, it appears to be sufficient for nucleating actin polymerization on the bacterial surface (Welch et al., 1997b, 1998; May et al., 1999). Analogous to the role of bacterial ActA, the WASP/Scar family of proteins has recently been shown to bind directly to the Arp2/3 complex in vivo to stimulate actin polymerization (Machesky and Insall, 1998; Machesky et al., 1999; Rohatgi et al., 1999; Winter et al., 1999a; Yarar et al., 1999).
The Arp2/3 complex has also been studied in both fission and budding yeasts. In yeasts, subunits of the complex have been localized to cortical actin patches, and temperature-sensitive alleles of Arp3p result in loss of actin patch motility (McCollum et al., 1996; Winter et al., 1997). Additionally, a mutant in S. cerevisiae Arp2p appears to have a defect in endocytosis but not exocytosis (Moreau et al., 1996, 1997).
In S. pombe, a mutant defective in Sop2p (the p41 subunit) function was first identified by its ability to suppress the temperature-sensitive growth defect of the profilin mutant cdc3-124 (Balasubramanian et al., 1996). Likewise, a mutation in S. pombe Arp3p is also able to suppress the mutant profilin, suggesting an interaction between profilin and the Arp2/3 complex (McCollum et al., 1996). These results are consistent with the original isolation of the complex on profilin-agarose (Machesky et al., 1994) and with in vitro cross-linking studies that indicate that Arp2p binds profilin (Mullins et al., 1998b).
The isolation and characterization of conditional mutants has proven to be very useful for understanding the function of essential proteins. In particular, S. pombe is well suited to studies involving cell cycle progression and actin organization because its growth and division cycles are highly characterized. In S. pombe cells, F-actin undergoes a well-defined series of rearrangements during the growth and division cycles and can be localized to three structures: actin cables, the medial ring, and cortical actin patches (Marks et al., 1986). In newly divided cells, actin is found primarily as cortical actin patches at the old (growing) ends of the cells. At new end take off, growth occurs from both ends of the cells and, accordingly, cortical actin patches are concentrated at both ends. At the initiation of mitosis, actin localization diminishes at the poles, and a medial ring, composed of actin and other proteins, assembles in the middle of the rod-shaped cell. Actin patches appear to translocate from the ends of the cells to the medial region during cell division and are thought to play a role in cell wall deposition (reviewed by Welch et al., 1994).
In this paper, we describe the characterization of a conditional mutant, arp2-1, which displays a defect in cortical actin patch distribution at restrictive temperature. The arp2+ gene encodes an essential actin-related protein with homology to the Arp2 family (reviewed by Frankel and Mooseker, 1996; Mullins et al., 1996). Analysis of arp2-1 mutant cells indicates a defect in the Arp2/3 sedimentation profile that results from loss of Arp2p and Arc18p from the complex at restrictive temperature. The temperature-sensitive Arp2p mutant also displays defects in ATP and Arp3p binding, suggesting a possible model for the disruption of the complex in these cells.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Methods
S. pombe and S. cerevisiae strains used in this study are listed in Table 1. S. pombe strains were grown in yeast extract (YE) medium or minimal medium with the appropriate supplements as described (Moreno et al., 1991). Crosses were performed on glutamate medium (minimal medium lacking ammonium chloride and containing 0.01 M glutamate). Transformations were performed by electroporation (Prentice, 1992). Genomic DNA was isolated with the use of the Nucleon MiY DNA extraction kit (Amersham Pharmacia Biotech, Piscataway, NJ). S. cerevisiae strains for two-hybrid analysis were grown on synthetic minimal medium with the appropriate supplements (Guthrie and Fink, 1991).
Table 1.
Strain list
| Strain | Genotype | Source or reference |
|---|---|---|
| S. pombe | ||
| KGY28 | h− 972 | P. Nurse |
| KGY246 | h−ade6-M210 ura4-D18 leu1-32 | P. Nurse |
| KGY249 | h+ade6-M216 ura4-D18 leu1-32 | P. Nurse |
| KGY783 | h−cdc3-124 leu1-32 his3-D1 ura4-D18 ade6-216 | Balasubramanian et al. (1994) |
| KGY1187 | h90arp2-1 mam2∷LEU2 leu1-32 ura4-D18 ade6-M210 | This study |
| KGY1361 | h−sop2-HA ade6-M210 ura4-D18 leu1-32 | This study |
| KGY1524 | h−arp2-HA ade6-M210 ura4-D18 leu1-32 | This study |
| KGY1525 | h−arp2-myc ade6-M210 ura4-D18 leu1-32 | This study |
| KGY1529 | h+sop2-HA arp2-1, mam2∷LEU2, leu1-32, ura4-D18, ade6-M210 | This study |
| KGY1929 | h+/h−arp2∷ura4/arp2+ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 | This study |
| KGY2224 | KGY246 carrying pRMH42arp2+ | This study |
| KGY2225 | KGY246 carrying pRMH42arp2-E316K | This study |
| KGY2226 | KGY246 carrying pRMH42arp2-T12A | This study |
| S. cerevisiae | ||
| KGY1400 | YPB2: MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 112 canRgal4-542 gal80-538 LYS2∷GAL1uas−LEU2TATA-HIS3 URA3∷(GAL417mersx3)-CyC1TATA-lacZ | Chevray and Nathans (1992) |
| KGY1897 | YPB2 carrying pBI-770 ARC18 and pBI-771ARP2 | This study |
| KGY1898 | YPB2 carrying pBI-770 ARC18 and pBI-771 | This study |
| KGY1974 | YPB2 carrying pBI-770 and pBI-771ARP2 | This study |
Cytology and Microscopy
All fluorescence microscopy was performed on a Zeiss (Thornwood, NY) Axioskop microscope with the use of the appropriate set of filters. To visualize DNA and the cell wall, cells were stained with DAPI and Calcofluor as described (Balasubramanian et al., 1997). For immunostaining, cells were fixed with methanol and stained with a 1:100 dilution of anti-actin N.350 mAb (Amersham Pharmacia Biotech) or with a 1:5 dilution of affinity-purified anti-Arp2 or anti-Arp3 (McCollum et al., 1996) antibodies followed by a 1:100 dilution of Alexa-conjugated goat anti-mouse or goat anti-rabbit immunoglobulin G (IgG) secondary antibody (Molecular Probes, Eugene, OR).
Cloning and DNA Sequence of arp2
arp2-1 mutant (KGY1187) cells were transformed with a pUR19-based S. pombe genomic library (Barbet et al., 1992) and selected for uracil prototrophy at 36°C. Plasmids were recovered from Ura+Arp2+ colonies and analyzed by restriction digests. The minimal fragment capable of rescuing arp2-1 was identified and sequenced with Thermosequenase (United States Biochemical, Cleveland, OH) with Redivue 33P Terminator (Amersham Pharmacia Biotech) and custom-synthesized oligonucleotide primers or M13 universal primer. Sequence data were analyzed by comparison with protein and nucleic acid sequences accessible through the National Center for Biotechnology Information (Bethesda, MD). Removal of the single predicted intron from the arp2 coding region was accomplished with the use of the Muta-Gene phagemid in vitro mutagenesis kit (Bio-Rad Laboratories, Richmond, CA), resulting in pKG1121. To determine the sequence of the arp2-1 allele, genomic DNA was isolated from the arp2-1 mutant strain KGY1187. The arp2-1 gene was PCR amplified with Pfu DNA polymerase (Stratagene, La Jolla, CA) in a PTC-100 programmable thermal controller (MJ Research, Watertown, MA) programmed as follows: 94°C, 1 min; 50°C, 2 min; 72°C, 3 min (30 cycles); 72°C, 10 min. The resulting PCR products were sequenced directly as described above. The arp2-T12A point mutant was created with the use of the Chameleon double-stranded mutagenesis kit (Stratagene) and confirmed by sequencing.
Construction of arp2::ura4+ Deletion Strain
A deletion construct was created by deleting the BglII-EcoRV fragment of the arp2+ gene that encodes amino acids 61–361 and replacing it with a 1.8-kilobase (kb) fragment carrying the ura4+ gene. This plasmid was linearized with BamHI, and the resulting 2.5-kb fragment was used to transform a diploid strain of genotype ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 h+/h− (KGY246/KGY249) to uracil prototrophy. Ura+ transformants were replica plated for 5 d at 1-d intervals to medium containing uracil to allow loss of autonomously replicating plasmids carrying the ura4+ gene. Putative stable integrants were then identified by growth on selective medium lacking uracil and subjected to Southern blot analysis to confirm the replacement of one copy of arp2+ with the ura4+ gene.
Generation of Anti-Arp2p Antibodies
The entire arp2+ coding region was cloned into bacterial expression vector pRSETB (Invitrogen, Carlsbad, CA) for production of fusion protein. The fusion protein was gel purified, and antibodies were produced in rabbits by Cocalico Biologicals (Reamstown, PA). Antibodies were affinity purified as described (Olmsted, 1981).
