Abstract
Background
Mature miRNAs can often be classified into large families, consisting of members with identical seeds (nucleotides 2 through 7 of the mature miRNAs) and highly homologous ∼21-nucleotide (nt) mature miRNA sequences. However, it is unclear whether members of a miRNA gene family, which encode identical or nearly identical mature miRNAs, are functionally interchangeable in vivo.
Methods and Findings
We show that mir-181a-1, but not mir-181c, can promote CD4 and CD8 double-positive (DP) T cell development when ectopically expressed in thymic progenitor cells. The distinct activities of mir-181a-1 and mir-181c are largely determined by their unique pre-miRNA loop nucleotides—not by the one-nucleotide difference in their mature miRNA sequences. Moreover, the activity of mir-181a-1 on DP cell development can be quantitatively influenced by nucleotide changes in its pre-miRNA loop region. We find that both the strength and the functional specificity of miRNA genes can be controlled by the pre-miRNA loop nucleotides. Intriguingly, we note that mutations in the pre-miRNA loop regions affect pre-miRNA and mature miRNA processing, but find no consistent correlation between the effects of pre-miRNA loop mutations on the levels of mature miRNAs and the activities of the mir-181a-1/c genes.
Conclusions
These results demonstrate that pre-miRNA loop nucleotides play a critical role in controlling the activity of miRNA genes and that members of the same miRNA gene families could have evolved to achieve different activities via alterations in their pre-miRNA loop sequences, while maintaining identical or nearly identical mature miRNA sequences.
Introduction
A large number of animal miRNAs have been identified and the genes encoding many of these small RNAs have been shown to play diverse functional roles in animals [1]. Each miRNA gene produces at least three small RNA species, including a long primary miRNA transcript (pri-miRNA), an intermediate ∼60-nt precursor miRNA (pre-miRNA), and a ∼21-nt mature miRNA, through sequential endonucleolytic maturation steps [2]. In vitro biochemical analyses have indicated that the mature ∼21-nt miRNAs, often the predominant RNA products of miRNA genes, can guide the RNA-induced silencing complex (RISC) to target mRNAs for repression. Pri-miRNA and pre-miRNA are considered to be transitory intermediates during miRNA biogenesis and are thought to play no direct role in gene repression, though they also contain sequences complementary to target element(s) in the 3′untranslated region (UTR) of their cognate target genes.
The deletion or mutation of specific miRNA genes can result in defects in all RNA species produced from the genes [3], [4]. It was known that precursor lin-4 RNA contains the mature lin-4 sequence, and genetic analyses were unable to definitively rule out the possible involvement of precursor lin-4 RNAs in gene regulation [3]. Loss-of-function phenotypes for miRNA genes, such as lin-4 and let-7, were rescued with genomic fragments encoding functional segments of pri-miRNAs, which can be processed into pre-miRNAs and mature miRNAs, but not with the mature miRNA alone [3], [4]. Therefore, the phenotypes observed for loss of miRNA genes cannot be definitively attributed to one of the RNA species made from these genes.
Interestingly, many miRNA genes can be classified into large families consisting of members with highly homologous ∼21-nt mature miRNAs and identical seed nucleotides (6,7), but divergent pre-miRNA loop nucleotides. According to computational and biochemical analyses [5]–[7], the members of a miRNA gene family should regulate a similar set of target genes and have the same biological function. Here we examine whether miRNA genes that encode identical and nearly identical mature miRNAs have the same function. We find that pre-miRNA loop nucleotides play critical roles in controlling the distinct activities of mir-181a-1 and mir-181c genes.
Results
Assay for measuring mir-181a-1 activity in DP cell development
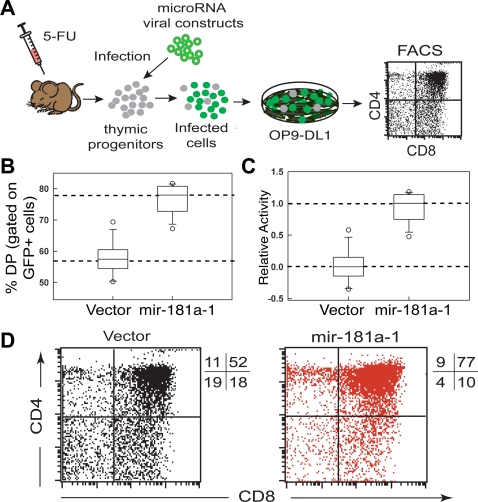
We used T cell development as a functional readout to determine the nucleotides and structural domains that are required for the function of mir-181 genes. It is known that mir-181a-1 plays important roles in T and B lymphocyte development [8]–[10], and can function as a “rheostat” to modulate the strength and threshold of T cell receptor (TCR) signaling [9]. Moreover, mature miR-181a is developmentally regulated during early T cell differentiation in the transition from CD4 and CD8 double-negative (DN) to CD4 and CD8 DP cells in the thymus [9], [10]. Using the OP9-DL1 co-culture assay (Fig. 1A), which can recapitulate the differentiation of DN progenitors into DP cells in vitro [11], we showed that ectopic expression of mir-181a-1 in DN thymic progenitor cells lead to a significant increase in the percentage of DP cells, from a median level of ∼57% in the control group to a median level of ∼77% in the mir-181a-1 expressing group (Fig. 1B–D). We have found that mir-181a-1 potentiates DN to DP cell development by targeting negative regulators in the Notch and pre-TCR signaling pathways (Mao T et al., manuscript in preparation). This assay allowed us to quantitatively measure the contribution of nucleotide sequences and structural domains to miRNA gene function via mutagenesis analyses.
