Abstract
To ensure successful replication, HIV-1 has developed a Rev-mediated RNA transport system that promotes the export of unspliced genomic RNA from nuclei to cytoplasm. This process requires the Rev responsive element (RRE) that is positioned in the viral transcript encoding Env protein, as well as in unspliced and singly spliced viral transcripts. We identified Purα, a single-stranded nucleic acid binding protein as a cellular partner for Rev that augments the appearance of unspliced viral RNAs in the cytoplasm. A decrease in the level of Purα expression by siRNA diminishes the level of Rev-dependent expression of viral RNA. Through its nucleic acid binding domain, Purα exhibits the ability to interact with the multimerization and RBD domains of Rev. Similar to Rev, Purα associates with RRE and in the presence of Rev forms a complex with slower electrophoretic mobility than those from Rev:RRE and Purα:RRE. The interaction of Purα with RRE occurs in the cytoplasm where enhanced association of Rev with RRE is observed. Our data indicate that the partnership of Purα with Rev is beneficial for Rev-mediated expression of the HIV-1 genome.
Keywords: Purα, HIV-1, Rev
During the course of HIV-1 infection, three classes of viral RNAs are produced. These include 9 kb unspliced RNAs which are packaged into progeny virions as genomic RNA that can also serve as a transcript for protein products of gag/pol genes [Schwartz et al., 1990; Purcell and Martin, 1993]. The second class is singly spliced mRNAs of approximately 4 kb that encode the Vif, Vpr, and Vpu/Env proteins. Finally, the third group is doubly spliced 2 kb transcripts that are responsible for the production of Tat, Rev, and Nef [Purcell and Martin, 1993]. To facilitate transport of unspliced or partially spliced pre-mRNA into nuclei, HIV-1 has developed an efficient mechanism that is orchestrated by the viral protein Rev and its association with a 351 nucleotide RNA sequence named Rev Responsive Element (RRE) that spans the Env gene and is present in unspliced and singly spliced RNAs [Malim et al., 1989; Zapp and Green, 1989; Pollard and Malim, 1998]. The association of Rev and RRE in the nucleus promotes engagement of several other cellular proteins including CRM1/Exportin-1 and RanGTP that eventually leads to the exit of Rev-associated RNA cages from the nucleus [Fornerod et al., 1997; Henderson and Per-cipalle, 1997; Neville et al., 1997; Stade et al., 1997; Nakielny and Dreyfuss, 1999; Kjems and Askjaer, 2000; Daelemans et al., 2005].
The cellular protein, Purα, which has a strong affinity for associating with single-stranded nucleic acid containing the GGC(A)GGA(C) sequence, exhibits diverse biological activities on RNA transcription, cell cycle, neuronal cell differentiation, and DNA replication [Gallia et al., 2000; Khalili et al., 2003; Zhang et al., 2005; Johnson et al., 2006; Knapp et al., 2006; Shimotai et al., 2006]. Earlier studies revealed that some of the activity of Purα is regulated upon its association with small RNA molecules [Hereault et al., 1995; Tretiakova et al., 1998; Gupta et al., 2003]. The interaction of Purα with the mRNA sequence from exon 3 of the mouse VSM α-actin which is placed in the 5′ untranslated region of a reporter mRNA suppresses its translation [Kelm et al., 1999].
Recent studies indicate that Purα participates in dendritic transport of mRNAs and associates with ribosomes [Ohashi et al., 2000, 2002; Li et al., 2001; Johnson et al., 2006]. Purα was isolated from RNA containing granules in dendrites together with a group of RNA binding proteins including hnRNP-U, Purβ,PSF,DDX1, DDX3, SYNCRIP, TLS, NonO, HSPC117, ALY, CGI-99, Staufen (Tang et al., 2001) three FMRPs, EF-1α, mRNAs for CaMKIIα and Arc. These RNP complexes were associated with kinesin [Kanai et al., 2004]. Purα was also identified in mRNA/Protein complexes containing mStaufen, Fragile X mental retardation protein (FMRP) and myosin Va and associated with rough endoplasmic reticulum [Ohashi et al., 2002]. Unique features and functions of Purα raise the possibility that Purα may be involved in post- transcriptional regulation of HIV-1 gene expression.
Here we provide evidence that Purα potentiates HIV-1 gene expression from unspliced RNAs and augments Rev/RRE-mediated stimulation of gene expression from viral mRNAs containing intron, probably facilitating their translocation, and targeting them to sites of translation or virion assembly.
MATERIALS AND METHODS
Cell Culture and Transfection
U-87MG (ATCC HTB14), a human glioblastoma cell line, was grown in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum. Transfection of U-87MG cells was performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) or the calcium phosphate precipitation method [Graham and van der Eb, 1973]. Leptomycin B (LMB, Sigma-Aldrich, St. Louis, MO) treatment was performed 24 h after transfection at 2 nM concentration for 16 h.
Plasmids
pIIIAR was previously described [Rosen et al., 1988]. pcRev was a generous gift from Dr. Roger Pomerantz (Tibotec, Inc., Yardley, PA). The standard PCR cloning technique was used to construct the following expression plasmids, with the templates, primers, and cloning backbone vectors shown in parentheses (restriction enzyme sites underlined): pcDNA6/myc-His-A-Rev 1-116 and deletion mutants, pGEX-5X-1-Rev 1-116 and deletion mutants (pcRev; forward for Rev 1-116, Rev 1-83, Rev 1-67, Rev 1-50, Rev 1-33, Rev 1-18: 5′-TAGGCAGAATTCATGGCAGGA AGA-3′; forward for Rev 86-116 5′-GCAGAATTCGCCATGGAG GAT TGTGGA-3′, forward for Rev 68-116 5′-TCAGAATTCGCCATGGAGCCT GTG CCT-3′; reverse for Rev 1-116, Rev 86-116, Rev 68-116: 5′-CTAACACTCGAGTTCTTT AGCTCC-3′; reverse for Rev 1-83: 5′-CTAACACTCGAGAAGAGTAAGTCT-3′; reverse for Rev 1-67: 5′-CTAACACTCGAGAGATCGTCCCAG -3′; reverse for Rev 1-50: 5′-CTAACACTCGAGTCTCTGTCTCTC-3′; reverse for Rev 1-33: 5′-CTAACACTCGAG CCCCTCGGGGTT-3′; reverse for Rev 1-18: 5′-CTAACA CTCGAGTCTGA CTGCCTT-3′; pcDNA6/myc-His-A (Invitrogen) and pGEX-5X-1), pDs-Rev-Red1 (pcDNA6/myc-His-A-Rev, forward: 5′-TAGGCAGGATCC GAATTCATGGCAGGAAGA-3′, reverse: 5′-CCTAACAGGATCCCTGAGTTCTTTAGCTCC-3′, pDsRed1 (CLONTECH, Mountain View, CA); pBluescript II KS(+)-RRE 1-234 (pIIIAR, forward 5′-CGCCAAGCTTGAATAGGAGCTTTGTTCC- 3′, reverse 5′-CTAGGATCCAGGAGCTGTTGATCCTTTAGG-3′, pBlue-script II KS(+)); pBluescript II KS(+)-RRE 34-110 (pIIIAR, 5′-TCACAAGCTTAGGAAGCACTAT GGGCGC-3′, reverse 5′-CTAGGATCCGCTGCTGCACTATACCAG-3′,pBluescript II KS(+)).