Electron Microscopy
Wild-type S. pombe cells (KGY28) were grown to midlog phase, harvested by vacuum filtration, transferred rapidly to a thin brass sample chamber, and then frozen in an HPM-010 high-pressure freezer (BalTech, Liechtenstein). The cells were then transferred under liquid nitrogen to a freeze substitution device in which their frozen water was dissolved at −90°C with acetone containing 0.1% anhydrous glutaraldehyde, as described previously (Demeter et al., 1995). The samples were warmed slowly to −40°C for transfer to liquid Lowicryl HM20 resin (Electron Microscopy Sciences, Fort Washington, PA) or to 0°C for transfer to LR White resin (Electron Microscopy Sciences). These resins were polymerized at −40°C or +43°C, respectively. Serial sections were cut with a diamond knife on a Leica (Deerfield, IL) Ultracut E microtome and picked up on Formvar-coated, carbon-stabilized slot grids.
Grids with sections were incubated for 30 min in a blocking buffer containing PBS, 0.8% BSA, 0.1% fish skin gelatin, and 0.01% Tween 80. All antibodies were diluted in this blocking buffer. All rinses between antibody applications used PBS containing 0.1% Tween 80. Affinity-purified rabbit anti-Arp2p antibodies (100 μg/ml) were applied to the sections at a dilution of 1:20, incubated overnight at 4°C, and then rinsed three times. Goat antibodies to rabbit IgG labeled with 10-nm colloidal gold (British Biocell International) were diluted 1:20 and applied to sections for 2 h at room temperature, followed by three rinses in the same buffer. A mouse mAb against actin (Amersham Pharmacia Biotech) diluted 1:200 was then applied to the sections for another overnight incubation at 4°C. The sections were rinsed, and rabbit antibodies to mouse IgG labeled with 5-nm colloidal gold were applied to the sections for 2 h at room temperature. After rinsing, the specimens were fixed with 0.5% glutaraldehyde in PBS, followed by staining with 2% uranyl acetate for 8 min and Reynold’s lead citrate for 3 min. The sections were examined in a Phillips (Mahwah, NJ) CM10 electron microscope operating at 80 kV, and images were recorded on Kodak (Rochester, NY) 4489 film.
Epitope Tagging
A genomic copy of sop2 encoding three copies of a hemagglutinin (HA) tag (3xHA-KAN cassette [Bahler et al., 1998]) fused to the C terminus of Sop2p was generated by the two-step PCR-based method described previously (Wach et al., 1994; Wach, 1996; Bahler et al., 1998). The amplification product was gel purified and transformed into diploid cells (KGY246/KGY249) by the lithium acetate method (Keeney and Boeke, 1994). Transformants were plated on YE plates and allowed to recover overnight at 32°C and then replica plated to YE plates containing 25 μg/ml G418. Tetrads were analyzed for segregation of kanamycin resistance (G418R), and G418R colonies were further analyzed by Southern blot analysis to confirm integration of the 3xHA-KAN tag at the sop2 genomic locus. Detection of HA-tagged Sop2p was accomplished by immunoblotting with the use of an anti-HA mAb (12CA5, Boehringer Mannheim, Indianapolis, IN), followed by a peroxidase-conjugated goat anti-mouse IgG (1:10,000; Jackson Immunoresearch Laboratories, West Grove, PA) and ECL detection reagents (Amersham Pharmacia Biotech). Strains KGY1524 (Arp2–3xHA) and KGY1525 (Arp2–9xmyc) were generated as described above with the 3xHA-KAN or 9xmyc-KAN cassettes (Bahler et al., 1998). Detection of myc-tagged Arp2p was accomplished by immunoblotting with the use of an anti-myc mAb (9E10, PharMingen, San Diego, CA).
Sucrose Gradient Analysis and Immunoprecipitation
For sucrose gradient analysis, cells were grown to midlog phase in 200 ml of YE medium at 25°C and then shifted to restrictive temperature (36°C) for 7 h. Approximately 60 ml (OD595 = 0.5) of each culture was harvested, and native cell lysates were prepared in KCl buffer (150 mM KCl, 12 mM sodium pyrophosphate, 1 mM ATP, 5 mM DTT, 0.1 mM benzamidine, 1 mM PMSF, 30 mM imidazole, pH 7) by glass bead disruption (Gould et al., 1991). After a high-speed clearing spin, 200 μl of each lysate was layered on a 5–20% sucrose gradient (5 ml volume) prepared in KCl buffer. Gradients were ultracentrifuged at 40,000 rpm for 19 h at 4°C in a Beckman (Fullerton, CA) SW50.1 rotor. Molecular weight standards were fractionated in a parallel gradient. Detection of Arp2p, Arp3p, and Sop2HA was accomplished by immunoblotting with the use of anti-Arp2, anti-Arp3 (McCollum et al., 1996), and anti-HA antibodies followed by a peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG (1:10,000; Jackson Immunoresearch Laboratories) and ECL detection reagents (Amersham Pharmacia Biotech). For immunoprecipitation experiments, midlog-phase cells from strains KGY28, KGY1361, or KGY1529 were metabolically labeled with 1 mCi of 35S-Trans label (a mixture of methionine and cysteine; ICN Pharmaceuticals, Irvine, CA) for 4.5 h in minimal medium with supplements at 36°C. After sucrose gradient fractionation, the Arp2/3 complex was isolated from each fraction by incubation with anti-HA mAb (12CA5, Boehringer Mannheim) coupled to protein A–Sepharose beads (Amersham Pharmacia Biotech) at 4°C for 4 h. The beads were pelleted and washed extensively with NP-40 buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 4 μg/ml leupeptin, 100 μM Na3VO4, 1 mM PMSF, 2 mM benzamidine), NP-40 buffer with 0.5 M NaCl, and RIPA buffer (NP-40 buffer with 1% sodium deoxycholate, 0.1% SDS) and analyzed by SDS-PAGE and autoradiography.
Two-Hybrid Analysis
The S. cerevisiae ARP2 and ARC18 genes were PCR amplified from either a S. cerevisiae cDNA library (Y2HL [James et al., 1996]) or from genomic DNA with the corresponding primers and cloned as an in-frame fusion to the GAL4 DNA-binding domain in vector pBI-770 to construct two-hybrid bait plasmids. Prey plasmids were constructed in vector pBI-771 by cloning the genes as in-frame fusions with the GAL4 transcription activation domain (pBI-770 and pBI-771 modified from pPC62 and pPC86 [Chevray and Nathans, 1992]). The 1.2-kb fragment of ARP2 was amplified from cDNA with the primers 5′-CTAACAAAACTGTCGACTAATAATGGACCCACATAATCC-3′ and 5′-GGATCAATAAAGTCGACATCTATCTTGGACC-3′ and TaqPlus Precision DNA polymerase (Stratagene) in a PTC-100 programmable thermal controller (MJ Research) programmed as follows: 95°C, 1 min; 65-1°C per cycle, 1 min; 72°C, 5 min (15 cycles); 95°C, 1 min; 50°C, 1 min; 72°C, 10 min (30 cycles). The 0.5-kb fragment of the ARC18 gene was amplified with the primers 5′-CTCCCAGTATGTCGACAAATGCCTGCGTATCACTCG-3′ and 5′-CCTTGCACCGTCGACCTTTATAGAGATTTGTTC-3′. Bait and prey plasmids were cotransformed into S. cerevisiae host strain YPB2 according to the method described previously (Gietz et al., 1995), and transformants carrying both plasmids were selected on medium lacking leucine and tryptophan. Initiation of HIS3 transcription as assayed by growth on medium lacking histidine was regarded as evidence of bait and prey protein interaction. Activation of GAL4 transcription was assayed by β-galactosidase filter assays to confirm protein-protein interactions.
GST Fusion Proteins
The S. cerevisiae ARC18 coding region was cloned into the vector pGEX-2T (Amersham Pharmacia Biotech) to produce a GST-Arc18p fusion protein. The fusion protein was produced in bacterial cells after induction with 0.4 mM isopropylthio-β-galactoside and purified from bacterial lysates according to the method described previously (Frangioni and Neel, 1993). The coding region of S. cerevisiae ARP2 was cloned into pBluescript SK+ (Stratagene) and translated in vitro in the presence of [35S]methionine (35S-Trans label, ICN Pharmaceuticals) with the use of the TNT coupled reticulocyte lysate system (Promega, Madison, WI). Purified GST or GST-Arc18p bound to glutathione-agarose beads was mixed with 35S-labeled Arp2p in binding buffer (20 mM Tris-HCl, pH 7.0, 150 mM NaCl, 2 mM EDTA, 0.1% NP-40) and incubated at 4°C for 1 h. The beads were then washed three times in binding buffer, and the proteins were resolved by SDS-PAGE, treated with Amplify (Amersham Pharmacia Biotech), and exposed to film.
Photoaffinity Labeling
The S. pombe coding regions of arp2+, arp2-E316K, and arp2-T12A were subcloned into pRMH42 (Craven et al., 1998) to produce proteins tagged at the N terminus with one copy of the 6× His tag and two copies of the myc epitope. The expression of the tagged proteins is under the control of the attenuated nmt1 thiamine-repressible promoter present in pREP41 (Basi et al., 1993). The constructs were transformed into wild-type cells (KGY246), and the resulting transformants were grown to midlog phase in minimal medium containing thiamine at 25°C. The cells were then washed to remove the thiamine and incubated for 22 h at 25°C to induce expression of the tagged proteins. Cells were harvested and lysed under native conditions in NP-40 buffer. The myc-tagged Arp2 proteins were immunoprecipitated with the use of an anti-myc mAb (9E10, PharMingen) coupled to protein G–Sepharose beads (Amersham Pharmacia Biotech) at 4°C for 2 h. The beads were pelleted and washed two times with NP-40 buffer and four times with PA buffer (2 mM Tris, pH 7.4, 0.1 mM CaCl2) and resuspended in PA buffer. Photoaffinity labeling was carried out by mixing immunoprecipitated protein and 2 μM 8-azido-[α-32P]ATP (20 Ci/mmol; ICN Pharmaceuticals) in a total volume of 100 μl. Cold ATP was added to a final concentration of 500 μM in competition experiments. Samples were irradiated for 10 min with a UV Stratalinker 1800 (Stratagene) and then washed twice in PA buffer.