Figure 1. An in vitro assay for measuring the activities of miRNA genes on T cell differentiation.
(A) Schematics depicting the OP9-DL1 stromal co-culture assay for T cell differentiation. (B) Box-plots to summarize the effects of mir-181a-1 on the percentage of DP cells differentiated from DN progenitor cells. The results of a representative OP9-DL1 stromal co-culture assay (12 independent replicates for each construct) are shown. (C) Normalized box-plots. The activities of mir-181a-1 in DP cell development were normalized so that the empty vector (negative control) had a median activity of “0” and mir-181a-1 expressing vector (positive control) had a median activity of “1.” (D) Representative FACS plots showing the effects of mir-181a-1 on DP cell development (gated on infected GFP cells).
Nucleotides in the pre-miRNA stem region have varied contribution to mir-181a-1 activity
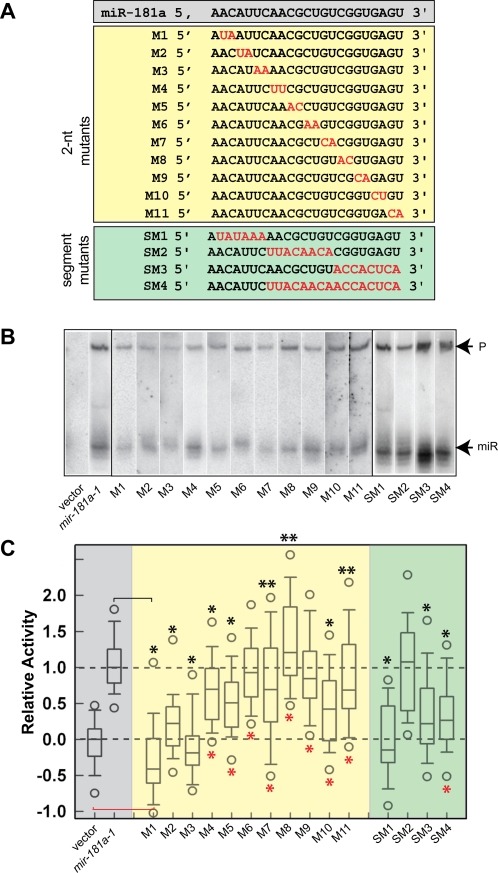
To determine which nucleotides of the mature miR-181a region are important for its function, we systematically mutated every set of 2-nt along its 23-nt mature miRNA region (Fig. 2A, yellow). The 2-nt sequences were altered to disrupt potential base pairings to cognate target sequences. To retain the structure of the miRNA stem-loop precursor, we simultaneously mutated the corresponding 2-nt on miR* strand, the complementary strand of the mature miRNA (Fig. S1). Thus, these “stem mutants” contain mutations on both the mature miRNA strands and the miR* strand, affecting the sequences of both pre- and mature miRNA species. Northern analyses of transfected BOSC23 cells demonstrate that the mature miR-181a can be produced from all the mutant constructs (Fig. 2B). The varied intensities of the mature miR-181a and its mutants may not indicate the differences in actual expression levels since different oligo nucleotide probes were used to detect each of the miR-181a mutant forms. When a shorter probe that matches perfectly to both wild-type miR-181a and M1 was used for Northern analyses, we found that the wild-type miR-181a and M1 mature miRNAs were made at comparable levels (Fig. S2).
Figure 2. Effects of the mutations in the stem region on mir-181a-1 activity in promoting DP cell development.
(A) Scanning mutations in the stem region of the mir-181a-1 gene. Two nucleotides (2-nt mutants, shaded yellow) or a stretch of nucleotides (segment mutants, green) in the mature miRNA region are altered (shown in red). Nucleotides are altered to disrupt the potential base pairing to target genes. Compensatory mutations are also generated on the miR* strand to maintain the secondary structure of the pre-miRNAs. (B) Expression and processing of wild-type mir-181a-1 and stem mutants. Specific probes that perfectly match the mature miR-181a and each of its mutant forms were used in hybridization to determine the expression of mature miR-181a and its stem mutant forms. (C) The effects of mir-181a-1 and its stem mutants on DP cell development. Normalized data from 3–5 independent T cell assays (each with 12 independent replicates, total 36–60 replicates) are pooled and graphed in the distribution box plots to summarize the distribution of the relative activities of mir-181a-1 (shaded grey), the 2-nt mutants (shaded yellow), and the segment mutants (shaded green) in DP cell development. Mann-Whitney Rank Sum Tests were performed to determine whether the activities of individual 2-nt mutants were statistically different from those of the control vector (red *, p<0.0001) and the mir-181a-1 vector (black *, p<0.0001; **, p<0.05) ( Table S1 for statistics). A representative OP9-DL1 stromal co-culture assay without normalization (12 independent replicates for each constructs) is also shown ( Fig. S5 ).