Construction of pcDNA3.1-SD4*-luciferase-RRE-SA7 and pcDNA3.1-SD4*-luciferase-SA7 was performed in few steps: First, pIIIAR was digested with BglII and XhoI and DNA fragment (1,276 bp) that contains the RRE (full-length) and splice acceptor 7 (SA7) sequences (corresponding to 7,650-8,926 nt of HIV-1 NCBI X01762) was removed and subcloned into pcDNA3.1 vector (Invitrogen) using BamHI and XhoI enzyme recognition sites producing pcDNA3.1-RRE-SA7 vector. Next, DNA fragment (562 bp), which contains sequences of the first intron of Rev and splicing donor 4 (SD4) (corresponding to 5,945-6,380 nt of HIV-1 NCBI X01762) was obtained by digestion of pIIIAR with SalI and KpnI restriction enzymes followed with blunting of overhangs. This fragment was subcloned into pGL3-Basic vector (Promega, Madison, WI), digested with HindIII and blunted, generating pGL3AR vector. Site-directed mutagenesis was then performed to mutate ATG of Rev present in pIIIAR and pGL3AR, using QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), pGL3AR vector as a template and following primers: forward 5′-CTTAGGCATCTCCTTAAGCAGGAAGAAGCGGAGAC-3′, reverse 5′-GTCTCCGCTTCTTCCTGCTTAAGGAGATGCCTAAG-3′. Site-directed mutagenesis resulted in pGL3AR* plasmid. Next, pGL3AR* was digested with XbaI, blunted then digested with NheI and fragment containing SD4* and luciferase gene was obtained (2,266 bp). Next, pcDNA3.1-RRE-SA7 was digested with KpnI, blunted and then digested with NheI. The 2,266 bp fragment flanked by the NheI and blunt end was subcloned into digested pcDNA3.1-RRE-SA7, resulting in pcDNA3.1-SD4*-luciferase-RRE-SA7. Finally, for pcDNA3.1-SD4*-luciferase-SA7, pcDNA3.1-RRE-SA7 was digested with HindIII, blunted and then digested with NheI restriction enzyme removing RRE sequence and keeping 756 nt fragment from pIIIAR (corresponding to 8,170-8,926 nt of HIV-1 NCBI X01762), encompassing SA7. Subcloning of the 2,266 bp fragment, flanked by the NheI and blunt end, generated pcDNA3.1-SD4*-luciferase-SA7. PLEGFP-Purα [Darbinian et al., 2006], pEBV-Purα and deletion mutants and pGEX1λT-Purα and deletion mutants were previously described [Johnson et al., 1995; Gallia et al., 1998]. All recombinant plasmids were verified by sequencing.
RNAi and siRNA Transfections
Purα-specific siRNA, ACAAGTACGGCGTGTTTAT, was derived from sequences corresponding to 772-790 of Purα mRNA (Dharamcon, Lafayette, CO). U-87MG cells were transfected with 50 nM Purα/siRNA using Oligofectamine kit (Invitrogen).
Luciferase and CAT Assay
For luciferase assay, cells were harvested at designated time points, and protein extracts (20 μg) were used to examine the level of luciferase activity with the dual-luciferase reporter assay system (Promega, Madison, WI). The pRLTK plasmid was used as an internal control for transfection efficiency. Chloramphenicol acetyltransferase (CAT) activity was determined by the established method [Gorman et al., 1982].
Protein Extracts and Western Blot Analysis
For whole cell extract preparation, cells were lysed for 30 min on ice in LB1 (50 mM Hepes, pH 7.5/150 mM NaCl/1.5 mM MgCl2/1 mM EGTA/10% glycerol/1% Triton X-100) buffer containing protease inhibitor cocktail (Sigma, P8340) and 0.2 mM Na-orthovanadate. Cell debris was removed by centrifugation at 14,000 rpm for 15 min at 4°C. Nuclear and cytoplasmic fractions were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce Biotechnology, Rockford, IL #78833). Western blot analysis was performed as described [Kaniowska et al., 2006].
Co-immunoprecipitation
U-87MG cells were transfected with pcDNA3.1-SD4*-luciferase-RRE-SA7 along with Purα and/or Rev expressing plasmids. Nuclear and cytoplasmic extracts were prepared and 100 μg of nuclear or 300 μg of cytoplasmic lysates were incubated with rabbit polyclonal antibody against Purα or normal rabbit serum as a negative control for immunoprecipitation in 500 of μl HNTG buffer overnight at 4°C. Immunocomplexes were precipitated by the addition of protein A-Sepharose beads, washed four times with rocking at 4°C in 1 ml of HNTG buffer, and resolved by SDS-PAGE followed by Western blotting using an anti-myc antibody for detection of Rev.
GST Fusion Proteins and GST Pull-Down Assay
For preparation of bacterially produced GST fusion proteins, we followed the methods previously described. Overnight cultures (100 ml) of Escherichia coli DH5α, were transformed with pGEX-5X1-Rev, pGEX1λT-Purα [Gallia et al., 1998]. GST pull-down assay was performed according to the procedures described previously [Kaniowska et al., 2006].
RNA Preparation and Northern Blot Analysis
Total RNA was isolated using RNeasy kit (Qiagen, Valencia, CA, #74104) according to the manufacturer’s directions. Preparation of cytoplasmic RNA was based on the protocol provided with RNeasy kit and using RLN buffer containing 50 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% (v/v) Nonidet P-40 (1.06 g/ml), 1,000 U/ml RNase inhibitor, 1 mM DTT. Ten micrograms of RNA were fractionated on 1.2% agarose, 0.4% formaldehyde, 1 ×morpholinepropanesulfonic acid (Mops) gel and transferred to a Hybond-N nylon membrane (Amersham, Piscataway, NJ). For the detection of CAT RNAs, the membranes were hybridized with [32P]-labeled CAT DNA probe obtained by digestion of pIIIAR plasmid with BgIII and NcoI (850 bp). For the detection of RRE containing RNAs, the filters were probed with a PCR-amplified and [32P]-labeled DNA fragment consisting of nucleotides 1-234 of the RRE. Radiolabeled DNA probes were prepared with Random Primed DNA Labeling Kit (Roche Molecular Biochemicals, Indianapolis, IN), followed by removal of unincorporated radionucleotides with MicroSpin™G-50 columns (Amersham). Relative levels of mRNA were determined by using the housekeeping gene GAPDH as an internal standard.
RNA Immunoprecipitation and RT-PCR
Nuclear and cytoplasmic protein extracts from cells transfected with pcDNA3.1-SD4*-luciferase-RRE-SA7 vector along with Purα and/or Rev expressing plasmids were prepared and precleared with protein A sepharose beads by 1 h incubation at 4°C. The resultant lysates were incubated with protein A sepharose beads and anti-T7 antibody against Purα. In control samples, normal mouse serum was added instead of anti-T7 antibody. For RT control Rev expressing extracts were immunoprecipitated with anti-myc antibody. After overnight incubation at 4°C, the beads were washed four times (3 min each) with HNTG buffer (20 mM HEPES, pH 7.5/150 mM NaCl/0.1% Triton X-100/10% Glycerol, protease inhibitor cocktail (Sigma, P8340) and 0.2 mM Na-orthovanadate) and resuspended in 150 μl of buffer 2 (50 mM Tris-HCl, pH 7.0/5 mM EDTA/1% SDS/10 mM DTT). Then RNA was extracted using phenol TE buffer pH 4.5 saturated/chloroform/isoamyl (25/24/1) and chloroform/isoamyl (24/1). Extracted RNA was precipitated by incubation with 7.5 M ammonium acetate/ethanol (0.5/2.5, V/V) and GlycoBlue (Ambion, Austin, TX) for 30 min at -70°C followed with washing in 70% ethanol, drying and rehydration in 20 μl of nuclease free water. Samples were treated with DNAse1 in the presence of RNase inhibitor (Roche) for 1 h at 37°C followed with phenol/chloroform extraction and precipitation as described above. RT reaction was performed using random primers (p(dN)6; Roche) and M-MuLV RT enzyme. For PCR, RRE specific primers and FailSafe™ PCR reagents (Epicentre, Madison, WI) were used. DNA products generated as a result of RT-PCR were analyzed by 2% agarose gel electrophoresis.