For immunoprecipitation studies, irradiated samples were washed twice in PA buffer, and the beads were then resuspended in 100 μl of SDS lysis buffer (Gould et al., 1991) and boiled for 3 min. NP-40 buffer was then added to a final volume of 1.2 ml. After a short centrifugation, Arp2p or Arp3p was immunoprecipitated from the denatured supernatant with the use of anti-Arp2p or anti-Arp3p antibodies, followed by incubation with protein A–Sepharose beads (Amersham Pharmacia Biotech). The beads were pelleted and washed two times with NP-40 buffer and analyzed by SDS-PAGE and autoradiography.
RESULTS
Phenotypic Characterization of the arp2-1 Mutant
In a screen designed to enrich for cells that diploidize as a result of failed cytokinesis (Balasubramanian et al., 1998), a recessive temperature-sensitive mutant was isolated that displayed a morphology typical of cells defective in actin cytoskeletal components. The temperature-sensitive phenotype segregated 2:2 in several backcrosses, indicating that this phenotype was due to a single mutation in the strain. At the permissive temperature of 25°C, mutant cells (hereafter referred to as arp2-1) divided normally and formed colonies that were indistinguishable from wild-type cells. When grown at the restrictive temperature of 36°C, however, arp2-1 cells displayed a dumbbell shape and eventually lysed.
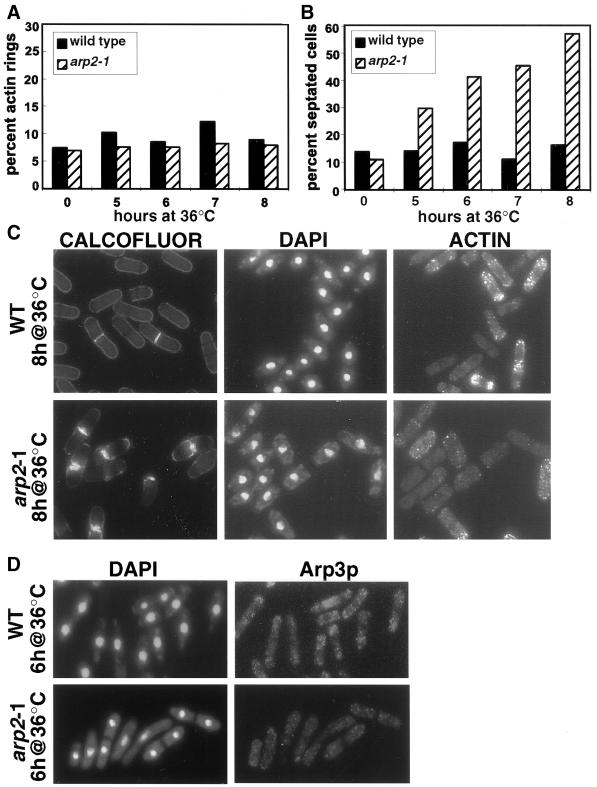
To more closely examine the temperature-sensitive phenotype, arp2-1 cells were grown to midlog phase under permissive conditions and then shifted to the restrictive temperature (36°C). For the first 4 h after the shift to 36°C, arp2-1 cells continued to divide normally and appeared indistinguishable from wild-type cells (our unpublished results). After ∼5 h at the restrictive temperature, however, septated cells began to accumulate in the arp2-1 culture compared with wild type (30% versus 14%; Figure 1B). At each subsequent time point, the percentage of septated arp2-1 cells increased, reaching more than 50% at 8 h (Figure 1B). Although initial septum formation in arp2-1 cells appeared normal, the medial region of the cells continued to accumulate excess cell wall material throughout the course of the experiment, a phenotype that was never seen in wild-type cells (Figure 1C). After 8–9 h at the restrictive temperature, the majority of arp2-1 cells were lysed, apparently as a result of cell separation after defective septum formation.
Figure 1.
The temperature-sensitive phenotype of arp2-1 cells. Wild-type (KGY28) or arp2-1 mutant cells (KGY1187) were grown to midlog phase at the permissive temperature (25°C) and then shifted to the restrictive temperature (36°C). Cells were harvested at the indicated times after the shift to 36°C, fixed, and stained with DAPI, anti-actin N.350 mAb, or Calcofluor to visualize DNA, actin, or septal material, respectively. (A) Percentage of wild-type and arp2-1 cells containing actin rings at indicated times after the shift to 36°C. (B) Percentage of wild-type and arp2-1 cells with a septum at indicated times after the shift to 36°C. (C) Phenotype of wild-type (WT) and arp2-1 cells after 8 h at 36°C. (D) Wild-type (WT) or arp2-1 cells were fixed after 6 h at 36°C and stained with DAPI and antibodies against Arp3p to visualize DNA and cortical actin patch distribution.
One possible explanation for the presence of abnormal septa in arp2-1 cells is a defect in actin ring formation, because proper assembly of the actin ring is required for septum formation (reviewed by Gould and Simanis, 1997). To determine whether this was the case, we examined the actin cytoskeleton in arp2-1 cells. At all time points during the experiment, we saw actin rings in ∼8% of cells (Figure 1A). We did not observe an increase in the percentage of cells with actin rings or the appearance of aberrant actin rings corresponding to the increase in septated cells. These results suggest that the increase in septated cells during the course of the experiment was more likely due to a defect in septation or cell separation than an increase or defect in actin ring formation.
Although the actin ring and actin cables appeared normal in arp2-1 cells, we observed a defect in another actin structure, the cortical actin patches. Unlike in wild-type cells, the actin patches appeared to be randomly distributed in the arp2-1 mutant rather than concentrated at the cell poles or at the medial region of the cell (Figure 1C). Additionally, staining of the actin patches in arp2-1 cells appeared slightly diminished compared with that in wild-type cells. To confirm the random distribution of actin patches in the mutant, we performed immunolocalization with antibodies against Arp3p, a protein previously shown to localize to the actin patches (McCollum et al., 1996). Similar to the results obtained with anti-actin staining, the actin patches recognized by the Arp3p antibody were distributed randomly throughout the cytoplasm in the arp2-1 mutant (Figure 1D). These results suggest that the arp2-1 mutant is unable to maintain properly polarized actin patches when grown at restrictive temperature.
Cloning and Sequence Analysis
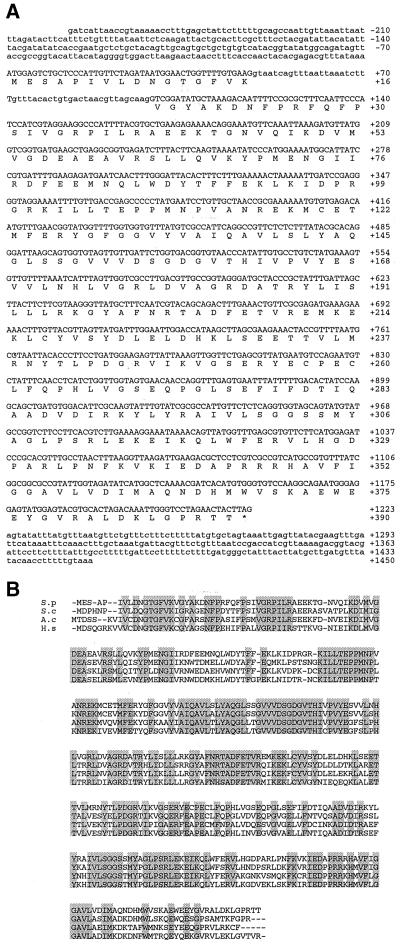
To gain insight into the role of the mutated protein, we isolated the arp2+ gene by complementation of the arp2-1 temperature-sensitive phenotype. Sequencing of the smallest complementing fragment revealed a 1.1-kb ORF, which is interrupted by a single intron and is predicted to encode a 390-amino acid protein. Comparison of the amino acid sequence to available protein sequences in the databases indicated that this protein is a member of the Arp2 family of actin-related proteins (Figure 2A). The predicted S. pombe Arp2 protein sequence is 44% identical (64% similar) to conventional actin but is 63–69% identical (75–79% similar) to Arp2 homologues in S. cerevisiae, human, chick, and Acanthamoeba (Figure 2B).
Figure 2.
Sequence analysis of arp2+. (A) Nucleotide sequence and predicted amino acid sequence of arp2+. The intron sequence is shown in lower case letters. (*) Stop codon. The GenBank accession number for arp2+ is AF095900. (B) Alignment of S. pombe Arp2p with Arp2 homologues. S.p, S. pombe Arp2p; S.c, S. cerevisiae Arp2p; A.c, Acanthamoeba castellanii Arp2p, H.s, human Arp2p. Identical amino acids are shaded. S. pombe Arp2p is 69% identical (79% similar) to S. cerevisiae Arp2p, 62% identical (75% similar) to Acanthamoeba Arp2p, and 63% identical (76% similar) to human Arp2p.
