Abstract
Spt5 is a conserved essential protein that represses or stimulates transcription elongation in vitro. Immunolocalization studies on Drosophila polytene chromosomes suggest that Spt5 is associated with many loci throughout the genome. However, little is known about the prevalence and identity of Spt5 target genes in vivo during development. Here, we identify direct target genes of Spt5 using fogsk8 zebrafish mutant, which disrupts the foggy/spt5 gene. We identified that fogsk8 and their wildtype siblings differentially express less than 5% of genes examined. These genes participate in diverse biological processes from stress response to cell fate specification. Up-regulated genes exhibit shorter overall gene length compared to all genes examined. Through chromatin immunoprecipitation in zebrafish embryos, we identified a subset of developmentally critical genes that are bound by both Spt5 and RNA polymerase II. The protein occupancy patterns on these genes are characteristic of both repressive and stimulatory elongation regulation. Together our findings establish Spt5 as a dual regulator of transcription elongation in vivo and identify a small but diverse set of target genes critically dependent on Spt5 during development.
Introduction
The production of functional mRNA involves multiple steps that include transcription initiation, elongation, and termination. The initiation step is thought to be the major regulatory point for most genes in vivo, despite that promoter-proximal stalled RNA polymerase II (RNAPII) has been detected at several loci including hsp70 [1], HIVLTR [2], c-myc [3], and c-fos [4]. Recently, it has been observed that promoters of many genes are poised for activation, as assessed by chromatin status and RNAPII occupancy in human Hela cells, mammalian embryonic stem cells, and Drosophila embryos [5]–[9]. These observations suggest that regulation at the level of transcription elongation may be more prevalent than previously thought. However, the distinct role of individual transcription elongation factors in critically regulating elongation of target genes remains unclear.
In recent years, biochemical studies have identified over a dozen proteins and a small nuclear RNA, which regulate RNAPII elongation in vitro [10]–[17]. Among these molecules, Spt5, an evolutionarily conserved protein, is the focus of this study. Spt5, together with Spt4 and Spt6, was initially discovered by genetic studies in Saccharomyces cerevisiae [18], and later found to be essential for transcription via modulating chromatin structure [19], [20]. A search for factors that inhibit transcription elongation in the presence of the nucleotide analog 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) led to the biochemical purification of Spt4 and Spt5 as a tight complex named DSIF (DRB Sensitivity Inducing Factor), which has both repressive and stimulatory activity on transcription elongation in vitro [21]. Spt5 has also been identified independently as a protein required for HIV-1 Tat trans-activation in fractionated extracts [22]. In addition, Spt5 interacts and modulates activity of factors implicated in mRNA maturation and surveillance [23]–[25].
At the phenotypic level, the in vivo role of Spt5 in multi-cellular organisms has been revealed through genetic analyses in C. elegans, Drosophila, and zebrafish. Inactivation of Spt5 through RNAi in C. elegans arrest embryonic development prior to gastrulation [26]. In Drosophila, a hypomorphic mutation in spt5 displays defects in the expression of segmental patterning genes [27]. At the molecular level, Spt5 localization on polytene chromosomes and chromatin immunoprecipitation studies reveal broad distribution of Spt5 at sites of active transcription as well as at the un-induced heat shock gene promoters [28]–[30]. In zebrafish, whereas a hypomorphic mutation in spt5 (the fogm806 allele) displays distinct defects in neuronal development [31], severe truncation (the fogsk8 allele) or deletion (the fogs30 allele) of the gene product causes broad deficits in embryonic development [32]. Together, these analyses establish functional significance of Spt5 in vivo, implicate intricate regulatory mechanisms that remain to be elucidated, and suggest that Spt5 may regulate a large number of target genes in vivo. Despite these insights, only the heat shock protein-encoding hsp70 and HIV viral genes, have been demonstrated to be regulated at the level of transcription elongation by Spt5 in intact cells [29], [33], whereas induction of the immediate early gene c-fos appears to depend on the stimulatory activity of Spt5 [34]. Thus, an important question remains: What other genes, if any, are critically dependent on Spt5 in vivo?
Here we report expression profiling of over 10,000 protein-coding genes using ∼24 hours post fertilization (hpf) zebrafish embryos. Our results identified a surprisingly small fraction (<5%) of genes that were differentially expressed between fogsk8 mutants and their wildtype (WT) siblings. Pathway analyses revealed that these differentially expressed genes are involved in diverse biological pathways that range from stress response to cell fate specification. A gene structure analysis showed that genes upregulated in fogsk8 embryos had a significantly shorter length, compared to the average gene length in zebrafish. Furthermore, in vivo chromatin immunoprecipitation (ChIP) on selected set of differentially expressed genes in 24 hpf WT embryos uncovered Spt5 target genes: among the target genes repressed by Spt5 (including gadd45b, c-fos, hsp70, smoothened, and follistatin), RNAPII occupancy was higher at the 5′ than 3′ end of these genes; among the target genes activated by Spt5 (including lunatic fringe and tropomyosin a), RNAPII occupancy was detected at both their 5′ and 3′ ends. ChIP analyses in fogsk8 mutant embryos identified gadd45b that exhibited significantly increased RNAPII occupancy at the 3′ end compared to that in WT embryos, whereas other target genes unexpectedly showed reduced RNAPII occupancy at both 5′ and 3′ ends. This observation highlights the dynamic and complex nature of elongation regulation in vivo. Taken together, our results establish Spt5 as a dual regulator of transcription elongation in vivo and reveal a small but diverse set of direct target genes.
Results
Spt5 protein is significantly reduced in 24 hpf fogsk8 mutant embryos
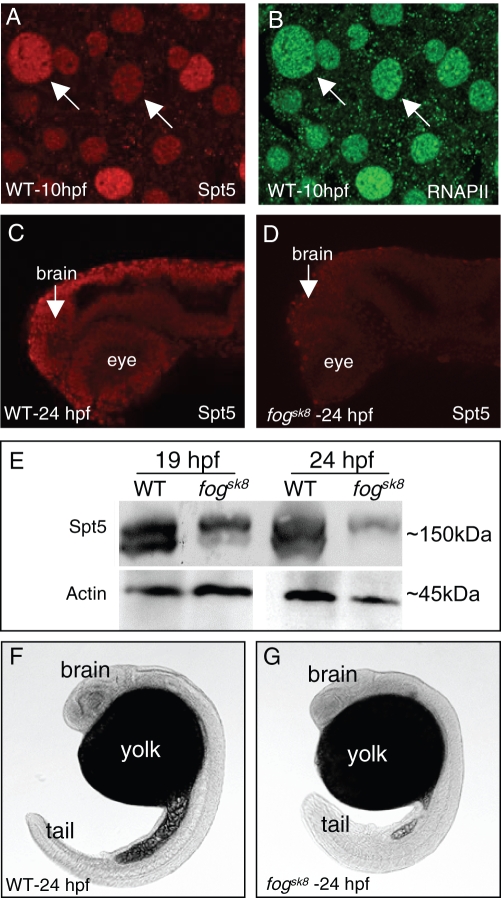
The foggy/spt5 transcript is broadly distributed throughout the developing embryo [31], [32]. To gain insights into the level and distribution of Spt5 protein, an antibody directed against the C-terminal end of the zebrafish Spt5 protein was generated. Double-labeling with 8WG16, an antibody that recognizes RNAPII, demonstrated prominent nuclear distribution of Spt5 (Fig. 1A–B). The fogsk8 mutation (Fig. 1F–G) is predicted to yield a non-functional zygotic gene product, and the maternal transcript is undetectable after 10 hours post fertilization (hpf) [32]. However, the status of the maternal protein was unclear. The sk8 mutant phenotype is first visible around 19 hpf, suggesting the presence of maternal Foggy protein at earlier stages of development. Immunostaining (Fig. 1C–D) showed that maternal Spt5 protein was significantly reduced in fogsk8 mutant embryos as compared to WT siblings. Western blotting (Fig. 1E) detected two bands around the predicted size of zebrafish Spt5 in WT embryos. Since Spt5 is known to be phosphorylated [14], [34]–[38] and methylated [39], these bands may represent forms of Spt5 with different post-translational modifications. Although the intensity of both bands was reduced in the fogsk8 mutant, the faster migrating form appeared more severely affected (Figure 1E). This result indicates that fogsk8 mutant at 24 hpf exhibits a significant reduction but not complete absence of Spt5 protein.
