Abstract
Mutations in PTEN-induced putative kinase 1 (PINK1) are a cause of autosomal recessive familial Parkinson's disease (PD). Efforts in deducing the PINK1 signaling pathway have been hindered by controversy around its subcellular and submitochondrial localization and the authenticity of its reported substrates. We show here that this mitochondrial protein exhibits a topology in which the kinase domain faces the cytoplasm and the N-terminal tail is inside the mitochondria. Although deletion of the transmembrane domain disrupts this topology, common PD-linked PINK1 mutations do not. These results are critical in rectifying the location and orientation of PINK1 in mitochondria, and they should help decipher its normal physiological function and potential pathogenic role in PD.
Keywords: parkin, Parkinson's disease, mitochondria, topology
Parkinson's disease (PD), the second most common neurodegenerative disorder, is a sporadic condition that can occasionally be inherited (1). The rationale for studying the rare genetic forms of PD is based on the phenotypic similarity between the familial and sporadic forms of the disease, implying that the two share important pathogenic mechanisms. Among the different gene products associated with familial PD (2), PTEN-induced putative kinase-1 (PINK1) is localized to the mitochondria (3–10), an organelle strongly linked to PD pathogenesis. Although recessively inherited PD mutations in PINK1 are found throughout the protein, they are most commonly found in the only recognized functional domain of PINK1 (3, 11, 12), which is a serine/threonine kinase domain similar to that in the Ca2+/calmodulin kinase family (13, 14). Overexpression of wild-type PINK1 can rescue the phenotype caused by PINK1 mutations in Drosophila (15, 16), supporting the notion that the mutated allele gives rise to a loss-of-function phenotype.
Human PINK1 is a 581-aa polypeptide with a predicted N-terminal mitochondrial targeting signal (MTS), consistent with the observation that PINK1 localizes to mitochondria (Fig. 1A) (3–10). Loss-of-function mutations in the gene encoding parkin (an E3 ubiquitin ligase) cause the most frequent forms of recessive familial PD (17). In Drosophila, parkin is thought to operate within the same molecular pathway as PINK1 to modulate mitochondrial morphology (15, 16, 18), an intriguing observation given the fact that parkin has been reported to essentially be cytosolic (19).
Fig. 1.
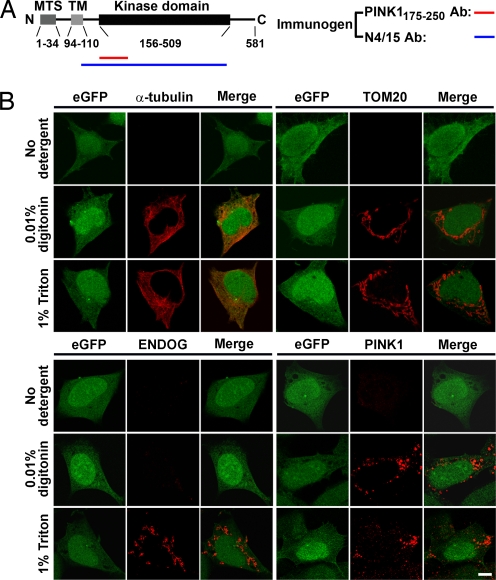
The kinase domain of mitochondrial PINK1 is accessible from the cytoplasm. (A) Schematic representation of PINK1 structure. The immunogens used to produce the antibodies (Abs) PINK1175–250 and N4/15 are indicated by the red and blue lines, respectively. (B) SH-SY5Y cells expressing PINK1-IRES-eGFP stably are immunolabeled for a cytosolic protein (α-tubulin), a cytoplasm-facing OMM protein (TOM20), and an IMS protein (ENDOG) (red signals). eGFP is used as a general cell marker (green signals). PINK1 is labeled using PINK1175-250 antibody. (Scale bar, 5 μm.)
Both the subcellular and submitochondrial locations of PINK1 and the authenticity of its reported substrates have been controversial. Although it is agreed that PINK1 is a mitochondrial-targeted protein, PINK1 has been reported to reside in the inner mitochondrial membrane (IMM) (6, 7, 9, 20), the mitochondrial intermembrane space (IMS) (6, 8, 9), the outer mitochondrial membrane (OMM) (7), and even the cytoplasm (21–24). Furthermore, the tumor necrosis factor type 1 receptor-associated protein (TRAP1) and the serine protease HtrA2/Omi have been identified as putative PINK1 substrates (8, 9). Although TRAP1 is localized mainly in the mitochondrial matrix, it has also been identified in the IMS (9) and at extramitochondrial subcellular sites (25). HtrA2/Omi is a mammalian serine protease that is localized in the IMS of healthy cells, but during apoptosis it is released into the cytosol (26). The subcellular and submitochondrial locations of PINK1 are critical to identifying its bona fide substrates and related signaling pathways. For this reason and in light of the aforementioned inconsistencies, we sought to reexamine the topology of PINK1 and to address more specifically where its kinase domain resides within mitochondria. We show here that PINK1 spans the OMM, with its kinase domain facing the cytoplasm. This topology relies on a transmembrane (TM) domain located just after the MTS (Fig. 1A), with the N-terminal end of PINK1 contained within the mitochondria. Common pathogenic PINK1 mutations have no effect on the topology of the protein. These new details on PINK1 topology may shed light on the molecular model underlying the genetic interaction of PINK1 and parkin and suggest that the loss of phosphorylation of cytosolic substrates or the loss of PINK1 interactions with cytosolic proteins may underlie PINK1-related neurodegeneration in familial PD.