The sequence of the arp2-1 mutant allele revealed a single g→a mutation, which is consistent with the type of mutations caused by nitrosoguanidine, the mutagen used to isolate arp2-1 (Balasubramanian et al., 1998). This g→a transition affects codon 316 and results in a substitution of lysine for the glutamic acid normally found at this position, a residue that is conserved among all Arp2 homologues that have been identified. Structural models of Acanthamoeba Arp2, based on comparisons with the structure of conventional actin, indicate that residue 316 is positioned in subdomain 3 of the protein (Kabsch et al., 1990; Kelleher et al., 1995). Residues in this subdomain of conventional actin have been implicated in profilin (Schutt et al., 1993) and nucleotide binding (Kabsch et al., 1990) and actin-actin interactions (Holmes et al., 1990).
arp2+ Is Essential for Viability
To determine whether arp2+ is an essential gene, we replaced most of the arp2+ ORF with the ura4+ gene and used this construct to replace one copy of arp2+ in a diploid strain. Southern blot analysis confirmed replacement of a single allele with the ura4+ cassette. Heterozygous diploids were allowed to sporulate, and tetrad analysis yielded two viable and two inviable progeny. In all tetrads, the viable colonies were Arp2+ Ura−, indicating that arp2+ is an essential gene.
arp2-1 Genetically Interacts with Cdc3-Profilin
The Arp2/3 complex was originally isolated by affinity chromatography on profilin-agarose (Machesky et al., 1994). In S. pombe, mutations in two of the subunits of the complex, Arp3p and Sop2p (p41), are able to suppress a temperature-sensitive mutation in Cdc3-profilin (Balasubramanian et al., 1996; McCollum et al., 1996), providing further evidence that the Arp2/3 complex interacts with profilin. Sequence comparisons of Arp2p with conventional actin predict that Arp2p should retain the ability to interact with profilin, because the residues implicated in actin-profilin binding are conserved in Arp2p (Kelleher et al., 1995). Indeed, in vitro cross-linking studies have recently been used to show that Arp2p is the subunit of the Arp2/3 complex that interacts with profilin in Acanthamoeba (Mullins et al., 1998b).
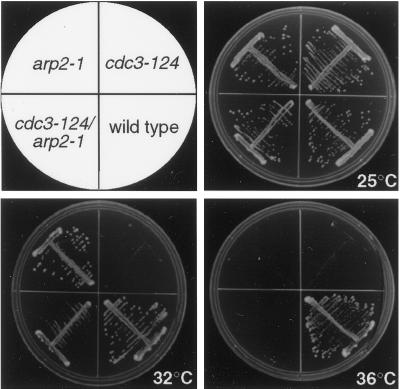
To determine whether Arp2p genetically interacts with profilin in S. pombe, we constructed a cdc3-124 arp2-1 double mutant and assessed the phenotype at various temperatures. At the restrictive temperature (36°C) for both parental strains, the cdc3-124 arp2-1 double mutant failed to grow (Figure 3). When the temperature was decreased to 32°C, however, the double mutant was capable of growth even though the cdc3-124 parent strain was inviable at this temperature (Figure 3).
Figure 3.
The arp2-1 mutation rescues the growth defect of cdc3-124 at semipermissive temperature. A double mutant, cdc3-124 arp2-1, constructed between the single mutants arp2-1 (KGY1187) and cdc3-124 (KGY783), was tested for growth in parallel with the single mutants at 25, 32, and 36°C.
Arp2p Localizes to Cortical Actin Patches
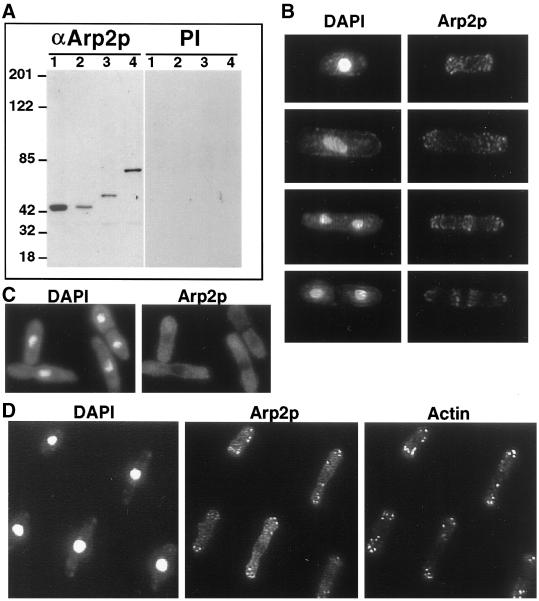
To determine whether Arp2p colocalized with other Arp2/3 complex members in S. pombe cells, we raised polyclonal antibodies against full-length Arp2p and affinity purified them. The anti-Arp2p antibody recognized a single band of an apparent molecular mass of 43 kDa on immunoblots of S. pombe whole cell lysates from either wild-type or arp2-1 cells, consistent with the predicted molecular mass of Arp2p (Figure 4A). From strains in which the endogenous arp2 locus was tagged with either three copies of the HA epitope or nine copies of the myc epitope, a single band was also recognized that corresponded to the predicted increase in molecular mass attributable to the epitope tag (Figure 4A).
Figure 4.
Immunolocalization of Arp2p. (A) Characterization of anti-Arp2p antibody. Lysates were prepared from wild-type (KGY28; lane 1), arp2-1 (KGY1187; lane 2), arp2-HA (KGY1524; lane 3), and arp2-myc (KGY1525; lane 4) strains and resolved by SDS-PAGE, then immunoblotted with anti-Arp2p or preimmune serum. (B) Wild-type cells (KGY28) were grown to midlog phase, fixed with methanol, and stained with a 1:5 dilution of affinity-purified rabbit polyclonal anti-Arp2p antibodies. S. pombe cells representing different stages of the cell cycle are shown stained with DAPI to visualize DNA and anti-Arp2p antibodies. Arp2p localizes to cortical actin patches at all stages of the cell cycle. (C) arp2-1 cells (KGY1187) were grown to midlog phase at 25°C and shifted to 36°C for 6 h. Cells were fixed with methanol and stained with affinity-purified anti-Arp2p antibodies as described above. Arp2p is diffusely distributed throughout the cytoplasm in the arp2-1 mutant. (D) Wild-type cells (KGY28) were grown at 25°C, fixed in methanol, and stained with a 1:5 dilution of affinity-purified rabbit polyclonal anti-Arp2p antibodies and a 1:50 dilution of monoclonal anti-actin antibodies.
In an asynchronously growing culture, Arp2p appeared to localize to the cytoplasm in a punctate pattern that correlated to cortical actin patch localization. In interphase cells, Arp2p was concentrated at the cell poles in a pattern mimicking the distribution of actin patches (Figure 4B). As cells underwent nuclear division, Arp2p staining appeared to move from the poles to the medial region of the cells, where it persisted until septation was completed (Figure 4B). Arp2p appeared to colocalize only with cortical actin patches; we did not observe staining of the medial actin ring or actin cables. We did not observe nuclear staining of Arp2p at any stage of the cell cycle. In arp2-1 mutant cells grown at the restrictive temperature, the protein was not detected in patches. Rather, it appeared to be diffusely distributed throughout the cytoplasm (Figure 4C).
The structure of actin patches in not well understood; however, a number of proteins, including short actin filaments, capping protein (Amatruda and Cooper, 1992), and cofilin (Moon et al., 1993), localize to the cortical patch in budding yeast (for review, see Welch et al., 1994). To more closely examine whether Arp2p is found at the cortical patch, we compared the localization of Arp2p and actin by two methods. First, we examined whether actin and Arp2p colocalized in wild-type cells by indirect immunofluorescence. These results demonstrated that all cortical patches recognized by the anti-Arp2p antibody were also recognized by the anti-actin antibody, indicating that Arp2p colocalizes with actin (Figure 4D). We did, however, occasionally observe cortical patches that were recognized by the anti-actin antibody but that were difficult to detect by the anti-Arp2p antibody. It is unclear whether this represents a subset of cortical actin patches that contain actin but not Arp2p or whether this result is due to difficulties of antibody accessibility.