Figure 1. Spt5 protein is significantly decreased in fogsk8 embryos.
(A–B) Whole mount immunofluorescence detection of Spt5 and RNAPII in zebrafish embryos, showing the nuclear localization pattern of Spt5 protein. (C–D) Whole mount immunofluorescence detection of Spt5 in WT and fogsk8 embryos at 24 hpf, showing overall decrease of Spt5 protein in fogsk8 embryos. Confocal images of lateral views are shown, with the anterior to the left and dorsal up. (E) Western blot of wild-type (WT) and fogsk8 embryonic extracts at 19 hpf and 24 hpf shows substantial decrease in maternal Spt5 protein in the mutant. (F–G) Light microscopic view of WT and sk8 embryos at 24 hpf, showing the gross morphological phenotype of the mutant embryos.
Expression profiling reveals a small number of differentially expressed genes in 24 hpf fogsk8 embryos
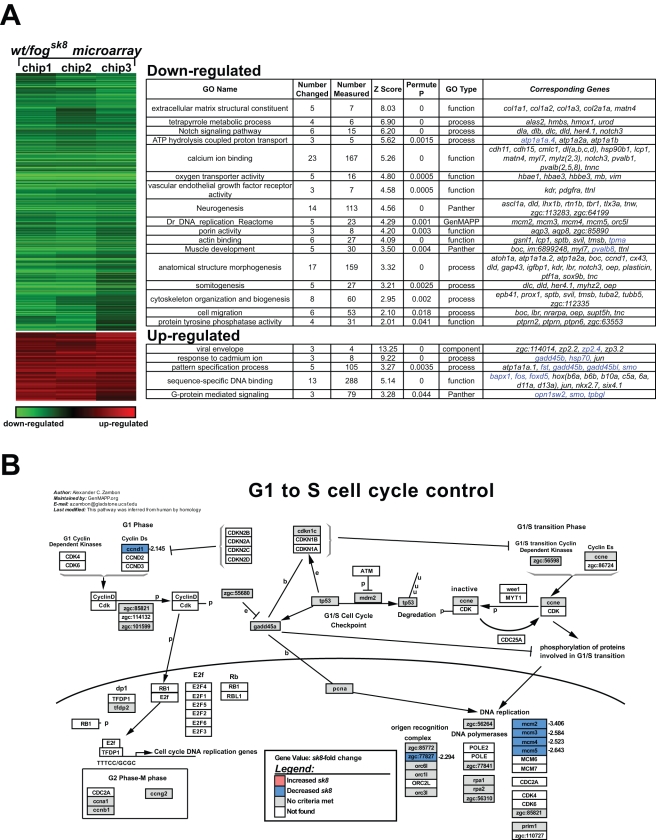
To identify potential downstream target genes of Spt5, we performed microarray analysis by hybridizing zebrafish Affymetrix oligoarrays, which contain over 10,000 protein-coding genes, with biotinylated cRNA probes from 24 hpf fogsk8 mutant and WT sibling embryos. 75% of the probe sets displayed above-background signal-to-noise ratio (as determined by microarray absent-present calls), suggesting that a majority of genes on the array have detectable expression at this stage. Interestingly however, only 4.5% of the genes were differentially expressed (absolute fold greater than 2, student t-test P<0.05) between fogsk8 and WT siblings. The 2-fold cutoff was used because an expression change of less than 2 fold was often not validated by quantitative PCR analysis (data not shown). Among the differentially expressed genes, 97 (∼1% of the total genes examined) were up-regulated (ranging from +2- to +10-fold), and 358 (∼3.5% of the total genes examined) were down-regulated (from −2- to −300-fold) in fogsk8 embryos (Fig. 2A). Many of the differentially expressed genes have previously been shown to display dynamic spatial and temporal expression patterns [40]. Four genes (gch, pvalb, hoxb6, dlx2) were validated through whole mount in situ analysis (Supplemental Fig. S1) and 28 other genes were further validated through quantitative RT-PCR analysis (Supplemental Tables S1 and S2).
Figure 2. Gene expression profiling reveals that the expression of a small percentage of genes is affected in fogsk8 mutant embryos.
(A) HOPACH cluster diagram of the microarray data are shown for individual replicates compared with the mean for 24 hpf embryos. 615 probe sets with fold change >2 and t test P-value <0.05 are displayed along with Gene Ontology terms and pathways over-represented as determined by a Fisher's exact test permutation based P-value. The intensity of down-regulation (green) and up-regulation (red) reflects the magnitude of fold change. Representative genes within the GO categories are shown in the last column. Genes subjected to further analyses in this study are represented in blue color in the last column. (B) G1 to S Cell Cycle pathway is shown as an example of GenMAPP pathway analysis. The pathway map shows genes that are regulated as a group (e.g. mcm genes) and genes that are not differentially expressed in fogsk8 embryos but are present on the array (grey color).
To identify biological pathways that might be regulated by Spt5, we employed GenMAPP [41] and associated tools [42] (http://www.genmapp.org/go_elite) to analyze pathway and gene ontology over-representation in our differentially expressed gene dataset. This analysis found that genes induced by environmental or cellular stress (such as gadd45b and hsp70) or encoding certain transcription factors (such as foxd5 and fos) were upregulated, whereas genes that are constituents of the extracellular matrix (e.g. collagen) and sodium/potassium ion transporters (e.g. atp1a1a) were down-regulated in fogsk8 embryos (Fig. 2A). The expression of genes previously suggested to be regulated at the level of elongation [10], including hsp70, fos, globin, tubulin, and opsin, were found altered in fogsk8 embryos. Moreover, the expression of multiple genes in the cell cycle pathway was reduced in fogsk8 embryos (Fig. 2B). In contrast, genes from major developmental pathways (e.g. fibroblast growth factor, wingless), although present on the array, were largely unaffected in fogsk8 embryos (Supplementary Table S3). In addition, RNA processing genes and protein biosynthesis genes (ribosome constituents) were under-represented in our differentially expressed gene dataset (data not shown). It is worth pointing out that many differentially expressed were novel, and were thus not assignable to specific GenMAPP pathways. Taken together, these results indicate that the lack of zygotic Spt5 protein only affects the expression of a small percentage of genes at 24 hpf, and these genes are involved in a number of distinct biological pathways.