We then ectopically expressed each of the mir-181a-1 “stem mutants” in DN thymocytes and examined their effects on DP T cell development using the OP9-DL1 co-culture assay (Fig. 1). By comparing the negative control (empty vector) to the positive control (wild-type mir-181a-1 expressing vector), it is clear that nucleotides in the stem region have different contributions to mir-181a-1 activity in promoting T cell development (Fig. 2C, yellow, Table S1 for statistics). The M1 and M3 seed mutants completely abolish mir-181a-1 activity, while the M2 mutant still display residual activity for promoting T cell development, demonstrating that nucleotides in the seed region play a critical role in the mir-181a-1 gene function. In comparison, 2-nt mutations outside the seed region have modest effects on mir-181a-1 activity. The M4, M5, M7, M10, and M11 mutants show a slight reduction in activity. The M6 and M9 mutants have no change in activity, while the M8 mutant show an increase in activity (Table S1 for statistic summary). Thus, nucleotides outside the seed region also contribute to mir-181a-1 function, but are more tolerant of nucleotide variations.
Since nucleotides outside the 5′ seed region have weaker effects on mir-181a-1 activity, we then created four additional stem mutants: the segment mutants (SM1–4) with longer stretches of mutations in the mir-181a-1 stem region (Fig. 2A, green). As shown by Northern blot analyses, these mutants are properly expressed and processed (Fig. 2B). As expected, altering the entire seed region (SM1) completely abolished mir-181a-1 activity in promoting DP cell development. Interestingly, segment mutants with 3′ 8-nt altered (SM3 and SM4) also exhibit significantly decreased activity, while the mutant with the center 8-nt mutation (SM2) has comparable activity to the wild-type mir-181a-1 (Fig. 2C, green, Table S1 for statistic summary).
Collectively, these findings demonstrate that the nucleotides in seed region are critical for mir-181a-1 activity—small alterations in the seed region cause dramatic decreases in activity. In comparison, the nucleotides in the 3′ end of the mature miRNA region have smaller contributions, and the nucleotides in the center of the mature miR-181a have little or no contribution to mir-181a-1 activity (Fig. 2). These findings confirmed the importance of the seed nucleotides, as shown previously by computational and biochemical analyses [5]–[7], thus validating the use of this assay to measure the activity of mir-181a-1 genes and to dissect the structural and functional relationships of mir-181 genes by mutagenesis. However, it is important to note that mutations in stem regions affect mature, precursor, and primary miRNAs, thus it is not possible to attribute the effects of the mutations on DP cell development to one of the RNAs made from the mir-181a-1 gene.
mir-181a-1, but not mir-181c, can promote DP cell development
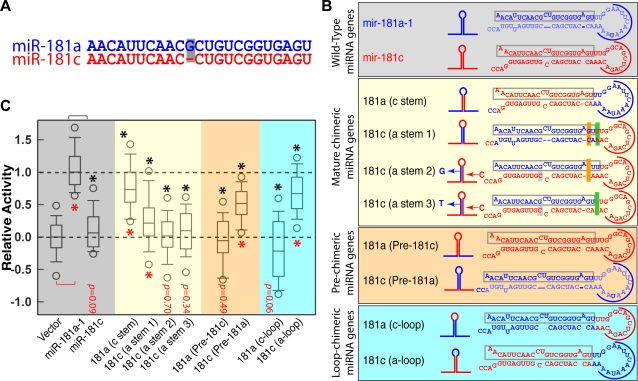
The members of the mir-181 family of genes produce four mature miRNAs: miR-181a, miR-181b, miR-181c, and miR-181d, from three putative polycistronic genes, mir-181a-1/b-1, mir-181a-2/b-2, and mir-181c/d, respectively (Fig. S3). The mature miRNAs of the miR-181 family, all of which have identical 5′ seed nucleotides, differ from one another by no more than 3-nt in either the center or the 3′ end of the mature miRNAs. Specifically, mature miR-181a differs from miR-181c by only one nucleotide in the center of the mature miRNA (Figs 3A, 3B). Thus, according to the “seed” hypothesis [5]–[7] and the results of “stem mutant” analyses (Fig. 2), it appears that mir-181a-1 and mir-181c should have similar activities in the co-culture assay.
Figure 3. The pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c on DP cell development.