Fluorescent Technique
U-87MG cells (1 ×105) were transfected with 5 μg of GFP-Purα or Rev-Red plasmids, alone or in combination, then seeded in poly-l-lysinecoated glass chamber slides, and after 16 h incubation, cells were fixed in 4% paraformaldehyde in 1 × PBS. Cells were then washed in PBS, and proteins were visualized for green or red fluorescence. Fluorescent images were captured using an inverted fluorescent Nikon microscope with deconvolution software (Slide-Book 4.0.1.34; Intelligent Imaging, Denver, CO).
RNA Electrophoretic Mobility Shift Assay
The RRE-containing RNA probe was in vitro transcribed using pBluescript IIKS(+) RRE 1-234 and pBluescript IIKS(+) RRE 34-110 plasmids as DNA template. In vitro transcription reaction was performed by incubating the above described template DNAs, T3 RNA polymerase, transcription optimized 5 × buffer, DTT 100 mM, recombinant RNasin ribonuclease inhibitor, rATP, rGTP, rCTP (2.5 mM each) and [α-32P]-UTP for 1 h at 37-40°C. For RNA-protein interaction studies 0.2 μM of total recombinant proteins GST, GST-Rev, GST-Purα were incubated with 100,000 cpm of [α-32P]-UTP RNA probe for 15 min at room temperature in 20 μl binding buffer containing 12 mM HEPES (pH 7.9), 4 mM Tris-HCl (pH 7.5), 60 mM KCl, 5 mM MgCl2, 0.8 mM dithiothreitol (DTT), 0.5 μg of poly[dI-dC] as a nonspecific competitor, 10% glycerol and RNase inhibitor. Reaction products were analyzed on a 1% agarose gel followed by autoradiography.
Antibodies
Anti-α-tubulin, clone B512 was obtained from Sigma-Aldrich. Anti-myc antibody was purchased from Invitrogen and anti-T7 tag antibody was from Novagen (Madison, WI). Rabbit polyconal anti-Purα antibody was raised against full-length Purα,cleaved from GST-Purα and injected into rabbits (Lampire Biological Laboratories, Pipersville, PA).
RESULTS
To investigate the effect of Purα on Rev activity, we constructed and utilized a luciferase-based reporter plasmid where the luciferase gene was placed within the HIV-1 intronic sequence flanked by splicing donor 4 and splicing acceptor 7 (Fig. 1A top). The translation start site of Rev, which is located 76 nucleotides upstream from the splice donor, was disrupted and the RRE was placed at the 3′ end of the luciferase gene within the intron. The control plasmid had no RRE sequence, yet encompassed all the other features of the reporter plasmid (Fig. 1A, bottom). As shown in Figure 1B, Purα enhanced the ability of Rev to stimulate luciferase activity directed by the RRE-containing plasmid and exhibited no stimulatory effect on the expression from the control RRE negative plasmid. Leptomycin, which targets the CRM1/Exportin-1 and inhibits Rev nuclear export [Nishi et al., 1994; Wolff et al., 1997] suppressed cooperativity of Rev and Purα, suggesting that Purα is important for Rev:RRE-mediated stimulation of HIV-1 expression. A decrease of approximately 40% in the level of endogenous Purα by siRNA (Fig. 1C) caused comparable levels of decline in the level of Rev-mediated activation of luciferase expression (Fig. 1D).
Fig. 1.
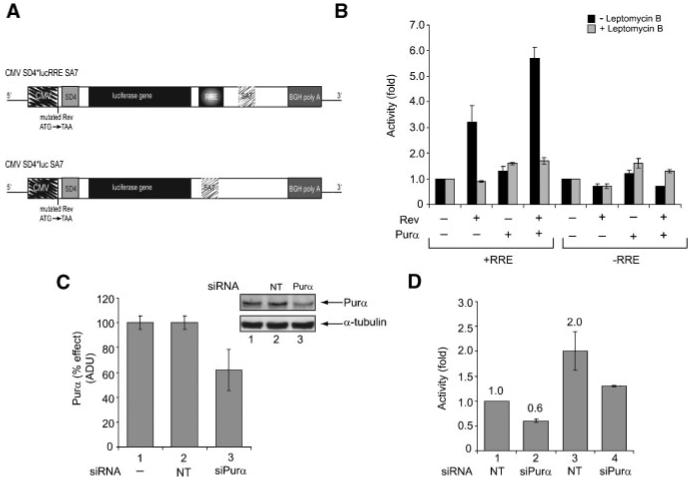
A: Structural organization of pcDNA3.1-SD4*-luciferase-RRE-SA7 and pcDNA3.1-SD4*-luciferase-SA7, a luciferase-based reporter construct. B: Effect of Purα on Rev-mediated luciferase activity in the presence of Leptomycin B (LMB) treatment. U-87MG cells were transfected with reporter vectors with or without RRE along with Rev and/or Purα expressing plasmids. One set of transfected cells was treated with LMB (2 nM, 16 h). Cells were harvested and luciferase activity was determined. Data represent fold increase relative to basal levels-reporter alone. C: Down-regulation of Purα expression in U-87MG cells by siRNA. Cells were transfected with non-target (NT) siRNA (lane 1) or Purα directed siRNA (lane 2). Western blot shows the levels of Purα and the housekeeping tubulin protein that served as a loading control. Efficiency of silencing is quantified. D: Effect of silencing of Purα gene expression on Rev/RRE-mediated luciferase activity. Rev expressing cells were transfected with pcDNA3.1-SD4*-luciferase-RRE-SA7 reporter plasmid and non-target siRNA or siPurα. Data represent fold increase relative to basal levels—Rev and luciferase-RRE. The experiments were repeated five times.
To demonstrate the effect of Purα on Revmediated RNA accumulation in the cytoplasm, total and cytoplasmic RNAs from cells transfected with pIIIAR [Rosen et al., 1988] along with pEBV-Purα and pCMV-Rev were analyzed by Northern blot using DNA probes derived from RRE or CAT sequences. The RRE probe detects 4.4 kb unspliced RNA whereas the CAT DNA probe identifies both 2.37 kb spliced and 4.4 kb unspliced RNA species. As seen in Figure 2A, a significant increase in the level of the unspliced form of cytoplasmic RNA was observed upon Rev expression. Interestingly, Purα also increased the level of unspliced RNA in the cytoplasm. Co-expression of both proteins resulted in an increase in the amount of unspliced RNA in the cytoplasm, more than that observed with Rev alone. Note that unspliced RNA in total RNA extract was also maximal when Rev and Purα were co-expressed. A similar pattern for 4.4 kb RNA was observed when DNA sequence from the CAT gene was used as a probe (Fig. 2B). The integrity of RNA preparation in various samples is shown in panel C. The level of hGAPDH in each sample served as an internal control (Fig. 2, panel D).
Fig. 2.