Genes that are upregulated in fogsk8 embryos are shorter in gene length compared to all genes examined
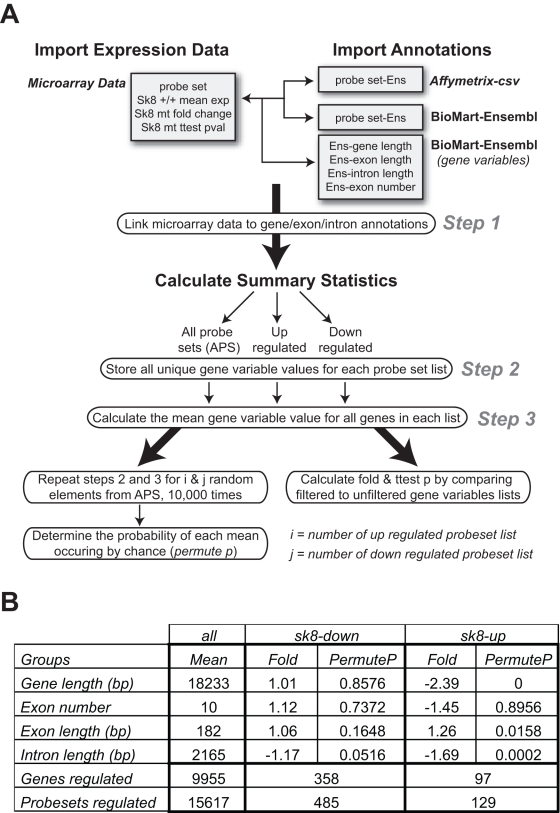
Little is known as to what signatures may subject a gene to Spt5 regulation. To determine if differentially expressed genes have common features in gene structures, we performed bioinformatics analysis on these gene sets. To assess the extent and overall likelihood of a difference between the length of fogsk8 –dependent genes and that of all other genes in the array, probe sets from the microarray were linked to gene structure data from the Ensembl database, the mean of different gene features (e.g., genomic length) was computed, and fold change and P values were calculated via a permutation based analysis (Fig. 3A).
Figure 3. Permutation analysis uncovers a significant shorter length of those genes that are up-regulated in fogsk8 mutant embryos.
(A) Algorithm design for assessing global changes in overall gene length. Unique Ensembl genes linked to probesets were obtained from the Affymetrix website and BioMart, and genomic gene, exon and intron length were also obtained from BioMart and summarized for up-, down-regulated and all array probesets (AAPs). Folds relative to AAPs linked genes were calculated as well as permutation based p-values by randomly selecting input probeset lists of the same size as the regulated sets from AAPs, over 10,000 iterations. (B) Table summarizing the permutation based analysis of the structural features of genes differentially expressed in fogsk8 embryos. Up-regulated genes have significantly shorter gene length, shorter intron length and longer exon length than other genes on the array. Down-regulated genes do not show significantly different gene structure requirements compared to other genes.
Using this method, we found that genes upregulated in fogsk8 embryos had on average ∼2.39 fold decrease in overall gene length and ∼1.69 fold decrease in intron length, as compared to all gene-linked probe sets on the array (P<0.05) (Fig. 3B). These differences were unlikely to occur due to chance, because a fold change larger than or equal to the gene length difference (2.39 fold) was not observed in 10,000 random permutations of all microarray probe sets, and that larger than or equal to the intron length difference (1.69 fold) only occurred twice over 10,000 random permutations (P = 0.0002), hence emphasizing the unlikelihood of such events occurring at random. Although this analysis also showed a significant increase in exon length for upregulated genes, the difference was relatively small (1.26 fold). None of the gene structure data differed between downregulated genes and all other genes on the array (Fig. 3B). Taken together, this analysis suggests that genes repressed by Spt5 tend to be shorter in length.
In vivo chromatin immunoprecipitation (ChIP) reveals Spt5 occupancy patterns on differentially expressed genes in 24 hpf WT embryos
As microarray experiments measure the steady-state levels of mRNA and Spt5 is involved in many aspects of mRNA stability by virtue of its association with many components of the transcriptional machinery , many of the genes differentially expressed in fogsk8 mutants might represent mRNA processing targets of Spt5. In order to better identify genes that are regulated at the elongation level at 24 hpf, we performed chromatin immunoprecipitation (ChIP) assays using antibodies against RNAPII and Foggy.
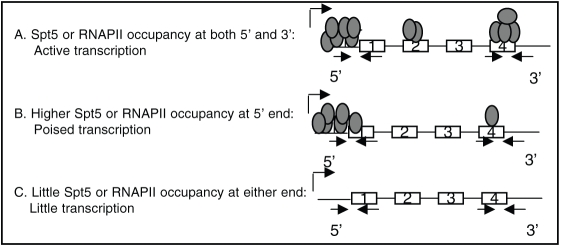
Based on the current model of regulation at the level of transcription elongation [16], [17], the following patterns of Spt5 and RNAPII occupancy are expected on their target genes (Fig. 4): A) Spt5 target genes that are in a state of productive transcription elongation shall display Spt5 and RNAPII occupancy at both 5′ and 3′ ends of their chromatin. B) Spt5 target genes that are repressed by a promoter-proximal transcription elongation block shall have minimal Spt5 and RNAPII occupancy at 3′ end but significantly more Spt5 and RNAPII occupancy at 5′ end. C) Genes that are not Spt5 targets or are transcriptionally inactive shall lack detectable Spt5 or RNAPII respectively throughout their chromatin.
Figure 4. Schemata showing predicted transcriptional status based on Spt5 and RNAPII occupancy.
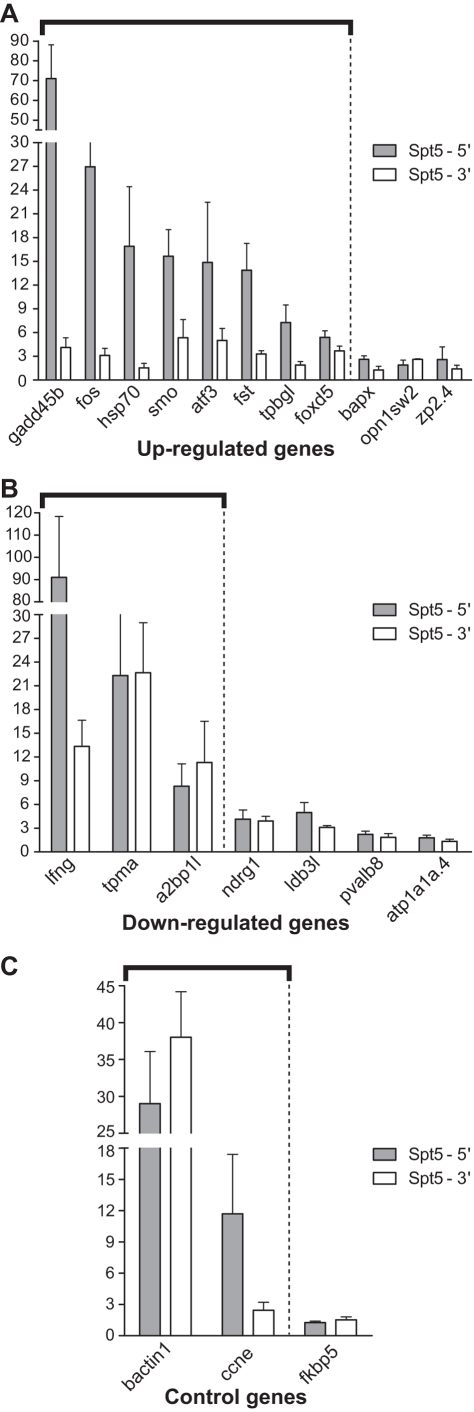
Based on the significance of expression fold changes, constraints on availability of reliable genomic structure (exon/intron) information, and known gene functionality, 11 upregulated genes, 7 downregulated genes, and 3 control genes were selected for ChIP analysis (Table 1 and Supplementary Table S4) in 24 hpf WT embryos. Among the 11 upregulated genes, 8 (gadd45b, fos, hsp70, smo, atf3, fst, tpbgl, foxd5) showed high Spt5 occupancy at 5′ end but little at 3′ end, whereas the remaining three genes did not show significant Spt5 occupancy on either end (Fig. 5A and Table 1). Among the 7 downregulated genes, 3 (lfng, tpma, a2bp1-like) showed significant Spt5 occupancy at both 5′ and 3′ ends, whereas the remaining 4 genes did not (Fig. 5B and Table 1). Genes that showed significant Spt5 occupancy are likely to be targets of Spt5, whereas those that did not have significant Spt5 occupancy were either transcriptionally inactive or not directly regulated by Spt5. Two (beta-actin 1 and cyclin E) of the three control genes showed significant Spt5 occupancy (Fig. 5C and Table 1), whose expression was largely unaltered in 24 hpf fogsk8 embryos, suggesting that these transcripts might be regulated by additional elongation factors.