(A) Nucleotide sequences of mature miR-181a and miR-181c. (B) Schematics and nucleotide sequences depicting the wild-type mir-181a-1 and mir-181c genes and corresponding precursors (shaded grey). Also shown are the chimeric miRNA genes, with the mature miRNAs, pre-miRNAs, and pre-miRNA loops swapped between mir-181a-1 and mir-181c, termed “mature-chimeric” (shaded yellow), “pre-chimeric” (shaded orange), and “loop-chimeric” (shaded blue), respectively. These mutant genes are designated as 181a(c stem), 181c(a stem), 181a (pre-181c), 181c (pre-181a), 181a (c-loop), and 181c (a-loop). Similar color codes are used in all figures. (C) The effects of the chimeric mir-181a-1/c genes on DP cell development. Normalized data from 3–7 independent T cell assays (each with 12 independent replicates for a total of 36–84 replicates) are pooled and graphed in the distribution box plots. Mann-Whitney Rank Sum Tests were performed ( Table S2 for statistics) to determine whether the activities of the chimeric miRNA genes are statistically different from those of the negative control vector (red *, p<0.0001) and/or mir-181a-1 positive control (black *, p<0.0001). A representative OP9-DL1 stromal co-culture assay without normalization is also shown ( Fig. S6 ).
To test this, we examined the ability of mir-181a-1 and mir-181c to promote DP cell development. Note that mature miR-181a and miR-181c have similar expression patterns during thymocyte development [9], [10], though the endogenous levels of miR-181c is about four to five times lower than that of miR-181a in the corresponding thymocytes, indicating that they are processed in thymocytes and may play roles in normal T cell development (Fig. S4). Thus, the thymocyte differentiation assay allows us to interrogate the functions of mir-181a-1 and mir-181c in a physiologically relevant mRNA and miRNA milieu. Interestingly, while the ectopic expression of mir-181a-1 results in a substantial increase in the generation of DP cells, the expression of mir-181c does not (Fig. 3C, grey), demonstrating that mir-181a-1 but not mir-181c can promote DP cell development (p<0.0001, Table S2).
miRNA genes encoding identical mature miRNAs can have distinct biological activities
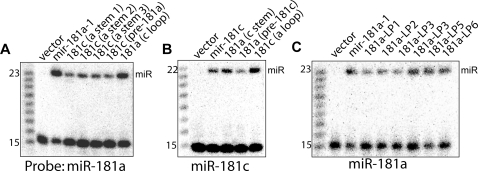
To examine whether the single nucleotide difference in the mature miRNA regions contributes to the distinct activities of mir-181a-1 and mir-181c, we generated “mature chimeric” miRNA genes by swapping the stem regions (miR and miR* duplexes) (Fig. 3B, yellow). The resulting “mature chimeric” miRNA genes, termed 181a (c stem) and 181c (a stem 1), should express mature miR-181c and miR-181a, respectively. We also generated two additional “mature chimeric”genes— 181c (a stem 2) and 181c (a stem 3)—by replacing mature miR-181c with mature miR-181a while maintaining the miR-181c complementary strand. Even though 181a (c-stem) is designed to produce mature miR-181c, we observed that this “mature chimeric” miRNA gene was still functionally active in promoting DP cell development, albeit with a median activity of ∼73% of the wild-type mir-181a-1 (Fig. 3C, yellow, Table S2). In contrast, the 181c (a stem 1) gene, which encodes mature miR-181a, had a median activity of only ∼21% of the wild-type mir-181a-1, and the 181c (a stem 2,3) genes had no significant activity (Fig. 3C, yellow, Table S2). These results demonstrate that the distinct activities of mir-181a-1 and mir-181c are not caused by the single nucleotide difference between their mature miRNA forms. Notably, these results demonstrate that miRNA genes encoding identical mature miRNAs, such as mir-181c and 181a (c-stem) that encode miR-181c, or mir-181a-1 and 181c (a-stem1, 2, 3) that encode miR-181a, can have distinct biological activities.
Pre-miRNAs and their loops determine the activities of the mir-181 genes
Since mir-181a-1 and mir-181c have divergent pre-miRNA flanking and loop sequences, we then tested whether their differences in activity are determined by their unique pre-miRNAs or by flanking sequences (Fig. 3B). We generated “pre-miRNA chimeric” genes by swapping the pre-miRNA regions between mir-181a-1 and mir-181c (Fig. 3B, orange). When tested in the OP9-DPL1 co-culture assay, the miRNA gene with pre-miR-181a, 181c (pre-181a), did promote DP cell development, albeit with a median activity of ∼52% of the wild-type mir-181a-1, while the miRNA gene with pre-miR-181c, 181a (pre-181c), had no activity (Fig. 3C, orange, Table S2). These results demonstrate that sequences specific to the pre-miRNAs play a key role in determining the distinct biological activities of the mir-181a-1 and mir-181c genes. However, pre-miRNA flanking sequences also contribute to the functions of the mir-181a-1 and mir-181c genes, since the activity of 181c (pre-181a) is lower than that of the wild-type mir-181a-1.