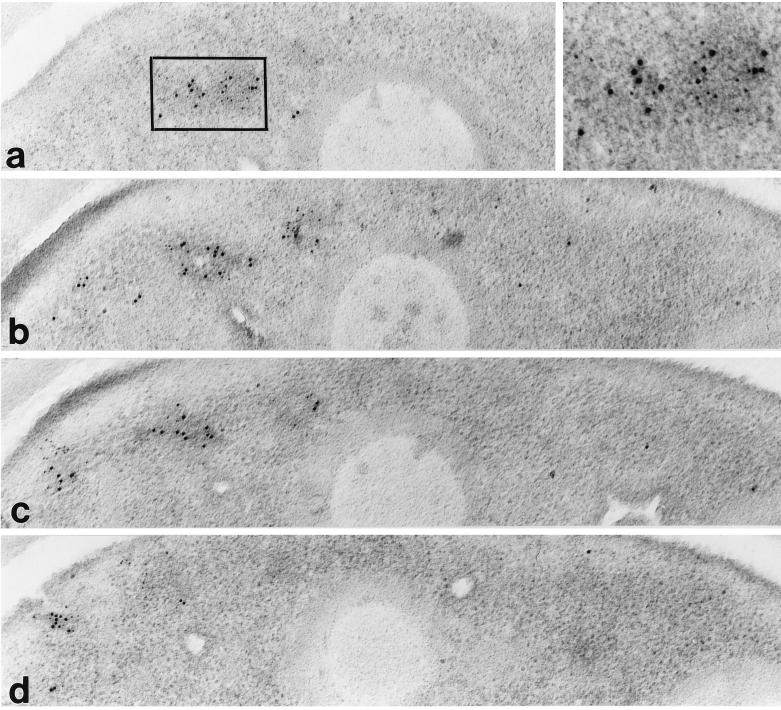
To further examine the colocalization of actin and Arp2p, wild-type cells were grown to midlog phase and then prepared for electron microscopy by double staining with antibodies against actin and Arp2p, followed by indirect labeling with 5- and 10-nm colloidal gold particles, respectively. When cells were examined, both 5- and 10-nm gold particles were found colocalized to patches in the cytoplasm (Figure 5). These patches were often found close to the plasma membrane. Examination of serial sections indicated that Arp2p and actin colocalized throughout the patch structure, and the pattern of Arp2p and actin localization with respect to each other appeared random in the patch (Figure 5).
Figure 5.
Arp2p and actin colocalize at the cortical actin patch. Serial electron micrographs from a S. pombe interphase cell showing a membrane-associated patch. The localization of actin is marked with 5-nm gold particles, and Arp2p is marked with 10-nm gold particles.
The Arp2/3 Complex Is Unstable in the arp2-1 Mutant
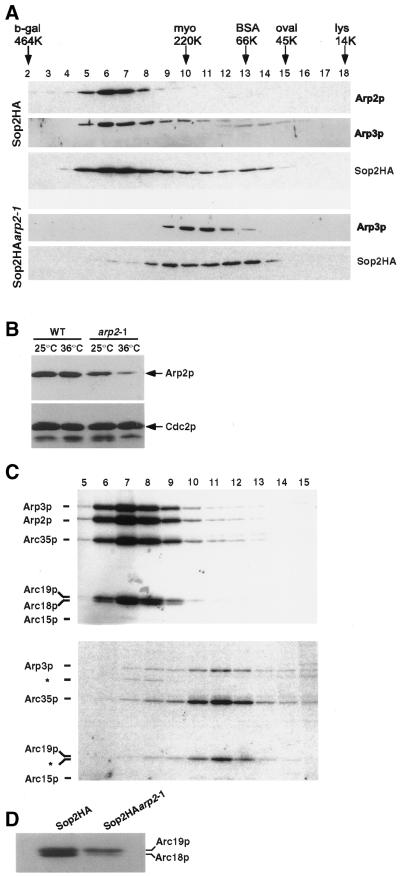
Because other Arp2p homologues have been described as components of the Arp2/3 complex (Machesky et al., 1994; Welch et al., 1997a; Winter et al., 1997), we performed sucrose gradient analyses to determine whether S. pombe Arp2p was also associated with this protein complex and whether the arp2-1 mutant allele had any effect on the integrity of the complex at the restrictive temperature. To facilitate isolation of the complex, we tagged the sop2+ ORF with sequences encoding three copies of the HA epitope at its 3′ end (see MATERIALS AND METHODS). This Sop2HA-tagged protein was capable of complementing a sop2 null strain; thus, we concluded that the tagged protein was fully functional (our unpublished results). Lysates from Sop2HA and Sop2HAarp2-1 cells grown for 6 h at 36°C were then prepared under native conditions and sedimented on sucrose gradients. After sedimentation, fractions were collected and analyzed by immunoblotting to determine whether Arp2p cosedimented with Arp3p and Sop2p, previously identified subunits of the S. pombe Arp2/3 complex (Balasubramanian et al., 1996; McCollum et al., 1996). As expected, the sedimentation profile of wild-type Arp2p was consistent with this protein being associated with a high-molecular-weight complex, and Arp2p cosedimented with both Arp3p and Sop2p in Sop2HA cells (Figure 6A), suggesting that these proteins are part of the same complex. In a parallel gradient prepared from Sop2HAarp2-1 cells, however, sedimentation of Arp3p and Sop2p was perturbed, and the profiles suggested that the complex was smaller in the arp2-1 mutant, migrating between molecular mass standards of 220 and 66 kDa, compared with the migration of the complex between molecular mass standards of 464 and 220 kDa in wild-type cells (Figure 6A). Although both Arp3p and Sop2p cofractionated in gradient fractions 9–13 collected from arp2-1 cells, we had difficulty detecting Arp2-1p in any gradient fraction by immunoblot analysis. The inability to detect Arp2-1p in these fractions was unexpected because we had detected Arp2-1 protein by immunoblot analysis of whole cell lysates before their centrifugation, albeit at somewhat reduced levels (discussed below). To determine which gradient fractions contained the mutant Arp2-1 protein, we then attempted to immunoprecipitate Arp2-1p with the use of anti-Arp2 antibodies from sets of pooled gradient fractions. From this analysis, we determined that a small amount of Arp2-1p was present in gradient fractions 5–8 and 9–13 and that the remaining Arp2-1p was spread among the other fractions at levels that are undetectable by immunoblot analysis (our unpublished results).
Figure 6.
The Arp2/3 complex is unstable in the arp2-1 mutant. (A) The sedimentation profile of the Arp2/3 complex is aberrant in arp2-1 cells. Lysates were prepared from Sop2HA (KGY1361) or Sop2HAarp2-1 (KGY1529) cells grown to midlog phase at 25°C and then shifted to 36°C for 6 h. After sedimentation, fractions were collected from the bottom (fraction 1) of a 5–20% sucrose gradient and then resolved by SDS-PAGE. Molecular size standards were run in parallel on an identical sucrose gradient. Fractions were analyzed for the presence of Arp2p, Arp3p, and Sop2HA by immunoblotting with anti-Arp2, anti-Arp3, or anti-HA antibodies. (B) Comparison of Arp2p levels at the permissive and restrictive temperatures. Cells were grown at 25°C or were grown at 25°C and then shifted to 36°C for 6 h. Denatured lysates were prepared from wild-type (KGY28) or arp2-1 (KGY1187) strains and resolved by SDS-PAGE. Arp2p and Cdc2p (loading control) were detected by immunoblotting with anti-Arp2p polyclonal antibodies or PSTAIR mAbs. (C) Immunoprecipitation of the Arp2/3 complex from gradient fractions. Sop2HA (KGY1361) and Sop2HAarp2-1 (KGY1529) cells were metabolically labeled with 35S-Trans label for 4.5 h at 36°C, and lysates prepared from these cells were fractionated over a sucrose gradient. The Arp2/3 complex was immunoprecipitated from each fraction with the anti-HA mAb 12CA5 and analyzed by SDS-PAGE and autoradiography. (D) Comparison of Arc19p and Arc18p levels in lysates from metabolically labeled Sop2HA (KGY1361) and Sop2HAarp2-1 (KGY1529) cells before fractionation over a sucrose gradient.
To determine whether the reduction in Arp2p levels in arp2-1 cells was a consequence of growth under restrictive conditions, we compared the levels of Arp2p from wild-type and arp2-1 cells grown at 25 or 36°C (Figure 6B). These results indicate that Arp2p levels were somewhat reduced at the permissive temperature in the arp2-1 mutant compared with wild-type cells and were further reduced after incubation at the restrictive temperature, suggesting that the mutant protein is likely less stable than the wild-type Arp2 protein (Figure 6B), a hypothesis confirmed below (see Figure 8C).
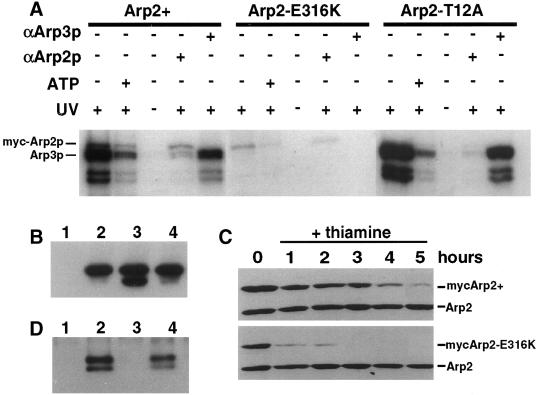
Figure 8.