Table 1. Summary of Genes Used for in vivo CHIP Analysis.
| No. | Gene Symbol | Gene Name | GO Terms* | Array/qRT-PCR Fold Change | Genomic length (kb) | Transcript length (kb) | No. of Exons | Degree of Spt5 occupancy | Degree of RNAPII occupancy |
| Genes Up-regulated in 24 hpf fogsk8 Mutant Embryos | |||||||||
| 1 | gadd45b | Growth arrest and DNA-damage-inducible, beta | Stress response | 6.34/6.07 | 1.6 | 1.2 | 4 | ++++ | ++++ |
| 2 | fos | v-fos FBJ murine osteosarcoma viral oncogene | Transcription Factor | 5.68/8.44 | 2.1 | 1.7 | 4 | ++++ | ++++ |
| 3 | hsp70 | Heat shock protein 70 | Stress response | 5.6/2.32 | 3.4 | 2.3 | 2 | +++ | +++ |
| 4 | fst | Follistatin | Pattern specification | 2.25/2.07 | 5.5 | 3.4 | 5 | +++ | ++ |
| 5 | smo | Smoothened homolog | Pattern specification | 2.02/2.12 | 5.6 | 3.0 | 12 | +++ | ++++ |
| 6 | atf3 | Activating transcription factor 3 | Transcription Factor | 2/1.89 | 6.1 | 2.6 | 4 | +++ | ++ |
| 7 | tpbgl | Trophoblast glycoprotein-like | Receptor | 3.24/2.37 | 2.6 | 1.7 | 2 | ++ | +++ |
| 8 | foxd5 | Forkhead box D5 | Transcription Factor | 9.83/10.77 | 4.1 | 1.1 | 2 | + | + |
| 9 | bapx | Bagpipe homeobox 1/ NK3 homeobox 2 | Transcription Factor | 4.62/4.62 | 0.5 | 0.3 | 2 | − | − |
| 10 | zp2.4 | Zona pellucida glycoprotein 2.4 | Viral envelope | 3.74/6.02 | 1.8 | 1.3 | 8 | − | − |
| 11 | opn1sw2 | Opsin 1, short-wave-sensitive 2 | Signal transduction | 2.72/7.81 | 2.6 | 1.3 | 5 | − | − |
| Genes Down-regulated in 24 hpf fogsk8 Mutant Embryos | |||||||||
| 1 | lfng | Lunatic fringe homolog | Signaling | 3/3.68 | 9.4 | 2.9 | 8 | ++++ | ++++ |
| 2 | tpma | Alpha tropomyosin | Actin binding | 2.86/3.41 | 4.5 | 1.3 | 10 | +++ | +++ |
| 3 | a2bp1l | ataxin 2-binding protein 1-like | Nucleotide binding | 10.69/7.83 | 18.5 | 1.1 | 12 | ++ | ++ |
| 4 | ndrg1 | N-myc downstream regulated gene 1 | Cell differentiation | 6.04/8.47 | 19.9 | 2.3 | 16 | − | + |
| 5 | ldb3l | LIM domain binding 3 like | Protein binding | 3.64/3.44 | 39.3 | 1.6 | 12 | − | − |
| 6 | pvalb8 | Parvalbumin 8 | Calcium ion binding | 17.62/12.23 | 174.7 | 0.8 | 5 | − | − |
| 7 | atp1a1a.4 | ATPase, Na+/K+ transporting, alpha 1a.4 | Sodium\potassium ATPase | 3.37/62.85 | 7.5 | 3.2 | 22 | − | − |
| Genes Unchanged in 24 hpf fogsk8 Mutant Embryos | |||||||||
| 1 | bactin1 | Beta Actin 1 | Cytoskeleton | 1.26/−1.8 | 3.6 | 1.7 | 6 | +++ | ++++ |
| 2 | ccne | Cyclin E | Cell cycle | 1/1.87 | 9.8 | 2.0 | 12 | ++ | − |
| 3 | fkbp5 | FK506 binding protein 5 | Protein folding | 1/1.04 | 9.9 | 2.4 | 11 | − | − |
GO/MAPP program-suggested gene function.
Figure 5. In vivo ChIP analysis using Spt5 antibody on differentially expressed genes reveals putative target genes directly bound by Spt5.
(A–C) Spt5 occupancy on upregulated, downregulated, and control genes. Y- axis values in the graphs represent fold Foggy occupancy over fold pre-immune serum occupancy levels. Arbitrary cutoff for detectable Foggy fold occupancy over control serum is set at 5. Thick black bars highlight the genes that display significant Spt5 occupancy at either 5′ or 3′ end of the chromatin. The results shown are the average of at least three independent replicates. Error bars are SEM values. Note that the scales are different for each of the graphs.
ChIP analysis using the 8WG16 antibody in 24 hpf WT embryos reveals RNAPII distribution patterns on the differentially expressed genes
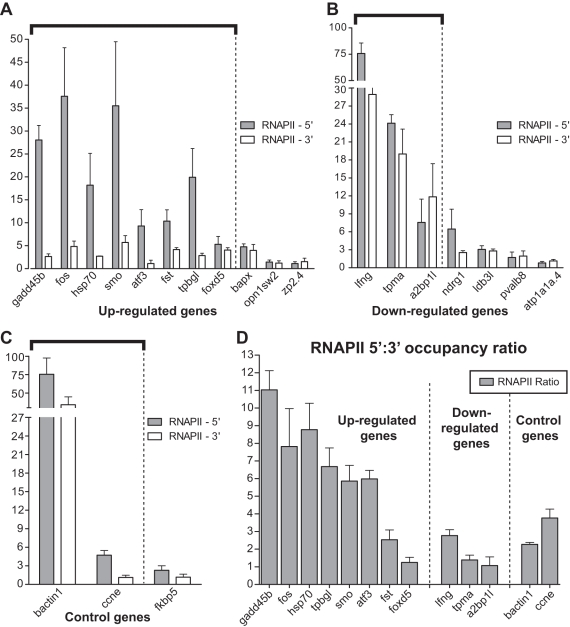
We next examined RNAPII distribution in 24 hpf WT embryos to determine the transcriptional status of the genes. The 8WG16 antibody was used in our experiments, as it has been successfully used to recognize RNAPII occupancy on the chromatin at both the 5′ and 3′ ends of genes in other species [43]. In general, the RNAPII occupancy pattern correlated well with that of Spt5. Among those that were upregulated in the fogsk8 mutant embryo and also displayed significant Spt5 occupancy (indicating that their expression is repressed by Spt5), more RNAPII was detected at 5′ than 3′ end (Fig. 6A, 6D, and Table 1). This finding is consistent with the idea that the genes repressed by Spt5 are in a poised state of transcription. Among those that were downregulated in the fogsk8 mutant embryo and also displayed significant Spt5 occupancy, significant occupancy of RNAPII was also detected at both 5′ and 3′ ends (Fig. 6B, 6D, and Table 1), indicating that these genes require the stimulatory activity of Spt5 for productive transcription in 24 hpf WT embryos. The genes that exhibited little Spt5 occupancy also showed little RNAPII occupancy, confirming that these genes are not transcriptionally active in 24 hpf WT embryos. Taken together, our results identify gadd45b, fos, hsp70, tpbgl, smo, atf3, fst, and foxd5 as target genes that are repressed by Spt5 at the elongation step, and lfng, tpma, and a2bp1-like as target genes that are activated by Spt5 at the elongation step. Though genes such as fos and hsp70 have been known to be regulated by elongation, our results identify multiple critical developmental regulators such as smo (regulating Hedgehog signaling) [44], fst (regulating Transforming Growth Factor beta signaling) [45], and lfng (regulating Notch signaling) [46] as novel elongation targets of Spt5.