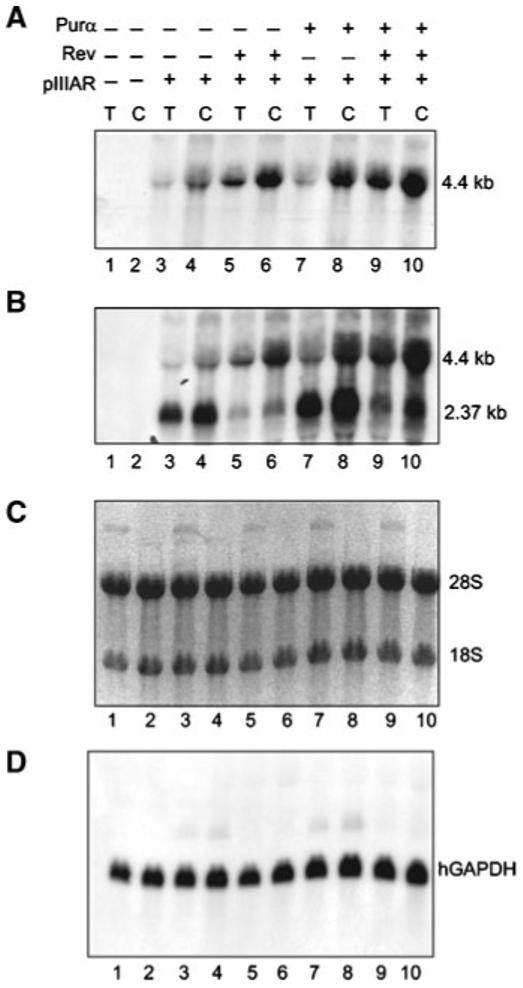
Effect of Purα on Rev-mediated accumulation of intron-containing unspliced RNA in the cytoplasm of cells. Total (T) and cytoplasmic (C) RNA from U-87MG cells transfected with pIIIAR, pEBV-Purα and pCMV-Rev were analyzed by Northern blot. Probes were RRE (A), CAT (B) and hGAPDH (D). Panel C demonstrates the RNA integrity and the ribosomal RNA bands 28S (5.0 kb) and 18S (1.9 kb) after transfer to a membrane.
Earlier studies have shown that Rev predominantly accumulates in the nucleolus with a fraction shuttling between the nucleus and cytoplasm [Cullen et al., 1988; Cochrane et al., 1990; Wolff et al., 1995]. To investigate the subcellular localization of Purα and Rev in our cell culture system, we constructed plasmids expressing Purα and Rev in fusion with green and red fluorescent proteins, respectively. Activity of Rev-Red fusion protein was verified by transfection assay using a reporter plasmid that is responsive to Rev activity. As seen in Figure 3A, similar to previous observations, Rev fusion protein had a stimulatory effect on reporter plasmid containing RRE, and this effect was augmented by expression of GFP-Purα. Co-transfection of cells with GFP Purα and/or Rev-Red expressing plasmids allowed visualization of the corresponding proteins in the cells. As shown in Figure 3B, Rev was detected within nucleoli with some accumulation in the perinuclear region (Fig. 3B). Purα was predominantly localized in the perinuclear region and cytoplasm of cells (Fig. 3C), although, a trace of Purα was also detected in the nucleoli. In cells co-transfected with both plasmids, some levels of Purα and Rev were found in the perinuclear compartment (Fig. 3D).
Fig. 3.
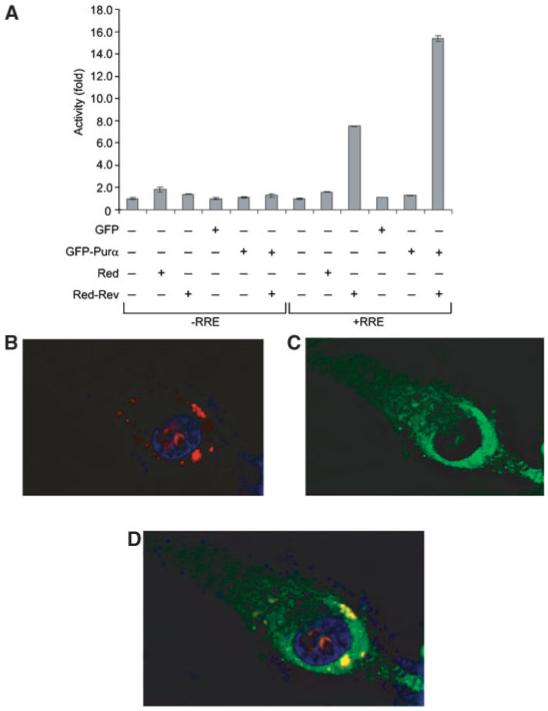
Subcellular localization of Rev and Purα in cells transfected with PLEGFP-Purα and pDs-Rev-Red1 plasmids. A: U-87MG cells were transfected with reporter vectors with or without RRE along with plasmids expressing fusion Red-Rev and GFP-Purα fusion fluorescent proteins in various combinations as denoted. Cells were fixed and red fluorescence from Rev (B) and green fluorescence from Purα (C) were detected by microscopy. Composite of the two colors demonstrates co-localization of Purα and Rev (D). DAPI Blue is for nuclear staining.
Co-localization of Purα and Rev prompted us to investigate the possible interaction of these two proteins. Results from the GST-based protein binding assay revealed the ability of Purα to interact with full-length Rev protein and highlighted the importance of the region of Rev that spans aa 18-50, which overlaps nuclear localization signal (NLS) and RNA binding domain (RBD) and multimerization domain (MD), in this interaction. Figure 4A (top panel) illustrates results from GST pull-down assay and the various mutants of Rev that were utilized in this experiment. The integrity of GST-Rev and its mutant variants were examined by SDS-PAGE (Fig. 4A, middle panel). Figure 4A (bottom panel) illustrates the linear organization of full-length Rev, its deletion mutants, and their binding ability to Purα. In a reciprocal study, we utilized a similar approach and identified a region of Purα located between amino acids 73-123 as a minimum domain that is recognized by Rev (Fig. 4B, top panel). Note that Rev, due to its modification, appears as a double band. Treatment with alkaline phosphatase or lambda protein phosphatase results in the disappearance of the top band (unpublished observations). The integrity of GST-Purα and its various mutants is shown in Figure 4B (middle panel). Figure 4B (bottom panel) also illustrates the structural organization of the full-length Purα, its mutant variants, and summarizes their binding abilities to Rev. The interaction of Purα and Rev was also detected by immunoprecipitation/Western blot analysis of the protein extracts expressing both proteins (Fig. 4C,D). Nuclear (100 μg) or cytoplasmic (300 μg) fractions were prepared from U-87MG cells transfected with pcDNA3.1-SD4*-luciferase-RRE-SA7 along with Purα and/or Rev expressing plasmids and the presence of proteins was verified by Western blot analysis using antibodies against Rev (Fig. 4C, top panel) and Purα (Fig. 4C, middle panel). Rabbit polyclonal antibody against Purα detects endogenous and overexpressed Purα (middle panel). Fractionation and equal loading was controlled by α-Tubulin detection (Fig. 4C, bottom panel). We immunoprecipitated protein complexes bound to Purα using anti-Purα antibody (Fig. 4D, lanes 1-10) in the presence of protein A Sepharose. After washing, protein complexes were separated by SDS-PAGE and analyzed by Western blot using anti-Myc antibody for detection of Rev. As shown in panel D, Rev was associated with endogenous Purα (lane 2) and overexpressed Purα (lane 4) in cytoplasmic fractions. Although, a faint band that points to the interaction of overexpressed Purα and Rev was also detected in nuclear fraction (lane 9).
Fig. 4.