Since pre-miR-181a-1 and pre-miR-181c differ mainly in their pre-miRNA loop nucleotides, we next swapped the pre-miRNA loops and examined the activity of loop chimeras in the OP9-DPL1 co-culture assay (Fig. 3B, blue). We found that 181c (a-loop) can promote DP cell development with a median activity of ∼67% of the wild-type mir-181a-1, while 181a (c-loop) is inactive in promoting DP cell development (Fig. 3C, blue, Table S2), demonstrating that the distinct biological activities of the mir-181a-1 and mir-181c genes are largely determined by the differences in their pre-miRNA loops.
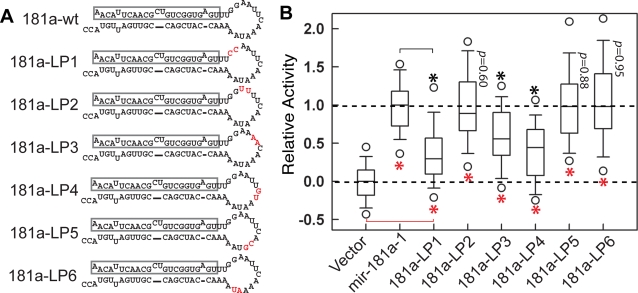
The mir-181a-1 activity is sensitive to nucleotide changes in its pre-miRNA loop
To further investigate the role of pre-miRNA loop nucleotides, we carried out scanning mutagenesis around the pre-miR-181a-1 loop (Fig. 4A) and found that dinucleotide mutations in the pre-miR-181a-1 loop had varied effects on mir-181a-1 activity (Fig. 4B). The 181a-LP1, 181a-LP3, and 181a-LP4 mutants had median activities of ∼29%, 55%, and 46% of the wild-type mir-181a-1, respectively (Fig. 4B, Table S3). In contrast, the 181a-LP2, 181a-LP5 and 181a-LP6 mutations did not significantly affect mir-181a-1 activity. The loop mutagenesis analyses further demonstrated that pre-miRNA loop nucleotides could also quantitatively influence the activity of the mir-181a-1 gene.
Figure 4. The activity of mir-181a-1 is sensitive to nucleotide changes in its pre-miRNA loop.
(A) Schematics of the pre-miR-181a-1 loop mutants. (B) The effects of pre-miR-181a-1 loop mutants on DP cell development. Normalized data from six independent T cell assays (each with 12 independent replicates for a total of 72 replicates) is shown. Mann-Whitney Rank Sum Tests were performed ( Table S3 for statistics) to determine whether the activities of the loop mutants were statistically different from those of the negative control vector (red *, p<0.0001) and/or the mir-181a-1 positive control vector (black *, p<0.0001). A representative OP9-DL1 stromal co-culture assay (12 independent replicates) without normalization is also shown ( Fig. S7 ).
mir-181a-1/c mutants and their wild-type genes produce mature miRNAs with the same 5′ ends
To understand the mechanisms by which pre-miRNA loop nucleotides may control the activities of miRNA genes, we characterized the effects of pre-miRNA loop mutations on mature miRNA biogenesis. According to computational and biochemical analyses [5]–[7], [12], seed nucleotides are essential for target gene recognition and repression. Since the seed nucleotides are localized at the 5′ end of mature miRNAs and mature miRNA often have polymorphic 3′ or 5′ ends as shown by miRNA cloning analyses [13], [14], a shift of just a few nucleotides in mature miRNA sequences during processing could change the seed nucleotides. To rule out the possibility that mutations in mir-181a-1/c cause shifts in the 5′ end of mature miRNAs and changes the seed nucleotides, we carried out primer extension analyses and showed that mature miRNAs produced from various mir-181a-1/c mutants have the same 5′ end as those produced from the corresponding wild-type mir-181a-1/c genes (Fig. 5A–C). These results demonstrate that mir-181a-1/c mutants do not cause changes in the 5′ ends of the mature miRNA sequences, eliminating the possibility that mir-181a-1/c mutants affect the activities of the mir-181a-1 or mir-181c genes by controlling the fidelity of the 5′ ends of the mature miRNAs that are produced.
Figure 5. Mature miRNAs produced from the mir-181a-1/c mutants have the same 5′ ends.
The 5′ ends of mature miR-181a (A) and miR-181c (B) produced from mir-181a-1/c domain swapping mutant genes, and the 5′ ends of mature miR-181a (C) produced from mir-181a-1 loop mutant genes were determined by primer extension analyses. Synthetic miR-181a or miR-181c oligo nucleotides in single nucleotide increments (15 nt–22/23-nt) were radio-labeled and used as size ladders. The upper band represents the major cDNA product of miR-181a (23-nt) or miR-181c (22-nt), and the lower band represents radio-labeled probes for the mature miRNAs.