Figure 6. Distinct patterns of RNAPII occupancy at putative Spt5 target genes.
(A–D) ChIP data showing RNAPII occupancy in 24 hpf WT embryos on the genes that are up-regulated in fogsk8 embryos (A), genes that are down-regulated in fogsk8 embryos (B), control genes (C), and the ratio of RNAPII occupancy at 5′ vs 3′ end (D). Y-axis values in the graphs represent fold RNAPII occupancy over fold mouse control serum occupancy. Arbitrary cutoff for detectable RNAPII fold occupancy over control serum is set at 5. Thick black bars highlight the genes that display significant RNAPII occupancy at either 5′ or 3′ end of the chromatin. The results shown are the average of at least three independent replicates. Error bars are SEM values.
Analysis of RNAPII occupancy on putative Spt5 target genes in 24 hpf fogsk8 embryos
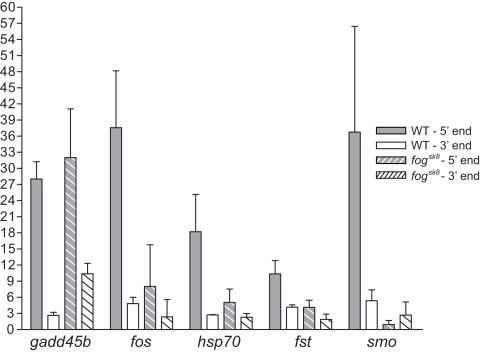
Based on the above results and current model of transcription elongation, one might expect, in the fogsk8 embryos, an increased occupancy of RNAPII at 3′ end of the genes whose transcription elongation is repressed by Spt5 in WT embryos. Alternatively, depletion of Spt5 may lead to a decreased association of RNAPII with the 5′ end of genes, due to a reduced level of promoter-proximal pausing, as has been observed in the case of NELF depletion [8]. To test these predictions, in vivo ChIP was carried out using RNAPII antibody in 24 hpf fogsk8 embryos. Due to limited availability of fogsk8 embryos, we focused on five genes, hsp70, fos, gadd45b, fst, and smo. The patterns of RNAPII occupancy in 24 hpf fogsk8 embryos were shown in Fig. 7. For most genes tested, we observed significantly decreased RNAPII occupancy at 5′ end and no increase of occupancy at 3′ end, in comparison to that in WT embryos, consistent with the second prediction. One gene, gadd45b, however, fitted with the first prediction: RNAPII occupancy at its 5′ end was similar between WT and fogsk8 mutant embryos, but RNAPII occupancy at its 3′ end was significantly increased in fogsk8 embryos as compared to WT (Fig. 7). Reasons behind such differential patterns of RNAPII occupancy at different target genes were not clear (see Discussion). Taken together, our in vivo expression profiling and ChIP analyses indicate that Spt5 exerts dual activity in regulating mRNA expression and RNAPII occupancy in vivo and in a gene-specific manner.
Figure 7. RNAPII occupancy on direct target genes of Spt5 in fogsk8 mutant embryos reveals unexpected complexity and increased 3′ occupancy at the gadd45b locus.
ChIP with RNAPII antibody in WT and fogsk8 embryos on gadd45b, fos, hsp70, fst, and smo genes show that in fogsk8 embryos, RNAPII occupancy is significantly increased at 3′ end of gadd45b without increase at 5′ end. Other genes tested showed reduction of RNAPII occupancy at 5′ ends but no change at 3′ ends in fogsk8 embryos compared to WT. The results shown are the average of at least three independent replicates. Error bars are SEM values.
gadd45b mRNA is globally increased leading to increased protein products in fogsk8 embryos
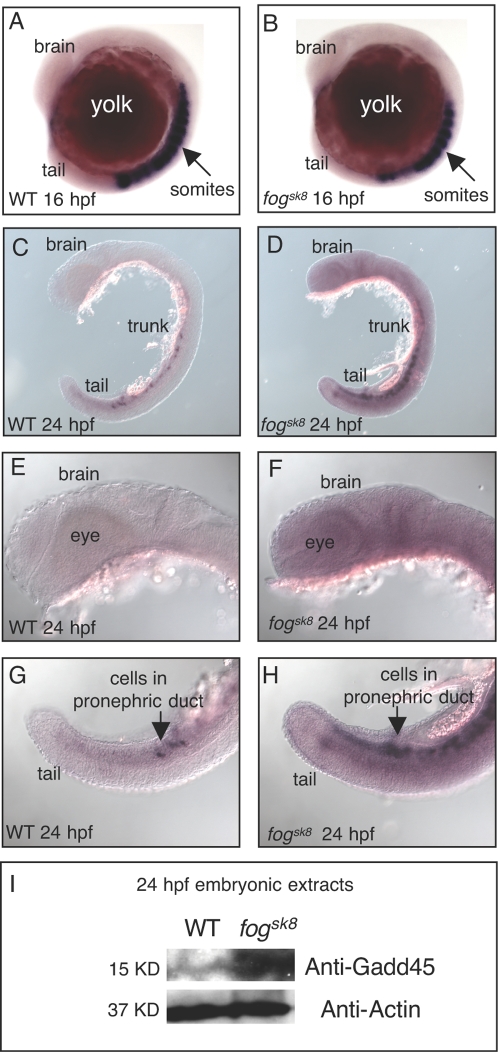
The one gene that did show increased RNAPII occupancy in the fogsk8 mutant, gadd45b, is regulated by multiple important cellular signaling pathways including NF-kB and JNK [47] and is involved in zebrafish somitogenesis [48]. To probe the significance of Spt5 regulation of this gene, we asked whether and where in the fogsk8 embryos the de-repression of gadd45b results in overall mRNA increase, through whole-mount in situ hybridization. In 16 hpf WT embryos, gadd45b was strongly expressed in the developing somites, and such expression was largely normal in the fogsk8 mutant (Fig. 8A–B). In 24 hpf WT embryos, gadd45b expression was detected in cells of pronephric ducts and weakly in posterior paraxial mesoderm and minimally elsewhere (Fig. 8C, 8E, 8G), revealing the dynamic nature of gadd45b expression in development. In 24 hpf fogsk8 embryos, a global increase of gadd45b RNA (Fig. 8D, 8F, 8H), and protein (Fig. 8I) was detected, thus demonstrating the significance of Spt5 regulation of this gene.