Results and Discussion
To define the submitochondrial location of the PINK1 kinase domain that extends from amino acids 156–509 (Fig. 1A), we performed immunocytochemistry on human PINK1-transfected neuroblastoma SH-SY5Y cells treated with either digitonin or Triton X-100 to respectively permeabilize the plasma membrane only or all cellular bilayer membranes (27). SH-SY5Y cells transfected stably with a cDNA plasmid expressing full-length human PINK1-IRES-eGFP were probed with antibodies raised against specific subcellular proteins (Fig. 1B) and against an epitope of the PINK1 kinase domain located between amino acids 175–250 (Fig. 1A). In the absence of detergent, none of the antibodies generated specific immunoreactivity (Fig. 1B). Cells permeabilized with 0.01% digitonin showed positive immunofluorescence for α-tubulin, a cytoplasmic protein, and for TOM20, an OMM protein, but not for endonuclease-G (ENDOG), an IMS protein (Fig. 1B), or for cytochrome c oxidase subunit II (COX II), an IMM protein (data not shown). These results indicate that the digitonin permeabilization allowed the antibodies to gain access to the cytoplasm but not to the inside of the mitochondria. Cells permeabilized with 1% Triton X-100 showed positive immunofluorescence for all of the antibodies (Fig. 1B), indicating that the OMM was disrupted, thereby allowing the antibodies to gain access to the inside of the mitochondria. By applying these permeabilization protocols, we found that the anti-PINK1175–250 antibody generated a positive immunofluorescence signal in both digitonin- and Triton X-100-treated cells (Fig. 1B), suggesting that the epitope recognized by this Ig is accessible when the OMM is intact. Because the validity of this interpretation depends on the specificity of the anti-PINK1 antibody, it is critical to mention that PINK1 immunoreactivity was abolished by incubation of the PINK1175–250 antibody with excess PINK1 blocking peptide [supporting information (SI) Fig. S1]; a similar result was obtained upon incubation of the anti-TOM20 antibody with an excess of recombinant TOM20 (Fig. S1). Human PINK1 has been localized by immunohistochemistry in both the mitochondria and the cytosol of transfected cell lines (22, 23). Consistent with this dual subcellular distribution, we observed a punctate PINK1 immunoreactivity in both digitonin- and Triton X-100-treated cells (Fig. 1B) that colocalized with the mitochondrial fluorescent probe MitoTracker and that was superimposed on a diffuse PINK1 immunofluorescence over the cytoplasm (Fig. S1C). Cells transfected with an empty vector did not show any specific immunofluorescent signal (data not shown). Thus, our results support the notion that at least a portion of PINK1 is mitochondrial and that, at this location, the PINK1 kinase domain is exposed to the cytoplasm. Similar results were obtained with neuronal-like MES23.5 clonal cells (data not shown).
Given the above results, we then tested the strength of the PINK1 association with mitochondria compared with a series of mitochondrial proteins (Fig. 2A). Consistent with previous studies (6), we found that human PINK1 can only be partially solubilized or not at all by extensive detergent or sodium carbonate treatments of purified mitochondria from SH-SY5Y cells stably overexpressing the full-length human PINK1, respectively (Fig. 2B). These results suggest that PINK1 is a resident mitochondrial protein, physically integrated within mitochondrial membrane(s).
Fig. 2.
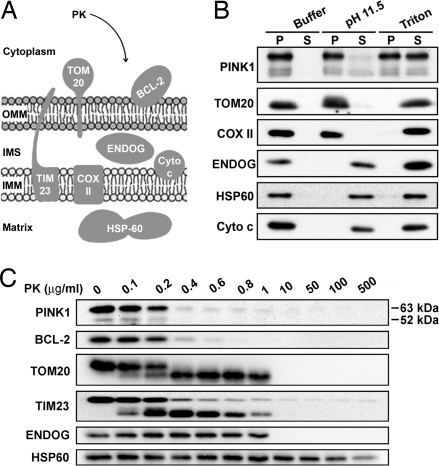
PINK1 is a mitochondrial membrane integral protein whose kinase domain localizes at OMM. (A) Schematic representation of the submitochondrial compartment protein markers used in this study. (B) Mitochondria isolated from SH-SY5Y cells expressing PINK1 stably are subjected to carbonate extraction. Then, pellets (particulate fraction, P) and supernatants (soluble fraction, S) are analyzed by Western blotting using antibodies raised against PINK1 (PINK1175–250), HSP60, ENDOG, peripheral membrane proteins (Cyto c), and membrane integral proteins (TOM20 and COX II). (C) Mitochondria isolated from SH-SY5Y cells expressing wild-type PINK1 stably are incubated with or without PK before analysis by immunoblotting using antibodies against PINK1 (PINK1175–250), BCL-2, TOM20, TIM23, ENDOG, and HSP60.