The effects of mir-181a-1/c mutations on the levels of mature miRNA
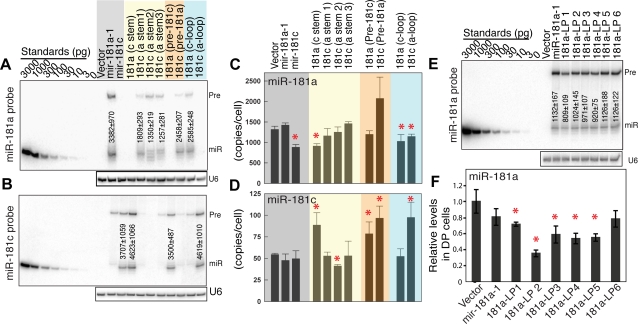
We then investigated whether mir-181a-1/c mutations cause changes in the levels of mature miRNAs made in BOSC 23 and DP cells (Fig. 6), and if so, whether these changes correlate with the effects of loop mutations on the activities of miRNA genes. BOSC 23 cells do not express endogenous mature miR-181a or miR-181c, thus facilitating accurate measurement of mature miRNA levels produced from the mutant constructs. Quantitative Northern blot analyses were used to define the levels of mature miR-181a and miR-181c, as well as the sizes of the mature miRNAs and the levels of the pre-miRNAs produced from various mir-181a-1/c mutant constructs in BOSC 23 cells (Fig. 6A, 6B, 6E, and S8). Since it is difficult to obtain sufficient numbers of infected DP thymocytes for Northern blot analyses, we carried out miRNA qPCR analyses to determine the number of copies of mature miR-181a and miR-181c in DP cells transduced with mir-181a-1/c mutant viruses (Fig. 6C, 6D, 6F). Comparing the effects of mir-181a-1/c mutations on the levels of mature miRNA in two different cell types may also reveal whether mir-181a-1/c loop mutations cause differential miRNA processing in different cell types.
Figure 6. The effects of mir-181a-1/c mutants on mature miRNA expression in BOSC 23 and DP cells.
(A, B) The copy numbers of mature miR-181a (A) and miR-181c (B) expressed in BOSC 23 cells transfected with the same amounts of vectors expressing different mir-181a-1/c mutants determined by quantitative Northern blot analyses ( Fig. S8 for standard curves and quantification, and Table S4 for statistics). (C, D) The copy numbers of mature miR-181a (C) and miR-181c (D) expressed in DP thymocytes transduced with viral vectors expressing mir-181a-1/c mutants determined by miRNA qPCR analyses ( Tables S5 and S6 for statistics). (E) The copy numbers of mature miR-181a expressed in BOSC 23 cells transfected with the same amounts of viral vectors expressing various mir-181a-1 loop mutants determined by quantitative Northern blot analyses ( Fig. S9 for standard curves, and Table S7 for statistics). (F). The copy numbers of mature miR-181a expressed in DP thymocytes transduced with viral vectors expressing various mir-181a-1 loop mutants, determined by miRNA qPCR analyses ( Table S8 for statistics). Statistical significance was determined by an unpaired two-tailed student's t test (compared to the control vector, red *, p<0.05). Representative blots of four or more independent quantitative Northern blot analyses are shown (A, B, E).
We find that mature miR-181a levels in either DP or BOSC 23 cells expressing the mir-181a-1/c mutants have no apparent correlation to the activities of mutant genes (Figs 3, 4, 6, S8, and S9). Ectopic expression of mir-181a-1 and mir-181c results in comparable levels of mature miR-181a and miR-181c in BOSC 23 cells (Figs. 6A, 6B, and S8), but not significant increases in mature miR-181a or miR-181c levels in the infected DP cells (Fig. 6C, 6D, 6F). Since mir-181a-1 activity can be effectively abolished by mutations in the seed region (Fig. 2), these findings suggest that we have probably measured the activity of the mir-181a-1 gene in T cell development, and that mir-181a-1 may exert its biological function without a significant increase in the mature miRNA level (Fig. 2, 3, 4, 6C). Nevertheless, we cannot rule out that a small but undetectable increase in the mature miR-181a level is sufficient for mir-181a-1 activity. In another case, the 181a-LP2 mutant exhibited the same activity as wild-type mir-181a-1, but DP cells infected with this mutant expressed ∼50% less mature miR-181a (Figs. 4B, 6E, 6F). Finally, 181a-LP3, 181a-LP4, and 181a-LP5 produced similar levels of mature miR-181a in DP cells and BOSC 23 cells (Fig. 6E, 6F, and S9), but these mutants had distinct activities in promoting DP cell development (Fig. 4B). The 181a-LP5 mutations had no effect on mir-181a-1 activity, while 181a-LP3 and 181a-LP4 mutations caused a ∼45% and 56% reduction in median activity, respectively.