Figure 8. Global increase of gadd45 mRNA and increased Gadd45b protein in fogsk8 mutant embryos.
(A–H) Whole-mount in situ hybridization using the gadd45b antisense probe in WT and fogsk8 embryos at 16 hpf and 24 hpf. gadd45b expression is not increased at 16 hpf but shows widespread increase at 24 hpf in fogsk8 embryos. (I) Western blot analysis of WT and fogsk8 embryo extracts with commercially available anti-Gadd45b antibody and anti-Actin antibody shows significant increase of Gadd45b protein in fogsk8 embryos at 24 hpf.
Discussion
Spt5 has a demonstrated dual activity in regulating transcription elongation in vitro [21], and may regulate a large number of genes in vivo [28], [29], [49]. However, the prevalence and identity of genes whose expression is critically dependent on Spt5 are poorly defined in any in vivo system. In this study, we have carried out expression profiling of over 10,000 genes using a zebrafish zygotic null mutant, fogsk8. Maternally deposited foggy/spt5 gene products have sustained fogsk8 embryos to a developmental stage when sufficient amount of tissues are experimentally available and foggy/spt5 gene products become significantly depleted. Our results show that a surprisingly small fraction of genes (<5%) are differentially expressed between WT and fogsk8 embryos at 24 hpf. In vivo ChIP analyses with both Spt5 and RNAPII antibodies on a select set of genes further identify targets that are regulated either by the repressive or by the stimulatory activity of Spt5 at the level of transcription elongation. The observation that Spt5 associates with control genes confirm that Spt5 occupancy does not equate to functional necessity. Moreover, RNAPII occupancy patterns observed in fogsk8 embryos emphasize the dynamic and complex nature of transcription regulation by Spt5 in a gene-specific manner in vivo.
The prevalence of target genes that require Spt5 regulation in vertebrate development
Previous immunolocalization studies using Drosophila polytene chromosomes show that non-productively elongating RNAPII, together with Spt5, labels many chromosomal sites [28], [29]. Recent genome-wide analyses also find significant RNAPII occupancy at the 5′ end of many genes, whether or not their transcripts can be detected [5]–[7]. These studies imply that most, if not all, genes may be dependent on Spt5 for transcription regulation. In contrast to this view, our survey of over 10,000 genes at one developmental stage (24 hpf) reveals less than 5% of differentially expressed genes in fogsk8 embryos. Several possibilities may explain why such a small fraction of genes show altered expression in fogsk8 embryos: First, as localization does not always reflect functional dependence, it is possible that, although a much larger number of genes are bound by Spt5, only a fraction of them are critically dependent on its activity. Indeed, our ChIP analyses have found that Spt5 are associated with two genes whose expression is largely unchanged in fogsk8 embryos. Second, despite significant reduction of Spt5 protein level in fogsk8 embryos, some maternal proteins still remain, which could mask the identification of those genes whose expression is not particularly sensitive to Spt5 protein reduction. Although removal of maternal Spt5 will allow the test of this hypothesis, it may potentially lead to early death of embryos, rendering the experiments impossible to carry out. Third, since this expression profiling experiment cannot distinguish the relative contribution of maternal versus zygotic mRNA to the steady state mRNA levels, it remains possible that maternal mRNAs that are not yet degraded by 24 hpf could mask changes of Spt5-dependent zygotic gene expression. Finally, since only one developmental stage is examined in this study, it remains possible that a different set of genes and/or a much larger number of them may show altered expression in the mutant at other developmental stages. Interestingly, when we analyzed the expression of 21 candidate target genes in a hypomorphic mutation (the fogm806 allele) at 48 hpf, 15 of the genes tested were unchanged while four (fos, ndrg1, atp1a1a.4 and pvalb) showed similar trend as in fogsk8 mutant (Supplemental Table S5). This observation suggests that the fogm806 allele is likely to affect even smaller number of genes. Taken together, we suggest that, at the late segmentation stage of zebrafish development, only a small fraction of genes is critically dependent on Spt5 for transcription regulation, even though a large number of genes may recruit Spt5 to their chromatin.
Target genes that are positively or negatively regulated by Spt5
In addition to previously known categories of inducible genes, our pathway analysis revealed that Spt5 target genes belong to diverse biological pathways and functions. Further studies are required to bridge the gap between the known function of the genes to the mechanism of their gene regulation at the elongation level, particularly to developmentally regulated genes. Our bioinformatics analyses reveal that genes repressed by Spt5 are significantly shorter in gene length. Two possibilities, not mutually exclusive, may account for this observation: First, as one distinct advantage of regulating transcription elongation is to allow rapid gene induction in response to extrinsic signals, genes that are regulated in this manner may have been selected during evolution to have shorter length, in order to further accommodate the need for accelerated transcription. Interestingly, genes with shorter introns and overall shorter gene length are hallmarks of immediate early (IE) genes. Since only three of the genes up-regulated in the fogsk8 appear to be well- characterized IE genes (gadd45b, fos and jun), further studies are required to determine if other genes represented in this category also function as IE genes. Second, since the severe depletion of Spt5 in fogsk8 embryos lead to deficits of not only the repressive but also the stimulatory activity, it is possible that the genes with longer length are not up-regulated in fogsk8 embryos because of their dependency on both the repressive as well as the stimulatory activity of Spt5.
Our expression profiling together with in vivo ChIP analyses at one developmental stage have identified two non-overlapping sets of target genes: those that are negatively regulated by Spt5 and those that are positively regulated by Spt5. Are there indeed two distinct populations of genes that differentially require Spt5 activity in vivo? We suggest not. First, this study found that hsp70 belongs to the group that is negatively regulated by Spt5. Under heat stimulus, however, it has been previously shown that hsp70 induction also depends on Spt5 [32]. Thus, one gene may require both the repressive and the stimulatory activity of Spt5, depending on developmental states or surrounding environmental signals. Spt5 is shown to be phosphorylated by the positive elongation kinase P-TEFb [14], [34], [36]–[38], and such phosphorylation is proposed to act as a switch that converts Spt5 from a repressor to an activator [34]. Although such model remains to be tested in vivo, it supports the idea that Spt5 and transcription elongation are dynamically regulated in vivo, and same set of genes may be regulated by the repressive and stimulatory activity of foggy/Stp5, depending on the in vivo states.
Dynamics and complexity of regulated transcription elongation in vivo
Our ChIP analyses in fogsk8 embryos reveal decreased RNAPII occupancy along the chromatin of four genes (fos, hsp70, fst, and smo) among total five genes tested. Since the transcripts of these genes are increased in fogsk8 embryos at 24 hpf, together with the demonstrated RNAPII and Spt5 occupancy on these genes in WT embryos, one possible explanation is that increased 3′ RNAPII occupancy may have occurred at an earlier developmental stage, coinciding with the onset of transcript increase from these genes. Because of Spt5 depletion, promoter-proximal association of RNAPII was reduced subsequently in fogsk8 mutant embryos, resulting in observed decrease of RNAPII occupancy at 24 hpf. Indeed, it has been shown in yeast that Spt4-5 complex is important for stabilizing RNAPII on the chromatin template under normal and DNA-damage inducing conditions [50]. Similar reduction of promoter-proximal associated RNAPII has also been observed after NELF knockdown in Drosophila [8].