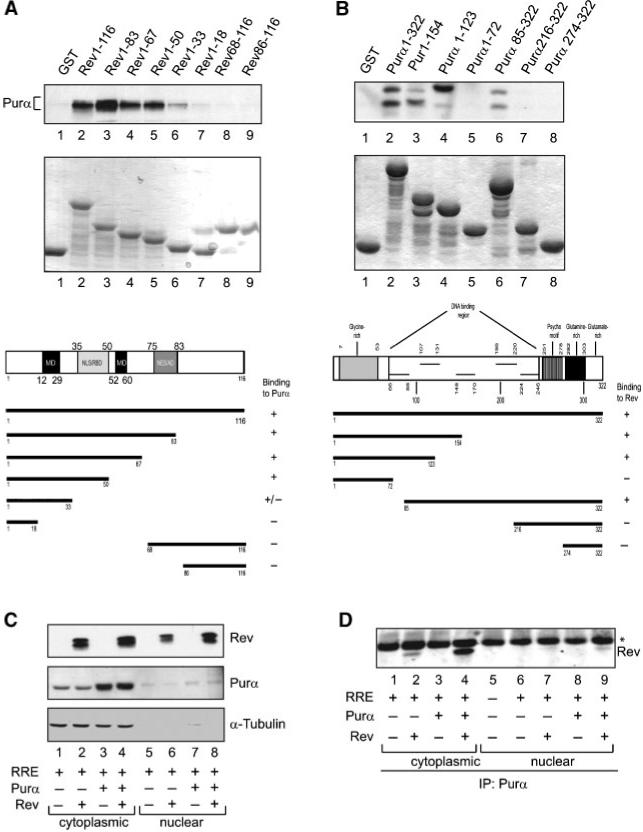
Interaction of Purα with Rev. A: Binding of Rev to Purα and localization of the interaction domain of Rev. Protein extract from cells expressing Purα was used in GST pull-down assay. The binding of Purα to purified GST-Rev and its various deletion mutants bound to GST beads (0.5 μM) was assessed by Western blot analysis with antibody against Purα (top panel). GST alone was used for control. The schematic structural organization of Rev is illustrated and binding abilities of GST proteins are summarized (bottom panel). B: Binding of Purα to Rev and mapping of the interaction region of Purα. Protein extracts from cells expressing Rev were incubated with GST-Purα or its deletion mutants (0.5 μM) in GST pull-down assay for identification of the region of Purα that binds to Rev (top panel). The schematic representation of structure of Rev protein and binding activity of GST proteins are illustrated (bottom panel). Integrity of fusion GST proteins used in assays is demonstrated (A,B, middle panels). C: U-87MG cells were transfected with pcDNA3.1-SD4*-luciferase-RRE-SA7 along with Purα and/or Rev expressing plasmids. Nuclear and cytoplasmic extracts were prepared and the presence of proteins was verified by Western blot analysis using antibodies against Rev (top panel) and Purα (middle panel). Rabbit polyclonal antibody against Purα detects endogenous and overexpressed Purα (middle panel). For loading and fractionation control, α-Tubulin was used (bottom panel). D: Rev and Purα were co-immunoprecipitated in cytoplasmic and nuclear lysates. Nuclear (100 μg) or cytoplasmic (300 μg) fractions prepared from cells as described in panel C, were immunoprecipitated using anti-Purα antibody (lanes 1-9). Immunoprecipitated protein complexes were washed, separated by SDS-PAGE followed by Western blot analysis using anti-Myc antibody for detection of Rev.
To investigate the relevance of Purα:Rev interaction to the observed functional cooperativity of these two proteins, we examined the effect of Rev on the RRE-containing reporter in the absence and presence of full-length and mutant Purα with no binding ability to Rev. As shown in Figure 5A, the activity of Rev was enhanced in the presence of full-length Purα, but not mutant Purα encompassing the C-terminus (216-332) of this protein. Of note, full-length Purα, but not its mutant 216-322, showed a modest increase in the level of luciferase gene expression (Fig. 5B). While full-length Purα was able to cooperate with full-length Rev (aa 1-116), it exhibited no cooperativity with the Rev mutant 1-33 which showed reduced binding activity to Rev (data not shown). These observations provided evidence that physical association between Purα and Rev may be important, at least in part, for their coordinated effect on the expression of the viral genome.
Fig. 5.
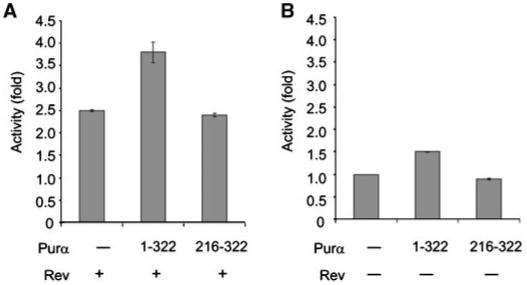
Mutant Purα with no binding activity to Rev is unable to enhance Rev-mediated RRE directed reporter gene expression. Cells were transfected with luciferase-RRE reporter along with plasmids expressing Rev and full-length Purα or mutant Purα which exhibited no binding activity to Rev. Data represent fold increase relative to basal levels-luciferase-RRE and Rev (A). Full-length Purα demonstrates modest effect on luciferase expression in the absence of Rev (B).
As Purα has a great affinity for binding to single-stranded RNAs containing GGCG sequence, a feature that is found in HIV-1 RRE, in the next series of experiments we evaluated the ability of Purα to bind to RRE and its impact on the interaction of Rev with RRE. We performed RNA electrophoretic gel mobility shift assays using RRE RNA probe and recombinant mutant GST-Rev, GST-Purα, and GST proteins. Two regions of the RRE spanning nucleotides 1-234 and 34-110 (numbering according to Malim et al., 1989) were synthesized in vitro and used as a probe. As shown in Figure 6A, incubation of both labeled RNAs with GST-Purα or GST-Rev resulted in the formation of complexes with slower electrophoretic mobilities. Co-incubation of the probes with both Rev and Purα resulted in the formation of a higher molecular weight complex than those seen by either protein alone (Fig. 6A, compare lane 5 to lanes 3 and 4, and lane 10 with lanes 8 and 9). The specificity of in vitro RNA binding was tested in competition assay with excess unlabeled probes. The addition of unlabeled RRE-containing RNAs 1-234 and 34-110 to the reaction mixture abrogated binding of both proteins with the labeled full-length RNA probe (data not shown).
Fig. 6.
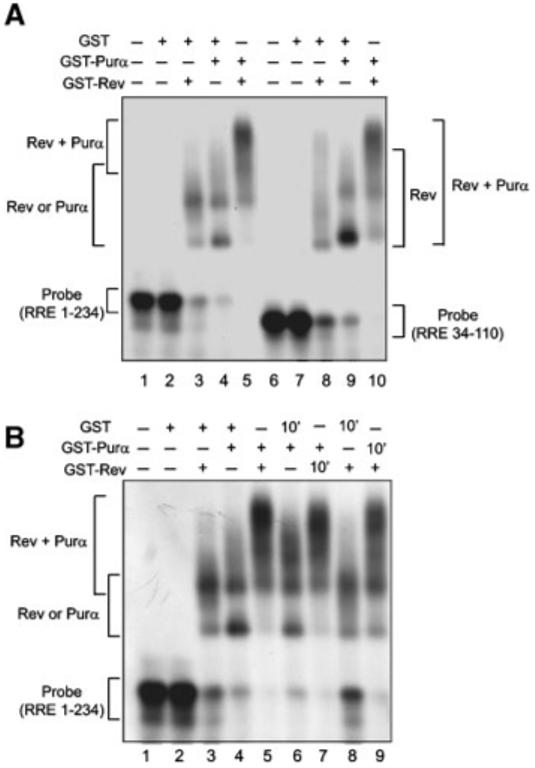
Interaction of Purα with RRE in vitro. A: Purα binds to RRE and enhances binding of Rev to RRE. GST, GST-Purα and GST-Rev proteins (100 nM) were assessed in RNA gel mobility shift assay with labeled RRE 1-234 (spanning stem I, stem loops IIA, IIB, IIC, III, IV, V) and RRE 34-110 (spanning stem loops IIA, IIB, IIC). B: Purα associates with RRE and with pre-existing Rev:RRE complex. Rev was incubated with labeled RRE for 10′ prior to the addition of Purα to the reaction (lane 7). Purα was incubated with labeled RRE 1-234 for 10′ prior to the addition of Rev to the reaction (lane 9). GST was used as a control in similar reactions (lanes 6 and 8).