In comparison, the levels of mature miR-181c in DP cells expressing mir-181a-1/c mutants also have no consistent correlation to their activities in DP cell development (Fig. 3, 6D). Ectopic expression of 181a (c-stem), 181a (pre-181c), and 181c (a-loop) resulted in significant increases in the levels of mature miR-181c in DP cells (Fig. 6D). However, while 181a (c-stem) and 181c (a-loop) can promote DP cell development, 181a (pre-181c) cannot (Fig. 3C), showing that increases in mature miR-181c in DP cells also do not always correlate with the activities of corresponding miRNA genes. Interestingly, mature miRNA expression from some miRNA genes might be differentially regulated in DP and BOSC 23 cells, but such differential regulation in mature miRNA processing also has little correlation to the activity of miRNA genes. For example, while mir-181c, 181a (c-stem), 181a (pre-181c), and 181c (a-loop) all produce similar levels of mature miR-181c in BOSC 23 cells (Figs. 6B and S8), ectopic expression of 181a (c-stem), 181a (pre-181c), and 181c (a-loop), but not mir-181c, resulted in significant increases in the levels of mature miR-181c in DP cells (Fig. 6D). However, among the ones that caused increases in mature miR-181c levels in DP cells, 181a (c-stem) and 181c (a-loop) can promote DP cell development, while 181a (pre-181c) cannot. Collectively, above analyses show that usually mature miR-181a and miR-181c levels in DP cells or BOSC 23 cells expressing the mir-181a-1/c and their mutants were the same within a factor of two, and in cases where different RNA levels were found, there was no consistent correlation with the activities of corresponding miRNA genes (Figs 3, 4, 6, S8, S9).
Discussion
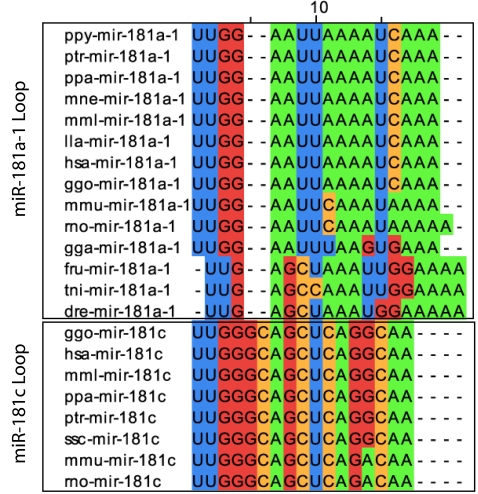
We have identified the nucleotide sequences and structural domains that are required for the function of mir-181a-1 and mir-181c through mutagenesis and domain-swapping analyses. We show that not only the nucleotides in the 5′ and 3′ ends of the stem but also those in the pre-miRNA loop are critical for mir-181a-1 activity. We find that mir-181a-1 and mir-181c have distinct activities in early T cell development, and the distinct activities of mir-181a-1 and mir-181c are controlled by their pre-miRNA loops (Fig. 3, 4), indicating that miRNA genes encoding identical or nearly identical mature miRNAs can exert different biological activities determined by their unique loop nucleotides. Interestingly, the pre-miRNA loop sequences of mir-181a-1 and mir-181c are divergent, but each is evolutionarily conserved in multiple animal species (Fig. 7), suggesting that members of the same miRNA gene families may have evolved to achieve distinct specificities or degrees of activity via alterations in their pre-miRNA loop sequences. However, mir-181a-1/c mutants do not change the 5′ ends of mature miRNAs produced (Fig. 5) and the levels of mature miRNAs produced from these genes have no consistent correlation with the activities of corresponding miRNA genes (Fig. 6). Clearly, our results demonstrate that pre-miRNA loop nucleotides have a key role in controlling miRNA gene function.
Figure 7. Phylogenetic comparison of pre-miR-181a-1 and pre-miR-181c loop sequences.
The full genus and species names and their abbreviations are as follows: Danio rerio, dre; Fugu rubripes, fru; Homo sapiens, hsa; Gallus gallus, gga; Gorilla gorilla, ggo; Lagothrix lagotricha, lla; Macaca mulatta, mml; Mus musculus, mmu; Macaca nemestrina, mne; Pan paniscus, ppa; Pongo pygmaeus, ppy; Pan troglodytes, ptr; Rattus norvegicus, rno; Sus scrofa, ssc; Tetraodon nigroviridis, tni.
When interpreting the above findings, it is critical to draw a distinction between the activity of a miRNA gene and the activity of a mature miRNA, which are often used interchangeably in the literature. In this study, we have examined the activities of miRNA genes, which may be contributed by one or more of the RNAs (mature miRNAs and their precursors) made from these miRNA genes, not the activities of mature miRNAs alone. Although siRNA duplexes have been shown to function as miRNA surrogates to target gene repression when transfected into mammalian cells [7], mature miRNAs delivered into cultured cells by transfection are diluted quickly during cell expansion, prohibiting their use in long-term T cell development culture assays. Thus, we were unable to quantitatively measure whether transfected mature miRNAs might be functionally equivalent to RNAs produced from miRNA genes. Furthermore, given the complex small RNA sorting pathways in animal cells [15], [16], we have not attempted to determine the efficiency of transfected miRNAs being incorporating into the pathways used by corresponding miRNA genes in vivo.