Among all the up-regulated genes subjected to ChIP analysis, gadd45b showed increased RNAPII occupancy at the 3′ end in the fogsk8 mutant embryos. It also exhibited highest Spt5 occupancy at the 5′ end in WT (Figure 5A). In addition, the onset of gadd45b mRNA increase in fogsk8 embryos may be rather close to 24 hpf, since no increase in gadd45b mRNA was detected in 16 hpf fogsk8 embryos. These observations may explain why gadd45b displays increased RNAPII occupancy patterns in fogsk8 embryos. The significantly up-regulated Gadd45b protein levels suggest that the produced gadd45b transcripts can be translated in vivo. In zebrafish, overexpression of gadd45b by mRNA injection at the 1-cell stage leads to somite segmentation defect and a down-regulation of myoD [48]. Since gadd45b was not misexpressed in fogsk8 embryos until late segmentation stages, fogsk8 embryos do not show obvious segmentation defects and no reduction in myoD transcript levels was observed in our expression profiling analysis. Thus, understanding the contribution of overexpressed gadd45b or any other target genes to the fogsk8 mutant phenotype would require a precise determination of temporal profiles of their expression in the fogsk8 mutant, followed by conditional knockdown or overexpression of these target genes with temporal control.
How might the control of gadd45b expression by Spt5 fit into the known regulation of gadd45b? Gadd45b is regulated by NF-kappaB [51] and TGF-beta [52] in mammalian cell cultures, though its in vivo regulation is poorly understood. Future studies are required to elucidate any potential interaction between Spt5 and the NF-kB and/or TGF-beta pathway members to understand the interplay between DNA binding transcription factors and elongation regulation of gadd45b. It has been previously suggested that gadd45 falls into the category of ‘preset’ genes whose upregulation is independent of changes in chromatin structure (by binding of transcription factors), or synthesis of new transcription factors, or stimuli-induced DNA-protein interactions [53]. Hence, it is tempting to speculate that the elongation control of gadd45b by Foggy/Spt5 is the switch that regulates these ‘preset’ genes. Transcription elongation control of such ‘preset’ genes might be crucial to integrate signals from multiple pathways (gadd45b controlled by NF-kB and TGF-b) to coordinate regulation of important signaling pathways that ultimately determine cell fate and survival.
Taken together, our findings provide the first comprehensive view of in vivo target genes whose transcription is critically dependent on Spt5 both positively and negatively during vertebrate development. Further studies are required to elucidate the complexities of elongation regulation through Spt5 in vivo, as well as to determine the significance of such regulation on individual target genes and their associated pathways.
Materials and Methods
Zebrafish strains and husbandry
Wild-type AB and fogsk8 embryos were obtained through natural spawning and were maintained and staged according to established procedures [54]. The use of zebrafish was approved by UCSF ethics committee.
Total RNA isolation and quantitative RT-PCR
Equal number of embryos (WT siblings vs. fogsk8 mutants) at 24 hpf was dechorionated manually or by Pronase K treatment. Trizol (Invitrogen) was used to extract total RNA that was DNase-treated (Qiagen) to minimize contamination by genomic DNA during qRT-PCR. To obtain cDNA, total RNA was reverse-transcribed using oligodT and Superscript (Invitrogen). Equal amount of cDNA was used in SybrGreen qPCR mastermix system (Applied Biosystems) with appropriate primers (Supplemental Table S2). All plates contained at least two-well duplicates for each primer and cDNA condition. Cycle values were averaged between the duplicates and normalized to control primer values. WT fold values were compared to fogsk8 mutant fold values and expressed as fold upregulated or downregulated in fogsk8 embryos. At least two independent replicates were performed for each primer set.
Microarray Data Analysis
Microarray hybridization was performed at the NINDS microarray consortium using zebrafish Affymetrix oligonucleotide arrays. Three independent replicates for each sample were performed. Gene intensity values for the Affymetrix Zebrafish Genome Array CEL files were obtained using the GC-RMA package from Bioconductor [55]. To identify genes with relative changes in steady-state mRNA levels between WT and fogsk8 mutants, a fold change was calculated from these log2 intensity values along with a student t test p value. Genes with 2 fold change in expression and p<0.05 were clustered using the program HOPACH (hierarchical ordered partitioning and collapsing hybrid), with uncentered correlation distance [56], [57]. Associations with Gene Ontology, GenMAPP [58] and PANTHER [59]pathways were obtained using MAPPFinder 2.0 [42], a part of the GenMAPP 2.1 [41] application package and further filtered and annotated using the program GO-Elite (http://www.genmapp.org/go_elite/go_elite.html). The raw image and processed expression of our microarray dataset is posted at NCBI's Gene Expression Omnibus database (GSE12826). To determine relative differences in gene length parameters, a custom python script was written that links microarray probe set data to gene structural information from the Ensembl database (PMID: 17148474) obtained through BioMart [60]. Probe set annotations to Ensembl genes obtained from BioMart were augmented with those directly from Affymetrix (http://www.affymetrix.com). In this script, fold changes were calculated by comparing the mean length of distinct gene features (genomic, exon and intron lengths/number) or gene expression between upregulated or downregulated Ensembl-linked genes compared to all genes on the microarray. For each parameter, a permutation p value was calculated by randomizing the selection of input probe sets 10,000 times and re-calculating and storing the mean fold changes for both up and down regulated sets to determine the probability that the original fold occurs by chance alone.
Chromatin Immunoprecipitation (ChIP) Assay
WT embryos at 24 hpf were dechorionated by Pronase treatment and washed thoroughly. Embryos were fixed in 1.5% formaldehyde for 20 minutes and then homogenized 25 times in 5× extraction buffer with protease inhibitors on ice. Sonication was carried out with Branson sonifier 250 on an ice bath for 10 times, 10 seconds each with 20 second breaks. Sonicated samples were then centrifuged at 14k rpm at 4°C for 30 minutes to separate the pellet from the supernatant. The supernatant was immunoprecipitated with the following antibodies: 8WG16 Ab-1∶40 (Covance Research Products), Anti-Spt5 antibody 1∶10, and control sera -Normal mouse serum-1∶100 (Sigma) and Rabbit pre-immune serum (1∶10) for 2–4 hours in the cold room. Protein A Sepharose (Sigma) beads were added to the samples and left on overnight. The next day, extensive washing with different buffers were performed before eluting the beads at 65°C for 20 minutes. Proteinase K and RNase were added to the eluates and reverse-crosslinked overnight at 65°C. Phenol/chloroform extraction followed by ethanol precipitation was performed. The immunoprecipitated DNA was immediately used for qPCRs with appropriate primers (Supplemental Table S4). All cycle values were first normalized to input values before comparing control samples to immunoprecipitated samples. These values were represented as fold occupancy over control values. The procedure was performed similarly for fogsk8 embryos.
Whole mount in situ hybridization and immunocytochemistry
Whole-mount in situ hybridization was performed as previously described [61]. Plasmid containing pBS-gadd45b was obtained from ZIRC, and antisense probe was made using T7 RNA polymerase (Ambion). Whole-mount immunocytochemistry was performed similarly with appropriate antibodies (8WG16 ab- 1∶100; Anti-zfSpt5 antibody – 1∶5000; CTD4H8 –AF488 – 1∶2500) and fluorescent secondary antibodies. Images were taken with Zeiss epi-fluorescent and confocal microscopes.