In attempts to assess the ability of Purα to associate with pre-existing Rev:RRE complex, RNA probe was first incubated with Rev for 10 min prior to the addition of Purα to the binding reaction. As shown in Figure 6 (panel B), the addition of Purα resulted in the formation of higher molecular weight complexes with RRE. Similar results were obtained in reciprocal experiments where Rev was added to the reaction after the incubation of RRE probe with Purα. These observations suggest that Purα binds to RNA sequences that are not bound to Rev and that Purα interacts with RRE RNA that is associated with Rev.
Next, we performed RNA immunoprecipitation assay to determine whether the association of Purα and Rev with RRE RNA occurs in the cytoplasm or in nuclei of the cells. The human astrocytic cell lines, U-87MG, was co-transfected with pcDNA3.1SD4*lucRRE-SA7 vector and plasmids expressing T7-tagged Purα and myc-tagged Rev, either alone or in combination. Cytoplasmic and nuclear fractions were prepared, and expression of Rev and Purα was determined by Western blot analysis (Fig. 7A and B, respectively). The quality of each fraction was tested by measuring the presence of α-tubulin (Fig. 7C). Next, RNA:protein complexes were immunoprecipitated using antibody that recognizes Purα (anti-T7) (Fig. 8A,B, lanes 1-5), Rev (anti-myc) (Fig. 8A,B, top, lane 7) or control normal mouse serum (nms). The immunocomplexes were extensively washed and their associated RNAs were prepared and subjected to reverse transcription (RT) reaction using random primers (Fig. 8A). RRE was subsequently amplified by polymerase chain reaction (PCR) using primers specific for RRE. The RRE fragment which was directly amplified from pcDNA3.1SD4*lucRRE-SA7 plasmid served as a positive control. Reactions lacking reverse transcriptase (-RT) served as negative controls in this study (Fig. 8B). A high intensity band, corresponding to RRE fragment was detected in cytoplasmic extracts from cells expressing both Purα and Rev proteins (Fig. 8A, lane 5, cytoplasmic fraction). In cells expressing only Purα, a weak signal pointing to the association of Purα with RRE was detected in nuclear fractions. RNA input was verified by extraction of RNA directly from nuclear and cytoplasmic fractions (without prior immunoprecipitation) following RT-PCR (Fig. 8C). Rev:Purα protein complexes were also detected in cytoplasmic fraction using IP-Western (data not shown). In addition, we repeated RNA immunoprecipitation experiments using anti-Purα antibody instead of anti-T7 antibody and detected association of endogenous Purα with RRE RNA and Rev in the cytoplasm (data not shown). These observations point to the in vivo association of Purα with RRE in the presence of Rev. Moreover, Purα:RRE and Purα:RRE:Rev complexes are present in the cytoplasmic compartment of cells.
Fig. 7.
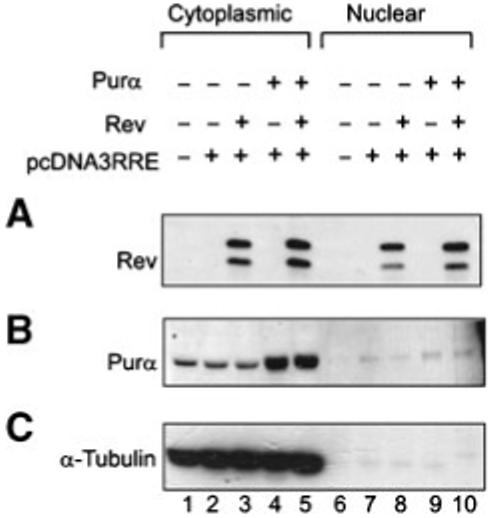
Cytoplasmic and nuclear presence of Purα and Rev. U-87MG cells were transfected with pcDNA3.1-SD4*-luciferase-RRE-SA7 along with Purα and/or Rev expressing plasmids. Nuclear and cytoplasmic extracts were prepared and presence of proteins was verified in Western blot analysis using antibodies against Rev (A) and Purα (B). For loading and fractionation control, tubulin was used (C).
Fig. 8.
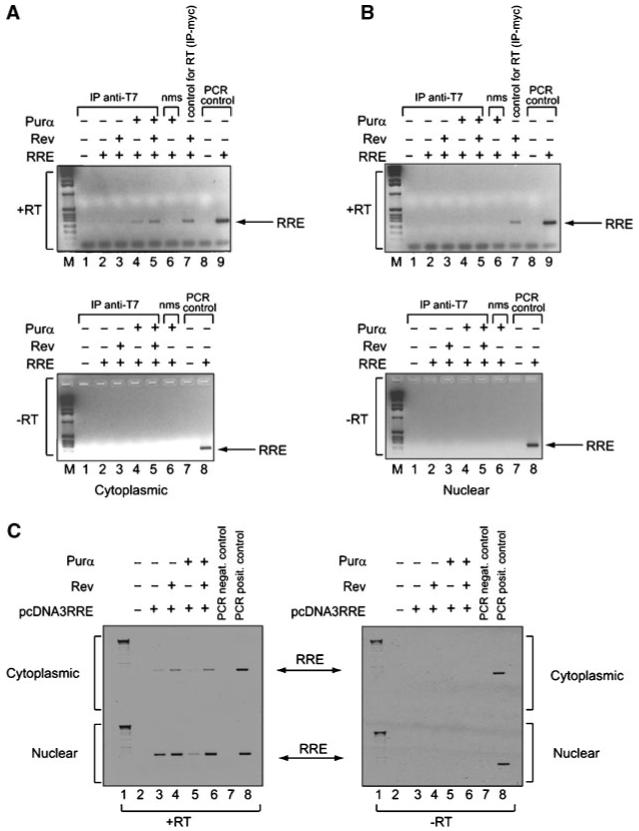
Interaction of Purα with RRE in vivo. Antibodies against Purα (anti-T7, lanes 1-5) and Rev (anti-myc, lane 7 in “+RT” panels) were used to immunoprecipitate interacting components from cytoplasmic (A) and nuclear (B) lysates from cells as described in Figure 7. Normal mouse serum (nms) was used as a negative control for immunoprecipitation (lane 6). RNA extracted from these complexes was subjected to RT-PCR with (“+RT”) and without (“-RT”) reverse transcriptase (RT) using RRE 1-234 specific primers. For PCR control, pcDNA3.1-SD4*-luciferase-RRE-SA7 and RRE 1-234 specific primers were used (lanes 8-9 in “+RT” panels and lanes 7 and 8 “-RT” panels). M: DNA marker. RNA input was verified by extraction of RNA directly from nuclear and cytoplasmic fractions (without prior immunoprecipitation) following RT-PCR (C).
DISCUSSION
Replication of HIV-1 is a highly regulated process and is dependent on cooperation between viral and host proteins. In an earlier study, we identified Purα as a potential partner for HIV-1 Tat that may facilitate Tat activation of the responsive promoters [Krachmarov et al., 1996; Chepenik et al., 1998; Gallia et al., 1999]. In this report we show that Purα enhances the action of Rev protein, the main regulator of export of unspliced viral mRNAs, and that depletion of endogenous Purα by siRNA diminishes Rev:RRE-mediated reporter activation. Similarly, silencing of Purα by siRNA during the course of HIV-1 infection of human primary culture of microglial cells caused nearly 25% suppression in viral replication pointing to the importance of Purα in HIV-1 gene expression and replication (data not shown).