Arp2p and Arp3p are ATP-binding proteins. (A) Arp2p and Arp3p were labeled by 8-azido-[α-32P]ATP. Wild-type cells carrying pRMH42 arp2+ (KGY2224), pRMH42 arp2-E316K (KGY2225), or pRMH42 arp2-T12A (KGY2226) were grown to midlog phase in minimal medium containing thiamine at 25°C, washed to remove the thiamine, and then incubated for 22 h at 25°C to induce expression of the myc-tagged Arp2 proteins. After cell lysis under native conditions, the myc-tagged Arp2 proteins were immunoprecipitated and then mixed with 2 μM 8-azido-[α-32P]ATP. Samples were subjected to UV irradiation to promote cross-linking between Arp2p and 8-azido-[α-32P]ATP. After irradiation, a fraction of each sample was denatured by boiling, and a second immunoprecipitation was performed with the use of either anti-Arp2 or anti-Arp3 antibodies to distinguish between myc-Arp2p and Arp3p. Cold ATP was added to a final concentration of 500 μM in competition experiments. Labeled proteins were resolved by SDS-PAGE and autoradiography. (B) Immunoprecipitation of myc-tagged Arp2 proteins from lysates used in ATP-binding assays with the 9E10 anti-myc mAb. (Lane 1) Wild type (KGY28); (lane 2) pRMH42 arp2+ (KGY2224); (lane 3) pRMH42 arp2-E316K (KGY2225); (lane 4) pRMH42 arp2-T12A (KGY2226). Immunoblots were probed with 9E10 anti-myc antibody to detect tagged Arp2. (C) After induction of myc-tagged wild-type Arp2 (KGY2224) and Arp2-E316K (KGY2225) in wild-type cells for 20 h at 32°C, thiamine was added back to the culture medium to repress expression of the tagged Arp2 proteins from the nmt1 promoter. Lysates were prepared from cells collected hourly after transcriptional repression, and the myc-tagged Arp2 proteins, along with endogenous Arp2p (loading control), were detected by immunoblotting with the use of anti-Arp2 antibodies. (D) myc-tagged Arp2 proteins were immunoprecipitated from native lysates and resolved by SDS-PAGE. (Lane 1) Wild-type (KGY28); (lane 2) pRMH42 arp2+ (KGY2224); (lane 3) pRMH42 arp2-E316K (KGY2225); (lane 4) pRMH42 arp2-T12A (KGY2226). The presence of coimmunoprecipitated Arp3p was detected by immunoblotting with the use of anti-Arp3p antibodies.
The observation that the complex sediments aberrantly in the arp2-1 mutant and the difficulty of detecting Arp2-1p in gradients prepared from arp2-1 cells suggested that the Arp2/3 complex may be unstable in these cells and that Arp2p may be lost from the complex. To more closely examine the effects of the arp2-1 allele on complex stability, cultures of either Sop2HA or Sop2HAarp2-1 cells were metabolically labeled with [35S]methionine for 4.5 h at 36°C, and lysates prepared from these cells were fractionated over a sucrose gradient. The Arp2/3 complex was then isolated from each fraction by immunoprecipitation with the use of the anti-HA mAb 12CA5 against Sop2HA. In Sop2HA cells, we were able to detect the Arp2/3 complex in fractions 6–10, consistent with our previous gradient fractionation experiments (Figure 6C). In metabolically labeled cells, five of the seven Arp2/3 subunits were easily detected. The Sop2p and Arc15p protein sequences, however, contain few methionine residues and therefore did not incorporate 35S-Trans label to a detectable level. Although Sop2HA could not be detected in this experiment, we assume that the protein is present in the complex because the antibody used for immunoprecipitation was directed to it. The results obtained from immunoprecipitations performed on fractions collected from the parallel Sop2HAarp2-1 gradient are much different. The majority of the Arp2/3 subunits were detected in fractions 10–12 (Figure 6C), again consistent with the previous fractionation experiment. However, in these fractions, both Arp2p and Arc18p appeared to be lost from the Arp2/3 complex, most likely accounting for its aberrant migration through the sucrose gradient (Figure 6C). Because Arc18p and Arc19p migrate close to each other, we confirmed the loss of Arc18p by examination of the original metabolically labeled lysates from Sop2HA and Sop2HAarp2-1 cells. Direct comparison of these lysates clearly shows that Arc18p is the subunit lost from the complex in Sop2HAarp2-1 cells (Figure 6D). There is a small amount of Arp2p, along with the other Arp2/3 subunits, detectable in fractions 7 and 8, suggesting that there is a small amount of intact complex present in these cells. The presence of this small fraction of intact Arp2/3 complex, however, does not appear to be sufficient to rescue the temperature-sensitive phenotype of the arp2-1 cells.
Arp2p Interacts Directly with Arc18p
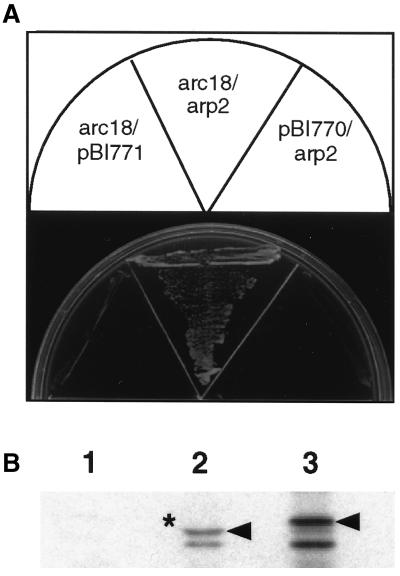
The simplest explanation for why Arc18p is lost from the complex in the arp2-1 mutant is that Arc18p binds directly to Arp2p. To test this possibility, we looked for interaction between the Arp2 and Arc18 proteins. Because the S. pombe arc18+ gene has not yet been cloned, we tested the S. cerevisiae Arp2p and Arc18p homologues for interaction first in the two-hybrid screen. Our two-hybrid results indicate that Arp2p and Arc18p are capable of interacting (Figure 7A). To determine whether this interaction was direct, we constructed a GST-Arc18p fusion protein and asked whether this fusion protein was capable of binding directly to in vitro translated Arp2p. These results showed that GST-Arc18p but not GST alone was able to bind Arp2p (Figure 7B). From this experiment, we conclude that Arp2p and Arc18p likely interact directly in the Arp2/3 complex and that upon incubation of arp2-1 cells at restrictive temperature, both Arp2p and Arc18p are lost from the Arp2/3 complex.
Figure 7.
Arp2p interacts directly with Arp18p. (A) YPB2 carrying pBI-770 ARC18 bait and pBI-771 ARP2 prey plasmids (KGY1897) was grown on medium lacking histidine to demonstrate interaction. KGY1898 and KGY1974 were used as controls. (B) GST-Arc18p fusion protein binds directly to Arp2p. Equal amounts of purified GST (lane 1) or GST-Arc18p (lane 2) bound to glutathione beads were mixed with in vitro translated Arp2p. After extensive washes, the proteins were resolved by SDS-PAGE and autoradiography. Lane 3 represents a sample of in vitro translated Arp2p before the binding reaction. Arrowheads indicate full-length Arp2p. (*) Compression attributable to GST-Arc18p and Arp2p having the same molecular weight.
Arp2p Binds ATP
As discussed previously, sequencing of the temperature-sensitive arp2-1 allele indicated a single point mutation that results in the substitution of lysine for glutamic acid at residue 316 of the Arp2 protein. Based on structural similarity to conventional actin, this residue is positioned in subdomain 3 of the Arp2 protein (Kabsch et al., 1990; Kelleher et al., 1995). Because residues in this subdomain have also been implicated in nucleotide binding (Kabsch et al., 1990), we asked whether the Arp2-E316K mutant protein was defective in its ability to bind ATP compared with wild-type Arp2p. Because the ATP cycle of Arp2p may be fundamental to its function, a defect in ATP binding might provide an explanation for the destabilization of the Arp2/3 complex in the arp2-1 mutant.
Actin and the actin-related proteins (Arps), along with heat shock protein 70 and sugar kinases, belong to a protein superfamily characterized by a conserved structural domain known as the actin fold (Kabsch and Holmes, 1995). The actin fold domain encompasses the ATP-binding pocket, and the residues required for ATP binding are highly conserved among these functionally diverse proteins (Kabsch and Holmes, 1995). Although ATP-binding potential is predicted for Arp2p, it has not yet been shown that any members of the Arp2 family of proteins do, in fact, bind ATP. To address this question, we used the ATP analogue 8-azido-[α-32P]ATP, a photoaffinity probe that has been used previously to label the nucleotide-binding site of actin (Huang et al., 1983; Hegyi et al., 1986).
To compare the affinity of Arp2p and Arp2-E316K for ATP, we created a point mutation in the arp2+ coding region that would result in an amino acid substitution in the predicted ATP-binding pocket of Arp2p. In conventional actin, the hydroxyl group of Ser-14 binds to the γ-phosphate of ATP (Kabsch et al., 1990), and the substitution of alanine for Ser-14 results in a 40–60-fold decrease in the mutant actin’s affinity for ATP (Chen et al., 1995; Orlova et al., 1997). Because threonine replaces Ser-14 in Arp2p and is predicted to maintain similar contacts to the γ-phosphate of ATP (Kelleher et al., 1995), we created a mutant with alanine substituted for Thr-14 (Arp2-T12A).
The coding regions for these arp2 constructs were then cloned behind the thiamine-repressible promoter nmt1 in vector pRMH42, which also carries one copy of the 6× His tag and two copies of the myc epitope to produce proteins tagged at the N terminus. The myc-tagged Arp2 proteins, with the exception of Arp2-E316K, were capable of rescuing the arp2-1 mutant at the restrictive temperature and the arp2 null strain (our unpublished results). Expression of the myc-tagged Arp2 proteins was induced in wild-type cells (KGY246), and the tagged Arp2 proteins were isolated from native lysates with anti-myc 9E10 mAb. Photoaffinity labeling was then carried out by mixing immunoprecipitated proteins and 8-azido-[α-32P]ATP and irradiating the samples to promote cross-linking between the ATP analogue and the myc-tagged proteins. Under these native lysis conditions, we expected that Arp3p would be coimmunoprecipitated with the myc-tagged Arp2 protein. Because Arp3p is also predicted to bind ATP, we distinguished between Arp2p and Arp3p by an additional immunoprecipitation step. After cross-linking to the ATP analog, a fraction of each sample was boiled to denature the proteins, and then Arp2p or Arp3p was immunoprecipitated from the sample with anti-Arp2p or anti-Arp3p antibodies.