To define the topology of the PINK1 kinase domain, we performed protease protection assays. Mitochondria isolated from SH-SY5Y cells expressing full-length human PINK1 were incubated with increasing concentrations of protease K (PK) in the presence or absence of Triton X-100. To verify the intactness of the isolated mitochondria, we analyzed the accessibility of PK to selected proteins of the OMM, the IMS, and the matrix (Fig. 2A). We found that at concentrations of 0.2–1.0 μg/ml PK, the mitochondrial peripheral protein BCL-2 and the cytoplasm-exposed C terminus of the OMM receptor TOM20 were degraded (Fig. 2C). In contrast, an IMS protein, ENDOG, and a mitochondrial matrix protein heat shock protein-60 (HSP60) were protease-resistant under these conditions (Fig. 2C). These data indicate that the isolated mitochondrial matrix remained impermeant to PK, at least at concentrations up to 1 μg/ml. Under these conditions, PINK1 immunoreactivity was lost in a PK-concentration-dependent manner, a pattern paralleled by that for BCL-2 (Fig. 2C), monoamine oxidase-B, another OMM protein (data not shown), and the C terminus of TOM20 (Fig. 2C), all of which face the cytosol. Note that we have confirmed the specificity of the key antibodies with an excess of their respective blocking peptides or protein recombinants (Fig. S2). The PK results were confirmed by using a mouse monoclonal antibody also raised against the PINK1 kinase domain (NeuroMab Clone N4/15) (Fig. S3B) (the validation of the specificity of N4/15 is shown in Fig. S3C), and with hemagglutinin (HA)-tagged human PINK1 detected with an anti-HA antibody (Fig. S3D). Similar results were also obtained in other neuronal-like cell lines, such as human neuroblastoma M17 (Fig. S3E) and mouse MES23.5 (Fig. S3B) cells, and with trypsin, another nonspecific protease (data not shown). In the presence of 1% Triton X-100, all of the studied proteins, including PINK1, were equally sensitive to PK proteolysis (Fig. S3F). Thus, the recognized epitope of the kinase domain in both main PINK1 species (≈63 and ≈52 kDa) seem to be accessible to general proteases, such as PK and trypsin, in intact mitochondria.
TIM23, an essential component of the mitochondrial protein import machinery located in the IMM, has been used as a marker to establish the localization of PINK1 inside the mitochondria (6). Yet, Fig. 2C shows a proteolytic profile of TIM23 similar to that of the OMM protein TOM20 and to the peripheral protein BCL-2. Although the C-terminal half of TIM23 (approximate residues 100–222) is indeed integrated into the IMM, residues 50–100 are exposed to the IMS and the N-terminal domain (residues 1–50) is exposed on the surface of the OMM (28). Thus, even though TIM23 is considered to be an IMM protein, this protein may display an OMM proteolytic profile with some region-specific antibodies, because of its N-terminal end projecting to the cytoplasm. This possibility explains why we found that the TIM23 and TOM20 proteolytic profiles were similar, because we used an antibody raised against TIM23 residues 5–126 (BD Transduction Laboratories), a result that is consistent with a previous study in mammalian cells (29).
It is worth noting that the cytoplasmic-facing orientation of the PINK1 kinase domain was not altered by common pathogenic mutations such as A217D, T313M (Fig. S3G), or L347P (data not shown), suggesting that the pathological effects of these mutated proteins are probably not related to a disrupted localization of PINK1 functional domain. The kinase function was also not necessary to establish PINK1 kinase domain topology, because expression in transfected SH-SY5Y cells of a “dead kinase” PINK1 mutant (K219M) had no effect on PINK1's orientation (data not shown).
We next explored whether the proposed PINK1 topology obtained by overexpression faithfully reflects that of the endogenous protein. PINK1-specific mRNA is detected in almost all tissues, with highest levels in heart, skeletal muscle, and testis (14, 30, 31). We also found detectable levels of PINK1 mRNA in both SH-SY5Y cells and mouse brain (Fig. 3A). Despite the detectable levels of PINK1 mRNA, we were unable to detect endogenous PINK1 protein in a variety of cell lines, including SH-SY5Y and primary ventral midbrain cultures and adult mouse and human brain tissues, despite trying 15 different anti-PINK1 antibodies (Table S1 and Fig. 3B, left lane). The only possible exception to these negative results was the protein extracts from HeLa cells. Indeed, the latter revealed faint immunoreactive bands on Western blots consistent with those corresponding to PINK1 species (Fig. 3C Right, first and second lanes). Similar observations were made by Exner and collaborators (10). These results suggested that available PINK1 antibodies are insufficiently sensitive for detecting low levels of endogenous PINK1.
Fig. 3.