Using reporter constructs containing luciferase gene sequence placed within intron alone or in combination with RRE, we demonstrated that Purα can function as a cofactor for Rev in activating gene expression from intronic sequences. Co-localization of Purα and Rev was observed within cytoplasm of cells expressing both proteins. Rev/RRE/Purα complexes were detected in vivo in the cytoplasmic compartment of cells also in RNA immunoprecipitation experiments. Thus Purα, which can also directly bind to RRE may act in post-transcriptional processing steps involving Rev function. In support of this notion, recent studies demonstrated that Purα is involved in dendritic transport of a subset of mRNAs and translation. Purα has been identified in mRNA/protein complexes containing mStaufen, FMRP, proteins that have been implicated to play a key role in targeting mRNAs to polyribosomes [Feng et al., 1997; Marion et al., 1999; Brown et al., 2001; Krichevsky and Kosik, 2001; Tang et al., 2001; Ohashi et al., 2002]. In addition, Staufen 1, which was found in HIV-1 Gag RNP complex, was implicated in regulating retroviral genomic encapsidation [Mouland et al., 2000; Chatel-Chaix et al., 2004]. Immunoprecipitation of these proteins with an anti-Purα antibody was abolished by RNase A treatment, suggesting an RNA dependence for Purα in the assembly of mRNPs [Ohashi et al., 2002]. These observations led to the hypothesis that Purα via binding to rRNA may mediate the association of mRNPs with polyribosomes. Interestingly, our earlier studies have revealed that Purα binds to RNAs that are homologous to 18S ribosomal RNA and to 7SL RNA [Tretiakova et al., 1998; Gallia et al., 1999, 2001]. Purα has also been isolated from kinesin-associated granules in dendrites together with a group of RNA binding proteins [Kanai et al., 2004], including DDX1, DDX3, staufen, PSF, EF-1α, which are involved in RNA splicing, nuclear RNA export and RNA translation and have been linked to retroviral replication [reviewed in Cochrane et al., 2006]. It was shown that Purα recognizes dendrite-targeting RNA motifs rich in G and U residues in the 5′ portion of BC1 RNA and links BC1 RNA to microtubules [Ohashi et al., 2000] and associates with ribosomes in neuronal cytoplasm [Li et al., 2001]. Non-coding BC1 RNA acts as a molecular scaffold for the formation of BC1 ribonucleoprotein particles (BC1 RNPs). These particles have been shown to be important for the dendritic delivery of mRNAs and factors regulating translation in dendrites. All of these observations are consistent with the notion that Purα via interaction with Rev and RRE may be involved in directing viral unspliced RNA association with RNP complexes to sites of translation.
Another interesting, although preliminary, observation comes from our experiments with RRE-negative construct showing that the level of cytoplasmic unspliced RNA in cells with expressing Purα and Rev is higher compared to that from cells with reporter construct alone, yet the level of protein remains comparable. Thus, one may speculate that in the presence of Rev, Purα and Rev may cooperate to enhance gene expression at the post-transcriptional level from intronic sequences, again supporting a role for Purα in post-transcription, that is, translation level. Our preliminary data also demonstrate that Purα can increase, albeit modestly, the level of cytoplasmic unspliced reporter RNAs (with or without RRE) and consequently, expression of the reporter protein in LMB insensitive manner. This observation along with the ability of Purα to associate with the RRE in the absence of Rev suggests that Purα may also be involved in CRM-1-independent nuclear export pathway that includes RNA helicase A (RHA), TAP, and Sam68 [Braun et al., 2001; Reddy et al., 2000].
The HIV-1 RRE has double helical, highly looped secondary structure with GG rich motifs. All known RNA molecules interacting with Purα in vivo contain such structures [Johnson et al., 2006]. It is possible that Purα through its interaction with Rev and RRE can act in remodeling of RNA cargo assisting DDX3, human RNA helicase, a member of DEAD-box family, which binds CRM1 and restructures cargo HIV-1 RNA to permit translocation [Yedavalli et al., 2004]. This is an interesting notion in light of earlier studies showing local helix-unwinding activity of Purα [Darbinian et al., 2001; Wortman et al., 2005].
Altogether, our results demonstrate that Purα participates in cytoplasmic processing of unspliced mRNAs containing intronic viral RNA sequences. Further, we demonstrate that Purα cooperates with Rev to stimulate expression of HIV-1 genes from unspliced mRNAs. Purα acts as a cellular factor, which by interaction with Rev alleviates molecular barriers in Rev function [Neumann et al., 1995; Brack-Werner 1999; Gorry et al., 2003]. Moreover, our results show that Purα is able to promote translocation and expression of unspliced mRNA in the absence of Rev and in LMB insensitive manner.
These observations together with the demonstration of Purα association with RNA transporting granules imply a new role for Purα in stabilization and transport of unspliced viral RNA to sites of translation or viral assembly for encapsidation and virion formation. Purα has been implicated in the transport of Map2 mRNA to sites of translation in neuronal dendrites [Johnson et al., 2006]. In light of earlier reports on the export of intron-containing mRNAs from nucleus and pre-mRNA splicing in neuronal dendrites [Brown et al., 2001; Rodrigues et al., 2001; Jin et al., 2003; Glanzer et al., 2005], it will be of particular interest to determine if Purα has any role in the intracellular transport, stability and translation of cellular pre-mRNAs.
ACKNOWLEDGMENTS
We wish to thank past and present members of the Department of Neuroscience and Center for Neurovirology for their continued support, insightful discussion, and sharing of reagents and ideas. We also wish to thank C. Schriver for editorial assistance. This work was made possible by grants awarded by NIH to KK and SA.
Grant sponsor: NIH.
REFERENCES
- Brack-Werner R. Astrocytes: HIV cellular reservoirs and important participants in neuropathogenesis. AIDS. 1999;13:1–22. doi: 10.1097/00002030-199901140-00003. [DOI] [PubMed] [Google Scholar]
- Braun IC, Herold A, Rode M, Conti E, Izaurallde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J Biol Chem. 2001;276:20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Chatel-Chaix L, Clément JF, Martel C, Bériault V, Gatignol A, DesGroseillers L, Mouland AJ. Staufen is part of the HIV-1 Gag Ribonucleoprotein complex and is involved in the generation of infectious viral particles. Mol Cell Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepenik L, Tretiakova AP, Krachmarov CP, Johnson EM, Khalili K. The single-stranded DNA binding protein, Pur-alpha, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene. 1998;210:37–44. doi: 10.1016/s0378-1119(98)00033-x. [DOI] [PubMed] [Google Scholar]
- Cochrane AW, Perkins A, Rosen CA. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: Relevance of nucleolar localization to function. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: From birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR, Hauber J, Campbell K, Sodroski JG, Haseltine WA, Rosen CA. Subcellular localization of the human immunodeficiency virus trans-acting art gene product. J Virol. 1988;62:2498–2501. doi: 10.1128/jvi.62.7.2498-2501.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daelemans D, Costes SV, Lockett S, Pavlakis GN. Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol Cell Biol. 2005;25:728–739. doi: 10.1128/MCB.25.2.728-739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinian N, Gallia GL, Khalili K. Helix-destabilizing properties of the human single-stranded DNA- and RNA-binding protein Puralpha. J Cell Biochem. 2001;80:589–595. [PubMed] [Google Scholar]