After UV irradiation, we found that both wild-type Arp2p and Arp3p were labeled by 8-azido-[α-32P]ATP, indicating that these actin-related proteins bind ATP as predicted (Figure 8A). The addition of cold ATP to the labeling reaction had the effect of a competitor of 8-azido-[α-32P]ATP and resulted in the reduction of labeled Arp2p and Arp3p, indicating a specific interaction between these proteins and ATP (Figure 8A). Detection of the labeled proteins required UV cross-linking (Figure 8A), and labeled proteins were not found in control lysates that lacked myc-tagged proteins (our unpublished results). As predicted, the mutant Arp2-T12A protein was also labeled by the ATP analogue, but to a much lesser degree than wild-type Arp2p (Figure 8A). Interestingly, the Arp2-E316K mutant protein had a reduced affinity for ATP compared with wild type Arp2p (Figure 8A). This reduction in labeling was not due to a reduction in the amount of immunoprecipitated myc-tagged Arp2-E316K, because comparisons of protein input levels indicated that the amounts of the tagged Arp2 proteins were nearly equal (Figure 8B). We did, however, detect more degradation of Arp2-E316K, suggesting that this protein may be less stable than wild-type Arp2p (Figure 8B). To directly compare the stability of wild-type Arp2 and Arp2-E316K, we performed a pulse-chase experiment by repressing expression of the tagged proteins in wild-type cells by adding thiamine to the culture medium. The relative levels of the myc-tagged Arp2 proteins in lysates prepared from cells collected at hourly intervals after repression of the nmt1 promoter were then determined by immunoblotting. These results clearly establish that the mutant Arp2-E316K protein turns over more rapidly than wild-type Arp2 protein after transcriptional repression (Figure 8C). These results are consistent with the reduction in Arp2p levels in the arp2-1 mutant (Figure 6B) and indicate that Arp2-E316K is less stable than wild-type Arp2p.
The most striking difference between wild-type Arp2p and Arp2-E316K was the failure to coimmunoprecipitate labeled Arp3p with Arp2-E316K (Figure 8A). To determine whether Arp3p failed to associate with Arp2-E316K or simply failed to be labeled by 8-azido-[α-32P]ATP in this reaction, we attempted to detect Arp3p by immunoblot analysis with the use of anti-Arp3p antibodies. We were unable to detect Arp3p coimmunoprecipitated with the myc-tagged Arp2-E316K by immunoblotting, suggesting that the interaction between Arp2p and other complex members is compromised by the E316K mutation (Figure 8D). We propose that in arp2-1 cells, Arp2-E316K is incorporated into the Arp2/3 complex in vivo and is stabilized by its interactions with other proteins at permissive temperature, but at restrictive temperature the E316K mutation disrupts this interaction, resulting in the loss of Arp2p (and Arc18p) from the complex and subsequent turnover of the mutant Arp2 protein.
DISCUSSION
The S. pombe arp2+ gene encodes an essential actin-related protein that belongs to the Arp2 family of actin-like proteins. As is the case with Arp2p homologues in other organisms, we have shown that S. pombe Arp2p is a subunit of a larger complex known as Arp2/3. Using both immunofluorescence and immunoelectron microscopy, we have demonstrated that Arp2p localizes to cortical actin patches but not to actin cables or the medial actin ring. We did not detect nuclear localization of Arp2p at any stage of the cell cycle. This localization pattern is similar to the localization patterns described for S. pombe Arp3p (McCollum et al., 1996), S. cerevisiae Arp2p (Moreau et al., 1996), and S. cerevisiae Arp3p (Winter et al., 1997). It differs from that of S. pombe Sop2p, which appears to localize to cortical actin patches and cable-like structures that extend the length of the cell (Balasubramanian et al., 1996).
A temperature-sensitive mutation in the arp2 gene results in actin cytoskeletal defects. The arp2-1 mutant arrests with randomly distributed cortical actin patches that show somewhat diminished staining with anti-actin antibodies compared with wild-type cells. In S. pombe, the distribution of actin patches changes throughout the cell cycle, with actin patches concentrated at areas of growth and cell wall deposition. Observation of live yeast cells with the use of GFP fusion proteins indicates that cortical actin patches are motile and that the distribution of patches within a single cell can change rapidly, suggesting that the rearrangement of actin patches throughout the cell cycle is accomplished by patch motility (Doyle and Botstein, 1996; Waddle et al., 1996). Thus, a likely explanation for the random distribution of actin patches in arp2-1 cells is a loss of actin patch motility. Loss of patch motility was observed directly in a conditional S. cerevisiae ARP3 mutant with the use of a GFP-Sac6 fusion protein that localizes to the actin patch, indicating that the Arp2/3 complex is required for patch motility in yeast (Winter et al., 1997). This mechanism may also provide an explanation for the thick, aberrantly formed septa in arp2-1 cells if the actin patches become immobile and continue to direct the localized secretion of cell wall material at the septum at restrictive temperature. Additionally, a reduction in actin patch motility may also provide a model for why the arp2-1 mutation suppresses cdc3-124. In this model, Cdc3-profilin retains partial function at semirestrictive temperature, resulting in delayed actin ring formation. This defect is suppressed in the cdc3-124 arp2-1 double mutant by a second delay in actin patch movement to the medial region of the cells, allowing a longer time for actin ring formation to occur.
The mechanism that drives cortical actin patch motility in yeast remains undefined. However, evidence exists that Arp2/3 plays a role in this process. By analogy to other models of Arp2/3 function in Listeria propulsion and actin dynamics at the leading edge of motile cells, it is possible that cortical actin patch movement is driven by actin polymerization (reviewed by Machesky, 1997; Welch et al., 1997c; Machesky and Gould, 1999). In this context, it is interesting to note the reduction in cortical actin patch staining in arp2-1 cells. It is possible that the arp2-1 mutation results in less polymerized actin at the patches compared with wild-type cells. Recent analysis of an actin point mutation (V159A) that results in a decrease in the rate of actin filament turnover in vivo, however, challenges this model, because cortical actin patch motility is apparently unaffected in this mutant (Belmont and Drubin, 1998; Belmont et al., 1999).
Whether the ATPase activity of Arp2p or Arp3p is essential for actin patch motility has yet to be determined. We have shown here the first evidence that Arp2p and Arp3p actually bind ATP with the photoaffinity probe 8-azido-[α-32P]ATP. This ATP analogue has been widely used to label other ATPases and interacts primarily with Lys-336 of actin (Hegyi et al., 1986), a residue conserved in Arp2p and Arp3p. Surprisingly, we found that the Arp2-E316K mutation resulted in a reduction in 8-azido-[α-32P]ATP labeling, suggesting that this mutation might affect ATP exchange. That the temperature sensitivity of the arp2-1 mutant is due to a reduction in ATP binding is an attractive model because the ATP cycle of Arp2p may be fundamental to its ability to interact with Arp3p and actin. This model is challenged, however, by our results that show that Arp2-T12A, which has a reduced affinity for ATP, is equally capable of binding Arp3p as wild-type Arp2p. For this reason, we favor a model in which the E316K mutation causes a general disruption in the functions that require residues in subdomain 3 of Arp2p. Based on comparisons with the structure of actin, these interactions probably include ATP and Arp3p binding (Kabsch and Vandekerckhove, 1992; Kelleher et al., 1995).
Random in vitro mutagenesis of the Drosophila melanogaster indirect flight muscle–specific actin gene Act88F also produced an E316K mutation. Characterization of this mutant demonstrated that Act88F-E316K accumulated to wild-type levels in vivo and assembled into nearly normal muscle structures but was functionally abnormal (Drummond et al., 1991a). Comparison of ATP affinity between Act88F-E316K and wild-type actin, as assayed by binding to ATP-agarose beads, showed no measurable differences (Drummond et al., 1992). Production of Act88F-E316K in vitro, on the other hand, resulted in a decrease in protein stability (Drummond et al., 1991b), similar to what we observed in vivo. Although we found a reduction in ATP binding by S. pombe Arp2-E316K compared with wild-type Arp2p, it remains possible that this difference reflects an inability of myc-tagged Arp2-E316K to be incorporated efficiently into the Arp2/3 complex. It is possible that Arp2p hydrolyzes ATP efficiently only as a mechanism of Arp2/3 function.