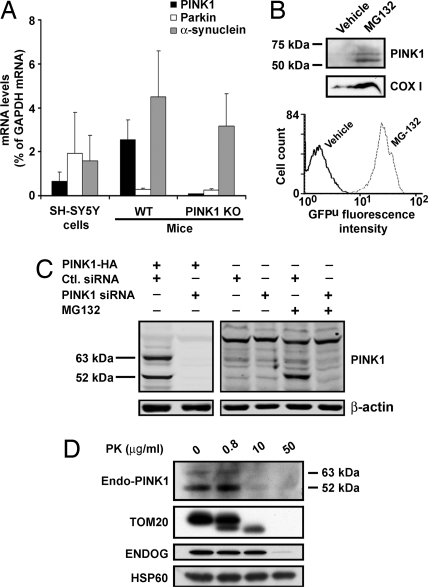
Topology of endogenous PINK1 using PINK1175–250 antibody. (A) mRNA levels of endogenous PINK1, parkin, and α-synuclein in SH-SY5Y cells and in PINK1 wild-type (WT) and knockout (KO) mouse brain. Total RNA extracted from each sample is quantified by real-time PCR (n = 3). Values represent means ± SD. PINK1 mRNA in KO mouse brain is below the limit of detection. (B) Using the antibody PINK1175–250, there is no detectable signal for endogenous PINK1 in SH-SY5Y cells (vehicle control, left lane), whereas endogenous PINK1 is detected in the presence of the proteasome inhibitor MG132 (right lane). Western blot analysis of mitochondrial fractions shows an increased level of COX I with MG132 treatment. Accumulation of GFPu, as shown by the increased GFP fluorescence intensity, indicates the successful inhibition of the proteasome pathway by MG132. (C) siRNA knockdown of PINK1. (Left) HeLa cells are cotransfected with either PINK1-HA/scramble control siRNA or PINK1-HA/PINK1 siRNA. Overexpressed full-length PINK1 and truncated ≈52-kDa PINK1 are diminished specifically by the PINK1 siRNA. (Right) Endogenous PINK1 is barely detected in whole HeLa cell lysates in the absence of MG-132, but it is clearly detected in the presence of MG-132. Endogenous PINK1 signals are markedly reduced by the PINK1 siRNA knockdown. (D) After MG132 treatment of SH-SY5Y cells, mitochondria are isolated and processed for PK protection assay as in Fig. 2.
The accumulation of selected mitochondrial proteins posttranscriptionally has been reported in cells treated with proteasome inhibitors (32). To increase the content of endogenous PINK1, we transfected SH-SY5Y cells stably to express a reporter consisting of a short ubiquitination signal sequence fused to the C terminus of GFP (GFPu) (33) (R. Kopito, Stanford University, Stanford, CA) and exposed these cells for 24 h to 2 μM proteasome inhibitor Z-Leu-Leu-Leu-al (MG132). Under these experimental conditions, which caused <10% cell death, and in agreement with the work of Margineantu and collaborators (32), we detected an increased signal for the cytochrome c oxidase subunit I (COX I) in Western blot analysis using protein extracts of purified mitochondria from SH-SY5Y cells (Fig. 3B). This strategy also enabled us to detect two main immunoreactive bands which coincided with the molecular mass of full-length and cleaved PINK1 (6, 21) (Fig. 3B). To confirm the identity of these bands, we used two different PINK1 siRNA constructs to knock down PINK1 mRNA (see Fig. S4 for siRNA validation). We were able to knock down PINK1 mRNA by ≈60% in SH-5YSY cells and by >80% in HeLa cells (Fig. S4 A and B), which was accompanied by a striking reduction in signal intensity on the Western blot of the PINK1 species (Fig. 3C). In contrast to the situation observed in PINK1 overexpression, here the ≈52-kDa species appear as the dominant form of PINK1 (Fig. 3C). After proteasome inhibition, a similar observation was reported by Takatori et al. (23), which led these authors to suggest that the ≈52-kDa PINK1 fragment may be more prone to proteasomal degradation. We found that the ≈52-kDa and ≈63-kDa endogenous species exhibited a proteolytic profile with PK similar to that found for the human overexpressed protein (Fig. 3D). These data thus support the fact that endogenous PINK1 is an OMM protein with its kinase domain facing the cytosol.
Most proteins destined for import into the mitochondria bear targeting information, typically at their N termini (34). The prediction programs iPSORT (http://biocaml.org/ipsort/iPSORT/), MitoProt II (ftp://ftp.ens.fr/pub/molbio/), and TargetP (http://www.cbs.dtu.dk/sevices/TargetP) predict a MTS within the N-terminal end of human PINK1 contained in amino acids 1–30, 1–34, or 1–76, respectively. Consistent with this notion, it has been shown that the first 93 amino acids (6) and the first 34 amino acids (20) suffice to target GFP to the mitochondria. To determine whether this region of PINK1 is required for its importation into mitochondria, we constructed cDNA plasmids expressing PINK1 truncated at its first 34, 91, or 150 amino acids (Fig. 4A). We found that, relative to their respective total cellular expression, ≈40 ± 3% of full-length PINK1 and ≈56 ± 1% of PINK1 lacking the first 34 amino acids (PINK1Δ1–34) were mitochondrial and displayed similar topologies (Fig. 4 B and C). With further N-terminal truncation, ≈26 ± 4% of PINK1Δ1–91 and only ≈6 ± 4% of PINK1Δ1–150 were mitochondrial (Fig. 4B). The differential subcellular localization of the products encoded by the PINK1 deletion constructs was confirmed with immunocytochemistry (Fig. S5). In keeping with these results, Haque et al. (22) and Takatori et al. (23) have also found that, upon deletion of amino acids 1–111 or 1–108, respectively, PINK1 could no longer be imported into the mitochondria. Collectively, these findings indicate that the N-terminal end of PINK1 contains the MTS and that its composition is complex and not limited to the first 34 amino acids (20).