Western Blot
Dechorionated embryos were fixed in 1.5% formaldehyde for 20 minutes and homogenized before addition of 2× SDS loading buffer. Equal amounts of the extracts were loaded on to the wells of the gel. Primary antibodies (Anti-Foggy antibody-1∶6000; Anti-Actin antibody – 1∶2000; Anti-Gadd45b antibody – 1∶100) and secondary antibody conjugated to horse-radish peroxidase (HRP) were used. The ECL kit (Amersham) was used to visualize the protein bands transferred from the blot to film.
Supporting Information
(0.88 MB DOC)
(0.03 MB DOC)
(0.04 MB DOC)
(0.08 MB DOC)
(0.05 MB DOC)
(0.04 MB DOC)
Acknowledgments
We thank NINDS Microarray Consortium and W. Liang for help with microarray experiments, J. Jeong for production of the anti-zfSpt5 antibody, S.Wadekar (H. Ingraham lab) and R. Raisner (H.Madhani lab) for technical advice on ChIP, D. Yelon for the fogsk8 mutant fish, and L. Reichardt for use of the confocal microscope. We thank M. Margeta, B. Lau, M. Wagle, B.R. Conklin, and members of the Guo laboratory for support, discussions, and comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the NIH grant NS042626. K. Krishnan was a recipient of a UCSF Graduate Student Research Award. N. Salomonis was supported by the J. David Gladstone Institutes and T32GM07175. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 3.Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 4.Collart MA, Tourkine N, Belin D, Vassalli P, Jeanteur P, et al. c-fos gene transcription in murine macrophages is modulated by a calcium-dependent block to elongation in intron 1. Mol Cell Biol. 1991;11:2826–2831. doi: 10.1128/mcb.11.5.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcription elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 11.Conaway JW, Conaway RC. Transcription elongation and human disease. Annu Rev Biochem. 1999;68:301–309. doi: 10.1146/annurev.biochem.68.1.301. [DOI] [PubMed] [Google Scholar]
- 12.Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- 13.Winston F. Control of eukaryotic transcription elongation. Genome Biol. 2001;2:1–3. doi: 10.1186/gb-2001-2-2-reviews1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DK, Yamaguchi Y, Wada T, Handa H. The regulation of elongation by eukaryotic RNA polymerase II: a recent view. Mol Cells. 2001;11:267–274. [PubMed] [Google Scholar]
- 15.Blackwell TK, Walker AK. Transcription elongation: TLKing to chromatin? Curr Biol. 2003;13:R915–916. doi: 10.1016/j.cub.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 17.Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 18.Winston F, Chaleff DT, Valent B, Fink GR. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984;107:179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bortvin A, Winston F. Evidence that Spt6 controls chromatin structure by a direct interaction with histone. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 21.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu-Baer F, Lane WS, Gaynor RB. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J Mol Biol. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- 23.Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, et al. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 25.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, et al. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jennings BH, Shah S, Yamaguchi Y, Seki M, Phillips RG, et al. Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr Biol. 2004;14:1680–1684. doi: 10.1016/j.cub.2004.08.066. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, et al. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 31.Guo S, Yamaguchi Y, Schilbach S, Wada T, Goddard A, et al. A regulator of transcriptional elongation, which is required for vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- 32.Keegan BR, Feldman JL, Lee DH, Koos DS, Ho RK, et al. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development. 2002;129:1623–1632. doi: 10.1242/dev.129.7.1623. [DOI] [PubMed] [Google Scholar]
- 33.Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol Cell Biol. 2002;22:1079–1093. doi: 10.1128/MCB.22.4.1079-1093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, et al. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 35.Stachora AA, Schafer RE, Pohlmeier M, Maier G, Ponstingl H. Human Supt5h protein, a putative modulator of chromatin structure, is reversibly phosphorylated in mitosis. FEBS Let. 1997;409:74–78. doi: 10.1016/s0014-5793(97)00486-9. [DOI] [PubMed] [Google Scholar]
- 36.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavoie SB, Albert AL, Handa H, Vincent M, Bensaude O. The peptidyl-prolyl isomerase Pin1 interacts with hSpt5 phosphorylated by Cdk9. J Mol Biol. 2001;312:675–685. doi: 10.1006/jmbi.2001.4991. [DOI] [PubMed] [Google Scholar]
- 38.Pei Y, Shuman S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J Biol Chem. 2003;278:43346–43356. doi: 10.1074/jbc.M307319200. [DOI] [PubMed] [Google Scholar]
- 39.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, et al. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 40.Thisse B, Pfumio S, Fürthauer MBL, Heyer V, et al. Expression of the zebrafish genome during embryogenesis. ZFIN on-line publication 2004 [Google Scholar]
- 41.Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, et al. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, et al. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osterlund T, Kogerman P. Hedgehog signalling: how to get from Smo to Ci and Gli. Trends Cell Biol. 2006;16:176–180. doi: 10.1016/j.tcb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Smith WC. TGF beta inhibitors. New and unexpected requirements in vertebrate development. Trends Genet. 1999;15:3–5. doi: 10.1016/s0168-9525(98)01641-2. [DOI] [PubMed] [Google Scholar]
- 46.Irvine KD. Fringe, Notch, and making developmental boundaries. Curr Opin Genet Dev. 1999;9:434–441. doi: 10.1016/S0959-437X(99)80066-5. [DOI] [PubMed] [Google Scholar]
- 47.Papa S, Zazzeroni F, Bubici C, Jayawardena S, Alvarez K, et al. Gadd45 beta mediates the NF-kappa B suppression of JNK signalling by targeting MKK7/JNKK2. Nat Cell Biol. 2004;6:146–153. doi: 10.1038/ncb1093. [DOI] [PubMed] [Google Scholar]
- 48.Kawahara A, Che YS, Hanaoka R, Takeda H, Dawid IB. Zebrafish GADD45beta genes are involved in somite segmentation. Proc Natl Acad Sci. 2005;102:361–366. doi: 10.1073/pnas.0408726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 51.De Smaele E, Zazzeroni F, Papa S, Nguyen DU, Jin R, et al. Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 52.Takekawa M, Tatebayashi K, Itoh F, Adachi M, Imai K, et al. Smad-dependent GADD45beta expression mediates delayed activation of p38 MAP kinase by TGF-beta. EMBO J. 2002;21:6473–6482. doi: 10.1093/emboj/cdf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graunke DM, Fornace AJJ, Pieper RO. Presetting of chromatin structure and transcription factor binding poise the human GADD45 gene for rapid transcriptional up-regulation. Nucleic Acids Res. 1999;27:3881–3890. doi: 10.1093/nar/27.19.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish, Brachydanio rerio. Eugene, OR: The University of Oregon Press; 1995. [Google Scholar]
- 55.Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;(Suppl):45–51. [PubMed] [Google Scholar]
- 56.Pollard KS, van der Laan MJ. A method to identify significant clusters in gene expression data. Cybernetics Informatics. 2002;II:318–325. [Google Scholar]
- 57.van der Laan MJ, Pollard KS. A new algorithm for hybrid clustering with visualization and the bootstrap. J Stat Planning Infer. 2003;117:275–303. [Google Scholar]
- 58.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 59.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucl Acids Res. 2007;35:D247–252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 61.Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, et al. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein Soulless/Phox2a. Neuron. 1999;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(0.88 MB DOC)
(0.03 MB DOC)
(0.04 MB DOC)
(0.08 MB DOC)
(0.05 MB DOC)
(0.04 MB DOC)