Consistent with this model, we have demonstrated with the use of sucrose gradient sedimentation and immunoprecipitation experiments that the Arp2/3 complex is unstable in arp2-1 cells. This phenotype is unique to the S. pombe arp2-1 mutant; sedimentation of the Arp2/3 complex is unaffected by the arp3-c1 and sop2-1 mutations (Balasubramanian et al., 1996; McCollum et al., 1996). The aberrant sedimentation profile of Arp2/3 from arp2-1 cells is due to the loss of Arp2p and Arc18p from the complex. We have also shown that the loss of Arc18p from the complex likely arises from its direct interaction with Arp2p. This result is in contrast to the model of Arp2/3 subunit interactions based on cross-linking studies, which predicts that Arc18p interacts solely with Arp3p (Mullins et al., 1997). Consistent with our results, recent analysis of an arc18 deletion strain in budding yeast demonstrated an increase in the amount of monomeric Arp2p by sedimentation analysis, suggesting that Arp2p and Arc18p interact in the complex (Winter et al., 1999b). Because Arc15p is not easily detected after cells are metabolically labeled with [35S]methionine, we were unable to conclude whether Arc15p was also lost from the complex at restrictive temperature. Interestingly, we were still able to immunoprecipitate the remaining Arp2/3 subunits after the loss of Arp2p and Arc18p, suggesting that a stable but smaller complex exists. Whether this smaller complex represents a biologically significant complex or a stage of Arp2/3 assembly remains to be determined.
These results are also consistent with our immunofluorescence studies of arp2-1 cells with antibodies against Arp2p and Arp3p. At the restrictive temperature, we were only able to detect diffuse cytoplasmic staining in arp2-1 cells with Arp2p antibodies, rather than localization at cortical actin patches, as seen in wild-type cells. In the same cells, however, we were able to detect Arp3p localization to cortical actin patches, indicating that at least this subunit of the Arp2/3 complex is localized properly in the absence of Arp2p. The mechanism by which Arp2/3 is localized to actin patches in S. pombe is unknown; however, our results suggest that the complex is probably anchored by a subunit other than Arp2p or Arc18p.
We suggest that endogenous S. pombe Arp2-E316K is stabilized in vivo by interactions with other Arp2/3 subunits at the permissive temperature but that the mutant protein undergoes a conformational change in subdomain 3 at the restrictive temperature that interferes with its ability to bind ATP and/or Arp3p. Disruption of the interaction between Arp2p and Arp3p by the mutation results in the separation of Arp2p and Arc18p from the cortical actin patch and subsequent turnover of the mutant Arp2 protein.
ACKNOWLEDGMENTS
We thank Dr. Dannel McCollum for initially identifying the arp2-1 mutant strain and Anna Feoktistova for constructing Sop2HA. We also thank Dr. Ian Hagan and Dr. Tony Carr for providing pRMH42 and Dr. Sean Hemmingsen for two-hybrid vectors. This work was supported by the Howard Hughes Medical Institute, of which K.L.G is an associate investigator.
REFERENCES
- Amatruda JF, Cooper JA. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J Cell Biol. 1992;117:1067–1076. doi: 10.1083/jcb.117.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR. In vivo functions of actin-binding proteins. Curr Opin Cell Biol. 1998;10:102–111. doi: 10.1016/s0955-0674(98)80092-6. [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bailly M, Macaluso F, Cammer M, Chan A, Segall JE, Condeelis JS. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J Cell Biol. 1999;145:331–345. doi: 10.1083/jcb.145.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Feoktistova A, McCollum D, Gould KL. Fission yeast Sop2p: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. EMBO J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, Sazer S, Gould KL. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Gould KL. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1997;283:494–506. doi: 10.1016/s0076-6879(97)83039-x. [DOI] [PubMed] [Google Scholar]
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Drubin DG. The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J Cell Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Orlova A, Drubin DG, Egelman EH. A change in actin conformation associated with filament instability after Pi release. Proc Natl Acad Sci USA. 1999;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Peng J, Pedram M, Swenson CA, Rubenstein PA. The effect of the S14A mutation on the conformation and thermostability of Saccharomyces cerevisiae G-actin and its interaction with adenine nucleotides. J Biol Chem. 1995;270:11415–11423. doi: 10.1074/jbc.270.19.11415. [DOI] [PubMed] [Google Scholar]
- Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RA, Griffiths DJ, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- Demeter J, Morphew M, Sazer S. A mutation in the RCC1-related protein pim1 results in nuclear envelope fragmentation in fission yeast. Proc Natl Acad Sci USA. 1995;92:1436–1440. doi: 10.1073/pnas.92.5.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Hennessey ES, Sparrow JC. Characterization of missense mutations in the Act88F gene of Drosophila melanogaster. Mol Gen Genet. 1991a;226:70–80. doi: 10.1007/BF00273589. [DOI] [PubMed] [Google Scholar]
- Drummond DR, Hennessey ES, Sparrow JC. Stability of mutant actins. Biochem J. 1991b;274:301–303. doi: 10.1042/bj2740301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Hennessey ES, Sparrow JC. The binding of mutant actins to profilin, ATP and DNase I. Eur J Biochem. 1992;209:171–179. doi: 10.1111/j.1432-1033.1992.tb17274.x. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Frankel S, Mooseker MS. The actin-related proteins. Curr Opin Cell Biol. 1996;8:30–37. doi: 10.1016/s0955-0674(96)80045-7. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gould KL, Moreno S, Owen DJ, Sazer S, Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Hegyi G, Szilagyi L, Elzinga M. Photoaffinity labeling of the nucleotide binding site of actin. Biochemistry. 1986;25:5793–5798. doi: 10.1021/bi00367a067. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Huang CK, Hill JM, Jr, Bormann BJ, Mackin WM, Becker EL. The photoaffinity probe 8-N3[alpha-32P]ATP labels the ATP-binding sites of rabbit neutrophil and skeletal muscle actin. FEBS Lett. 1983;159:145–149. doi: 10.1016/0014-5793(83)80434-7. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Holmes KC. The actin fold. FASEB J. 1995;9:167–174. doi: 10.1096/fasebj.9.2.7781919. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Vandekerckhove J. Structure and function of actin. Annu Rev Biophys Biomol Struct. 1992;21:49–76. doi: 10.1146/annurev.bb.21.060192.000405. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller JP, Helfman DM, Schroer TA. A vertebrate actin-related protein is a component of a multisubunit complex involved in microtubule-based vesicle motility. Nature. 1992;359:244–246. doi: 10.1038/359244a0. [DOI] [PubMed] [Google Scholar]
- Machesky LM. Cell motility: complex dynamics at the leading edge. Curr Biol. 1997;7:R164–R167. doi: 10.1016/s0960-9822(97)70079-4. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Gould KL. The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol. 1999;11:117–121. doi: 10.1016/s0955-0674(99)80014-3. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;31:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASP-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Reeves E, Wientjes F, Mattheyse FJ, Grogan A, Totty NF, Burlingame AL, Hsuan JJ, Segal AW. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 1997;328:105–112. doi: 10.1042/bj3280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Way M. Actin branches out. Nature. 1998;394:125–126. doi: 10.1038/28039. (News). [DOI] [PubMed] [Google Scholar]
- Marks J, Hagan IM, Hyams JS. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J Cell Sci Suppl. 1986;5:229–241. doi: 10.1242/jcs.1986.supplement_5.15. [DOI] [PubMed] [Google Scholar]
- May RC, Hall ME, Higgs HN, Pollard TD, Chakraborty T, Wehland J, Machesky LM, Sechi AS. The Arp2/3 complex is essential for the actin-based motility of Listeria monocytogenes. Curr Biol. 1999;9:759–762. doi: 10.1016/s0960-9822(99)80337-6. [DOI] [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Madania A, Martin RP, Winsor B. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998a;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Kelleher JF, Pollard TD. Actin’ like actin? Trends Cell Biol. 1996;6:208–212. doi: 10.1016/0962-8924(96)20017-0. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Kelleher JF, Xu J, Pollard TD. Arp2/3 complex from Acanthamoeba binds profilin and cross-links actin filaments. Mol Biol Cell. 1998b;9:841–852. doi: 10.1091/mbc.9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia I, Maciver SK, Morandini P. An actin-related protein from Dictyostelium discoideum is developmentally regulated and associated with mitochondria. FEBS Lett. 1995;360:235–241. doi: 10.1016/0014-5793(95)00111-l. [DOI] [PubMed] [Google Scholar]
- Olmsted JB. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981;256:11955–11957. [PubMed] [Google Scholar]
- Orlova A, Chen X, Rubenstein PA, Egelman EH. Modulation of yeast F-actin structure by a mutation in the nucleotide-binding cleft. J Mol Biol. 1997;271:235–243. doi: 10.1006/jmbi.1997.1163. [DOI] [PubMed] [Google Scholar]
- Prentice HL. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Schroer TA, Bingham JB, Gill SR. Actin-related protein 1 and cytoplasmic dynein-based motility: what’s the connection? Trends Cell Biol. 1996;6:212–215. doi: 10.1016/0962-8924(96)20014-5. [DOI] [PubMed] [Google Scholar]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Schwob E, Martin RP. New yeast actin-like gene required late in the cell cycle. Nature. 1992;355:179–182. doi: 10.1038/355179a0. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Waddle JA, Karpova TS, Waterston RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997a;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Holtzman DA, Drubin DG. The yeast actin cytoskeleton. Curr Opin Cell Biol. 1994;6:110–119. doi: 10.1016/0955-0674(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997b;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Welch MD, Mallavarapu A, Rosenblatt J, Mitchison TJ. Actin dynamics in vivo. Curr Opin Cell Biol. 1997c;9:54–61. doi: 10.1016/s0955-0674(97)80152-4. [DOI] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr Biol. 1999a;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- Winter D, Podtelejnikov AV, Mann M, Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- Winter DC, Choe EY, Li R. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci USA. 1999b;96:7288–7293. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, To W, Abo A, Welch MD. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]