Fig. 4.
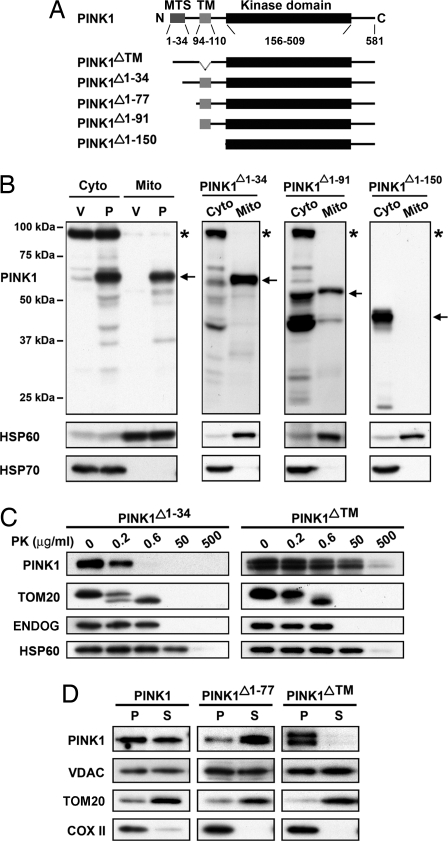
The TM domain anchors PINK1 to the OMM by using PINK1175–250 antibody. (A) Schematic representation of PINK1 constructs with various deletions. (B) SH-SY5Y cells transfected stably with empty vector (V), PINK1 (P), PINK1Δ1–34, PINK1Δ1–91, and PINK1Δ1–150 are harvested to prepare cytosol and mitochondrial fractions. All preparations are analyzed by Western blotting. The percentages of various PINK1 constructs, including all degraded forms, which localize to mitochondria, are quantified by using Scion Image software and expressed as mean ± SD (n = 3). Note that for PINK1Δ1–91, a large fraction of its cytosolic portion was degraded and for PINK1Δ1–150 films are exposed for a shorter time to avoid saturation (thus the nonspecific band is not seen). Arrows, full-length constructs. Asterisks, nonspecific bands. (C) Mitochondria are isolated from SH-SY5Y cells expressing PINK1Δ1–34 or PINK1ΔTM stably followed by PK treatment as in Fig. 2. (D) Mitochondria are purified from SH-SY5Y cells expressing PINK1, PINK1Δ1–77, or PINK1ΔTM stably followed by permeabilization of the OMM with digitonin. After centrifugation, IMM (pellet, P) is separated from the OMM (supernatant, S). The purity of the fractions is assessed by probing the prepared fractions with antibodies to VDAC, TOM20, and COX II.
Typically, the MTS is cleaved after import into the mitochondria by specific proteases (34), converting the full-length precursor polypeptide into the mature protein. In keeping with this concept, published studies on PINK1 have designated the ≈63-kDa form as the precursor and the ≈52-kDa form as mature PINK1 (6, 21). This interpretation has been corroborated by at least one study in which, by using an in vitro mitochondrial import assay, it was shown that the appearance of the ≈52-kDa fragment required a mitochondrial transmembrane potential (6), as typically seen with this type of protein maturation process (34). However, using protein extracts from transfected SH-SY5Y cells expressing the three N-terminal-truncated PINK1 species, the Western blot analysis pattern argues against the cleavage site being either at amino acid 34 or amino acid 77 as predicted, but rather between amino acids 91–101 (Fig. S6). This raises the interesting possibility that PINK1 mitochondrial processing is mediated by an atypical mitochondrial mechanism. Thus, it is likely that PINK1 cleavage does not reflect a canonical maturation process as initially thought.
In addition to a MTS, some mitochondrially targeted proteins contain in their N-terminal region a hydrophobic sequence called a stop-transfer signal to help anchor the imported polypeptide to the mitochondrial membrane (34). We thus analyzed the sequence of the N-terminal segment of PINK1 that follows the MTS and precedes the kinase domain (Fig. 1A) by using TopPred (ftp://ftp.ens.fr/pub/miobio), TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), and DAS (http://www.sbc.su.se/∼miklos/DAS/) transmembrane prediction programs. This analysis revealed a highly probable TM domain (>2 on the Kyte-Doolittle scale) of 16 residues located at amino acids 94–110. Using the PINK1175–250 antibody, deletion of this sequence (PINK1Δ91–117) did not prevent the import of the protein to the mitochondrion but conferred a PK proteolytic profile similar to that of the matrix protein HSP60 (Fig. 4C). Interestingly, on 10% SDS/PAGE, PINK1Δ91–117 resolved as a doublet and not as a single species like wild-type PINK1, indicating that once mislocated in the matrix, some of PINK1Δ91–117 is cleaved by mitochondrial matrix proteases or is subjected to a posttranslational modification that alters its physical properties. Although additional work is required to clarify this finding, our results indicate that the hydrophobic segment of PINK1 at amino acids 94–110 is crucial to anchor PINK1 to the mitochondrial membrane and to ensure that the kinase domain faces the cytoplasm.