- Darbinian N, White MK, Khalili K. Regulation of the Pur-alpha promoter by E2F-1. J Cell Biochem. 2006;99:1052–1063. doi: 10.1002/jcb.20872. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304 N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Safak M, Khalili K. Interaction of the single-stranded DNA-binding protein Puralpha with the human polyomavirus JC virus early protein T-antigen. J Biol Chem. 1998;273:32662–32669. doi: 10.1074/jbc.273.49.32662. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Darbinian N, Tretiakova A, Ansari SA, Rappaport J, Brady J, Wortman B, Johnson EM, Khalili K. Association of HIV-1 Tat with the cellular protein, Puralpha, is mediated by RNA. Proc Natl Acad Sci USA. 1999;96:11572–11577. doi: 10.1073/pnas.96.20.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallia GL, Johnson EM, Khalili K. Puralpha: A multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–3205. doi: 10.1093/nar/28.17.3197. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallia GL, Darbinian N, Jaffe N, Khalili K. Single-stranded nucleic acid-binding protein, Pur alpha, interacts with RNA homologous to 18S ribosomal RNA and inhibits translation in vitro. J Cell Biochem. 2001;83:355–363. doi: 10.1002/jcb.1247. [DOI] [PubMed] [Google Scholar]
- Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J. RNA splicing capability of live neuronal dendrites. Proc Natl Acad Sci USA. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman CM, Moffat LF, Howard BH. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry PR, Ong C, Thorpe J, Bannwarth S, Thompson KA, Gatignol A, Wesselingh SL, Purcell DF. Astrocyte infection by HIV-1: Mechanisms of restricted virus replication, and role in the pathogenesis of HIV-1-associated dementia. Curr HIV Res. 2003;1:463–473. doi: 10.2174/1570162033485122. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gupta M, Sueblinvong V, Raman J, Jeevanandam V, Gupta MP. Single-stranded DNA-binding proteins PURalpha and PURbeta bind to a purine-rich negative regulatory element of the alpha-myosin heavy chain gene and control transcriptional and translational regulation of the gene expression. Implications in the repression of alpha-myosin heavy chain during heart failure. J Biol Chem. 2003;278:44935–44948. doi: 10.1074/jbc.M307696200. [DOI] [PubMed] [Google Scholar]
- Henderson BR, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: The Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- Hereault Y, Chatelain G, Brun G, Michel D. RNA-dependent DNA binding activity of the Pur factor, potentially involved in DNA replication and gene transcription. Gene Expr. 1995;4:85–93. [PMC free article] [PubMed] [Google Scholar]
- Jin L, Guzik BW, Bor YC, Rekosh D, Hammarskjold ML. Tap and NXT promote translation of unspliced mRNA. Genes Dev. 2003;17:3075–3086. doi: 10.1101/gad.1155703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM, Chen PL, Krachmarov CP, Barr SM, Kanovsky M, Ma Z-W, Lee WH. Association of human Pur alpha with the retinoblastoma protein, Rb, regulates binding to the single-stranded DNA Pur alpha recognition element. J Biol Chem. 1995;270:24352–24360. doi: 10.1074/jbc.270.41.24352. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, Khalili K, Winckler B, Gordon J. Role of Puralpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res. 2006;83:929–943. doi: 10.1002/jnr.20806. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kaniowska D, Kaminski R, Amini S, Radhakrishnan S, Rappaport J, Johnson EM, Khalili K, Del Valle L, Darbinyan A. Cross-interaction between JC virus agnoprotein and human immunodeficiency virus type 1 (HIV-1) Tat modulates transcription of the HIV-1 long terminal repeat in glial cells. J Virol. 2006;80:9288–9299. doi: 10.1128/JVI.02138-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelm RJ, Jr., Elder PK, Getz MJ. The single-stranded DNA-binding proteins, Puralpha, Purbeta, and MSY1 specifically interact with an exon 3-derived mouse vascular smooth muscle alpha-actin messenger RNA sequence. J Biol Chem. 1999;274:38268–38275. doi: 10.1074/jbc.274.53.38268. [DOI] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Muralidharan M, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, Amini S, Gordon J. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol. 2003;23:6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J, Askjaer P. Rev protein and its cellular partners. Adv Pharmacol. 2000;48:251–298. doi: 10.1016/s1054-3589(00)48009-9. [DOI] [PubMed] [Google Scholar]
- Knapp AM, Ramsey JE, Wang SX, Godburn KE, Strauch AR, Kelm RJ., Jr. Nucleoprotein interactions governing cell type-dependent repression of the mouse smooth muscle alpha-actin promoter by single-stranded DNA-binding proteins Pur alpha and Pur beta. J Biol Chem. 2006;281:7907–7918. doi: 10.1074/jbc.M509682200. [DOI] [PubMed] [Google Scholar]
- Krachmarov CP, Chepenik LG, Barr-Vagell S, Khalili K, Johnson EM. Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Pur alpha. Proc Natl Acad Sci USA. 1996;93:14112–14117. doi: 10.1073/pnas.93.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Li Y, Koike K, Ohashi S, Funakoshi T, Tadano M, Kobayashi S, Anzai K, Shibata N, Kobayashi M. Pur alpha protein implicated in dendritic RNA transport interacts with ribosomes in neuronal cytoplasm. Biol Pharm Bull. 2001;24:231–235. doi: 10.1248/bpb.24.231. [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Marion RM, Fortes P, Beloso A, Dotti C, Ortin JA. The human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: Evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Neumann M, Felber BK, Kleinschmidt A, Froese B, Erfle V, Pavlakis GN, Brack-Werner R. Restriction of human immunodeficiency virus type 1 production in a human astrocytoma cell line is associated with a cellular block in Rev function. J Virol. 1995;69:2159–2167. doi: 10.1128/jvi.69.4.2159-2167.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- Ohashi S, Kobayashi S, Omori A, Ohara S, Omae A, Muramatsu T, Li Y, Anzai K. The single-stranded DNA- and RNA-binding proteins pur alpha and pur beta link BC1 RNA to microtubules through binding to the dendrite-targeting RNA motifs. J Neurochem. 2000;75:1781–1790. doi: 10.1046/j.1471-4159.2000.0751781.x. [DOI] [PubMed] [Google Scholar]
- Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TR, Tang H, Xu W, Wong-Staal F. Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA. Oncogene. 2000;19:3570–3575. doi: 10.1038/sj.onc.1203676. [DOI] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci USA. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen CA, Terwilliger E, Dayton A, Sodroski JG, Haseltine WA. Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc Natl Acad Sci USA. 1988;85:2071–2075. doi: 10.1073/pnas.85.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Felber BK, Benko DM, Fenyo EM, Pavlakis GN. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotai Y, Minami H, Saitoh Y, Onodera Y, Mishima Y, Kelm RJ, Jr., Tsutsumi K. A binding site for Pur alpha and Pur beta is structurally unstable and is required for replication in vivo from the rat aldolase B origin. Biochem Biophys Res Commun. 2006;340:517–525. doi: 10.1016/j.bbrc.2005.12.032. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Tang S, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of Staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Tretiakova A, Gallia GL, Shcherbik N, Jameson B, Johnson EM, Amini S, Khalili K. Association of Pur with RNAs homologous to 7SL determines its binding ability to the myelin basic protein promoter DNA sequence. J Biol Chem. 1998;273:22241–22247. doi: 10.1074/jbc.273.35.22241. [DOI] [PubMed] [Google Scholar]
- Wolff B, Cohen G, Hauber J, Meshcheryakova D, Rabeck C. Nucleocytoplasmic transport of the Rev protein of human immunodeficiency virus type 1 is dependent on the activation domain of the protein. Exp Cell Res. 1995;217:31–41. doi: 10.1006/excr.1995.1060. [DOI] [PubMed] [Google Scholar]
- Wolff B, Sanglier JJ, Wang Y. Leptomycin B is an inhibitor of nuclear export: Inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- Wortman MJ, Johnson EM, Bergemann AD. Mechanism of DNA binding and localized strand separation by Pur alpha and comparison with Pur family member, Pur beta. Biochim Biophys Acta. 2005;1743:64–78. doi: 10.1016/j.bbamcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Zapp ML, Green MR. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Pedigo N, Shenoy S, Khalili K, Kaetzel DM. Puralpha activates PDGF-A gene transcription via interactions with a G-rich, single-stranded region of the promoter. Gene. 2005;348:25–32. doi: 10.1016/j.gene.2004.12.050. [DOI] [PubMed] [Google Scholar]