In light of the above data, we next tested our hypothesis that PINK1 is inserted into the OMM by preparing submitochondrial fractions (i.e., mitoplasts) using digitonin to peel the OMM (35). After ultracentrifugation, the soluble fractions were enriched with OMM proteins, such as TOM20, and were free of IMM proteins, such as COX II, whereas the particulate fractions showed the reverse (Fig. 4D). Probing the submitochondrial fractions from cells expressing wild-type PINK1 with the PINK1175–250 antibody revealed PINK1 immunoreactivity in both submitochondrial fractions (Fig. 4D). However, in submitochondrial fractions from cells expressing a deletion of the TM (PINK1ΔTM), PINK1 was recovered entirely in the IMM-containing fractions, whereas in cells expressing a truncation of the first 77 amino acids (PINK1Δ1–77), PINK1 was recovered mainly in the OMM-containing fractions. These results suggest that PINK1 spans the OMM, but they do not allow for precise definition of the intramitochondrial topology of the PINK1 N terminus. This latter aspect will have to be elucidated by future studies.
As noted earlier, the localization of PINK1 is controversial (6–9, 20–24). Although the exact methodologies used by different groups (e.g., localization by using electron microscopy vs. immunoblotting, use of different methods of subcellular fractionation, use of different constructs and epitope tags, expression using transient vs. stable transfections, use of different region-specific antibodies, use of different protease-protection protocols) make it difficult to resolve the discrepancies among the reported localizations of PINK1, our results, which support a model in which PINK1 spans the OMM with the N-terminal end inside the mitochondria and the C-terminal kinase domain facing the cytosol, may help reconcile these views. The questionable specificity of the large number of available PINK1 antibodies may also have contributed to the confusion. Therefore, the data reported in Table S1 may represent a unique resource for researchers in the field to select a suitable immuno reagent for their work.
A topological model in which the C-terminal segment spans the OMM and projects to the cytoplasm, while the N-terminal tail is inside mitochondria, has also been documented for Mmm1 (36), a yeast protein required for normal mitochondrial morphology and mtDNA stability (37, 38). Despite the fact that no significant structural homology exists between Mmm1 and PINK1, there is a growing body of evidence that, like Mmm1, PINK1 influences mitochondrial morphology (10, 15, 16, 18). In this context, our data on PINK1 topology offer an important new piece of information supporting the idea that the role of PINK1, and in particular of its OMM-localized kinase domain, may involve phosphorylation of molecules involved in mitochondrial dynamics that project to or are in the cytoplasm. This PINK1 topology may also be relevant to the reported genetic interaction between PINK1 and the essentially cytosolic E3 ubiquitin ligase parkin (15, 16), which according to our model could now interact with the part of PINK1 that extends outside of the mitochondria.
Materials and Methods
Molecular Cloning.
For stable PINK1-overexpressing cells, full-length PINK1 cDNA (OriGene) was subcloned into the mammalian expression vector pIRES2-EGFP. A HA epitope (YPYDVPDYA) was inserted at the C terminus of PINK1 by PCR. PINK1 mutants (A217D, T313M, L347P, and K219M) were generated by using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). PINK1 deletions [PINK1Δ1–34; PINK1Δ1–49; PINK1Δ1–63; PINK1Δ1–77; PINK1Δ1–91; PINK1Δ1–117; PINK1Δ1–150; PINK1ΔTM (91–117-aa deletion)] were generated by PCR. For transient transfection, eGFP cDNA was removed from the constructs to only express PINK1. All constructs were confirmed by sequencing.
Cell Culture and Stable PINK1-Overexpressing Cells.
Human neuroblastoma SH-SY5Y (D. Yamashiro, Columbia University, New York), M17 (L. Petrucelli, Mayo Clinic, Jacksonville, FL), and HeLa and COS7 cells (American Type Culture Collection) were maintained in DMEM/Ham's F-12 medium (Invitrogen) with 10% FBS and 1% (vol/vol) penicillin-streptomycin. MES23.5 cells (M. LaVoie, Harvard University, Cambridge, MA) were maintained in DMEM/F12 medium containing 1% N-2 supplement and 2% FBS. For stable overexpression, PINK1-IRES-eGFP plasmids were transfected into cells by using FuGENE HD (Roche Applied Science). eGFP-positive cells were selected by the FACSVantage cell sorter (BD Biosciences, Franklin Lakes, NJ), and flow cytometric analysis was performed on a FACScan cytometer (BD Biosciences, Franklin Lakes, NJ).
Immunocytochemistry.
Cells were grown on glass coverslips, transiently transfected with various constructs by using either FuGENE HD or Lipofectamine 2000 (Invitrogen), and processed as previous described (39). Primary antibodies were: rabbit anti-PINK1 antibody (PINK1175–250) (Novus), mouse anti-myc antibody (Abcam), rabbit anti-ENDOG antibody (ProSci), and mouse anti-TOM20 (BD Biosciences, Franklin Lakes, NJ). Secondary antibodies were Igs of the appropriate species conjugated to either Alexa Fluor 594 or Alexa Fluor 488 (Invitrogen). For mitochondrial labeling, cells were incubated with MitoTracker Red CMXRos or MitoTracker Deep Red 633 (Invitrogen). For the liquid absorption tests, PINK1 control peptide was purchased from Novus and the recombinant TOM20 protein from Abnova.
Western Blot Analysis.
Western blot analysis was performed as described in ref. 40 by using the primary antibodies raised against PINK1175–250 (Novus), PINK1 N4/15 (NeuroMab), HA (Santa Cruz Biotechnology), Myc (Abcam), TOM20 (BD Biosciences, Franklin Lakes, NJ), and β-actin (Sigma Aldrich). Band quantification was performed by using Scion Image software, and all immunoblot data were from three independent experiments. In addition to the ECL method, the Odyssey Infrared Imaging system was used for fluorescence immunoblotting with IRDye dye-labeled secondary antibodies (LI-COR). For liquid absorption tests, PINK1 blocking peptide and recombinant TOM20 protein were obtained from the sources indicated above, in Immunocytochemistry. The ENDOG blocking peptide was from Prosci and the HSP60 blocking peptide from Santa Cruz Biotechnology.
Mitochondrial Fractionation and Carbonate Extraction.
These procedures were performed as described in ref. 40. The purity of each fraction was determined by using anti-HSP60 and anti-HSP70 antibodies. Cytoplasmic fractions were concentrated by using centricon YM-10 devices (Millipore) according to the manufacturer's instructions.
PK Treatment.
For PK digestion, fresh isolated mitochondria were resuspended in isotonic mitochondrial buffer [250 mM sucrose, 10 mM Hepes, 10 mM KCl, 2 mM MgCl2, 1 mM EDTA (pH 7.4)] and incubated with various concentrations of PK for 20 min on ice. Digestion was terminated with 5 mM phenylmethylsulfonyl fluoride. Mitochondrial proteins were separated by SDS/PAGE and proteins were detected by Western blot analysis.
RNA Extraction and Real-Time PCR Analysis.
Total RNA from HeLa cells, SH-SY5Y cells, SH-SY5Y cells stably expressing GFPu (GFPu SH-SY5Y), and mice and human tissues were prepared by using TRIzol (Invitrogen) according to the manufacturer's instructions and then reversely transcribed to cDNA with the SuperScript First-strand Synthesis System (Invitrogen). The resulting cDNAs were quantified by real-time PCR with TaqMan Gene Expression Assays as per the manufacturer's instructions (Applied Biosystems) by using an ABI Prism 7000 Sequence Detection system.
siRNA Down-Regulation of PINK1.
Silencer validated siRNA (ID: 1199, Ambion) and Hs_PINK1_4 HP validated siRNA (Qiagen) were selected for PINK1 knockdown. Two scramble siRNAs [Silencer negative control 1 siRNA (Ambion) and negative control siRNA (Cat 1022076, Qiagen)] with no known mammalian homology were used as negative controls. SH-SY5Y and HeLa cells were transfected with siRNAs (final concentration 100 nM) by using Lipofectamine 2000 (Invitrogen). After 24 h, sister cultures were used for real-time PCR analysis, and other sister cultures were treated with MG132 for another 24 h and then harvested for Western blot analysis.
Supplementary Material
Acknowledgments.
We thank Drs. Liza Pon (Columbia University) and Carla Koehler (UCLA) for their insightful comments on the manuscript and Dr. Jie Shen (Harvard University) for providing the PINK1 knockout mouse tissues. This work was supported by National Institutes of Health Grants NS42269, NS38370, NS11766, AG21617, A608702, HD83062, ES013177, DK58056; the U.S. Department of Defense (DAMD 17–03-1), the Parkinson's Disease Foundation, the Marriott Foundation, the Muscular Dystrophy Association, and the Bernard and Anne Spitzer Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802814105/DCSupplemental.
References
- 1.Vila M, Przedborski S. Genetic clues to the pathogenesis of Parkinson's disease. Nat Med. 2004;10(Suppl):S58–S62. doi: 10.1038/nm1068. [DOI] [PubMed] [Google Scholar]
- 2.Klein C, Lohmann-Hedrich K. Impact of recent genetic findings in Parkinson's disease. Curr Opin Neurol. 2007;20:453–464. doi: 10.1097/WCO.0b013e3281e6692b. [DOI] [PubMed] [Google Scholar]
- 3.Valente EM, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 4.Deng H, Jankovic J, Guo Y, Xie W, Le W. Small interfering RNA targeting the PINK1 induces apoptosis in dopaminergic cells SH-SY5Y. Biochem Biophys Res Commun. 2005;337:1133–1138. doi: 10.1016/j.bbrc.2005.09.178. [DOI] [PubMed] [Google Scholar]
- 5.Petit A, et al. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem. 2005;280:34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 6.Silvestri L, et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi S, et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 8.Plun-Favreau H, et al. The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat Cell Biol. 2007;9:1243–1252. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 9.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 Protects against Oxidative Stress by Phosphorylating Mitochondrial Chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Exner N, et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogaeva E, et al. Analysis of the PINK1 gene in a large cohort of cases with Parkinson disease. Arch Neurol. 2004;61:1898–1904. doi: 10.1001/archneur.61.12.1898. [DOI] [PubMed] [Google Scholar]
- 12.Rohe CF, et al. Homozygous PINK1 C-terminus mutation causing early-onset parkinsonism. Ann Neurol. 2004;56:427–431. doi: 10.1002/ana.20247. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima A, Kataoka K, Hong M, Sakaguchi M, Huh NH. BRPK, a novel protein kinase showing increased expression in mouse cancer cell lines with higher metastatic potential. Cancer Lett. 2003;201:195–201. doi: 10.1016/s0304-3835(03)00443-9. [DOI] [PubMed] [Google Scholar]
- 14.Unoki M, Nakamura Y. Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene. 2001;20:4457–4465. doi: 10.1038/sj.onc.1204608. [DOI] [PubMed] [Google Scholar]
- 15.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 16.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 17.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 18.Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darios F, et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12:517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- 20.Muqit MM, et al. Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J Neurochem. 2006;98:156–169. doi: 10.1111/j.1471-4159.2006.03845.x. [DOI] [PubMed] [Google Scholar]
- 21.Beilina A, et al. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haque ME, et al. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci USA. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takatori S, Ito G, Iwatsubo T. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci Lett. 2008;430:13–17. doi: 10.1016/j.neulet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 24.Weihofen A, Ostaszewski B, Minami Y, Selkoe DJ. Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum Mol Genet. 2008;17:602–616. doi: 10.1093/hmg/ddm334. [DOI] [PubMed] [Google Scholar]
- 25.Cechetto JD, Gupta RS. Immunoelectron microscopy provides evidence that tumor necrosis factor receptor-associated protein 1 (TRAP-1) is a mitochondrial protein which also localizes at specific extramitochondrial sites. Exp Cell Res. 2000;260:30–39. doi: 10.1006/excr.2000.4983. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki Y, et al. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Baro MR, Granger DA, Coleman RA. Mitochondrial glycerol phosphate acyltransferase contains two transmembrane domains with the active site in the N-terminal domain facing the cytosol. J Biol Chem. 2001;276:43182–43188. doi: 10.1074/jbc.M107885200. [DOI] [PubMed] [Google Scholar]
- 28.Donzeau M, et al. Tim23 links the inner and outer mitochondrial membranes. Cell. 2000;101:401–412. doi: 10.1016/s0092-8674(00)80850-8. [DOI] [PubMed] [Google Scholar]
- 29.Guo Y, et al. Tim50, a component of the mitochondrial translocator, regulates mitochondrial integrity and cell death. J Biol Chem. 2004;279:24813–24825. doi: 10.1074/jbc.M402049200. [DOI] [PubMed] [Google Scholar]
- 30.Taymans JM, Van den HC, Baekelandt V. Distribution of PINK1 and LRRK2 in rat and mouse brain. J Neurochem. 2006;98:951–961. doi: 10.1111/j.1471-4159.2006.03919.x. [DOI] [PubMed] [Google Scholar]
- 31.Blackinton JG, et al. Expression of PINK1 mRNA in human and rodent brain and in Parkinson's disease. Brain Res. 2007;1184:10–16. doi: 10.1016/j.brainres.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 32.Margineantu DH, Emerson CB, Diaz D, Hockenbery DM. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS ONE. 2007;2:e1066. doi: 10.1371/journal.pone.0001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 34.Pfanner N, Geissler A. Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 35.Hackenbrock CR, Miller KJ. The distribution of anionic sites on the surfaces of mitochondrial membranes. Visual probing with polycationic ferritin. J Cell Biol. 1975;65:615–630. doi: 10.1083/jcb.65.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondo-Okamoto N, Shaw JM, Okamoto K. Mmm1p spans both the outer and inner mitochondrial membranes and contains distinct domains for targeting and foci formation. J Biol Chem. 2003;278:48997–49005. doi: 10.1074/jbc.M308436200. [DOI] [PubMed] [Google Scholar]
- 37.Burgess SM, Delannoy M, Jensen RE. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kikuchi H, et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc Natl Acad Sci USA. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perier C, et al. Two molecular pathways initiate mitochondria-dependent dopaminergic neurodegeneration in experimental Parkinson's disease. Proc Natl Acad Sci USA. 2007;104:8161–8166. doi: 10.1073/pnas.0609874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.