It is also important to emphasize that limited conclusions can be drawn by correlating the changes in mature miRNA levels to the activities of corresponding mutant miRNA genes (Fig. 3, 4, 6). We believe that such analyses can only establish correlations, but not causal relationships, between the levels of mature miRNA expression and the activity of miRNA genes. However, strong and positive correlations would support that pre-miRNA loop nucleotides control the activity of miRNA genes by influencing mature miRNA levels, whereas negative or inconsistent correlations would disfavor such idea. Most importantly, miRNA genes make at least three RNA species, including pri-miRNA, pre-miRNA, or mature miRNA, prohibiting us from definitively attributing miRNA gene functions to one of these RNAs in the T cell assays. Moreover, the complex regulatory steps in the miRNA biogenesis pathways and during early T cell development may also compromise the robustness of such correlation analyses. Finally, we cannot determine whether alterations in pre-miRNA loop sequences affect all or only selected cognate targets. Since multi-target regulation is required for the mir-181a-1 function in T cells and the ectopic expression of a single miR-181a insensitive target is sufficient to block the function of the mir-181a-1 [9], it is possible that pre-miRNA loop nucleotides only contribute to the regulation of one or a few targets among those that are regulated by the mir-181a/c or their mutants.
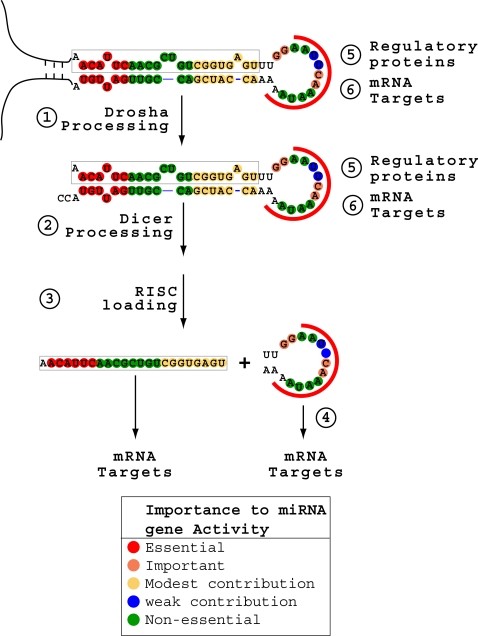
Nevertheless, based on the functional importance of various nucleotides in mir-181a-1 genes, we may postulate some mechanisms by which pre-miRNA loop nucleotides control the activities of miRNA genes (Fig. 8). Pre-miRNA loops might influence miRNA function by controlling the processing of pri-miRNAs into pre-miRNAs, the transport and sub-cellular localization of pre-miRNAs, the processing of pre-miRNAs into mature miRNAs, or the loading of mature miRNAs into RISCs [15]–[21]. For example, animal cells are thought to have multiple RISCs with distinct Argonatue proteins (AGO1–4). If pre-miRNA loops play a role in guiding small RNAs into various RISCs, which might have different efficiencies in gene silencing [15], [16], then mature miRNA levels would not necessarily correlate with the function of the miRNA genes. Moreover, functional miRNAs may be generated from pre-miRNA loops, since pre-miRNA loop-derived small RNAs have been found in miRNA cloning and deep sequencing analyses [22]. Finally, recent studies have shown that LIN-28 may recognize the pre-let-7 loop nucleotides and block the processing of human pri-let-7 RNAs into mature miRNAs in embryonic stem cells, suggesting that pre-miRNA loops may be recognized by LIN-28 or LIN-28-like RNA binding factors that control mature miRNA biogenesis [23]–[25]. However, these RNA binding factors would control the activity of miRNA genes by blocking mature miRNA biogenesis [23]–[25], which is inconsistent with the fact that the levels of mature miR-181a/c have no consistent correlations with the effects of corresponding mutations on the activities of the mir-181a-1/c genes (Figs 3, 4, 6). Therefore, pre-miRNA loop nucleotides may not control mir-181a-1/c activity through recognizing the LIN-28 or LIN-28-like RNA binding factors.
Figure 8. A “heat map” of the functionally important nucleotides in the pre-miR-181a-1 region according to mutagenesis analyses.
Color-code was used to illustrate the importance of the pre-miRNA nucleotides to the activity of the miR-181a-1 gene. Possible mechanisms by which pre-miRNA loop nucleotides control the activities of miRNA genes are listed.
Since pre-miRNA loop nucleotides — not the levels of mature miRNAs — were more critically linked to the function of miRNA genes, our findings raised an important question: which of the RNA species synthesized from miRNA genes is the functional one(s)? Definitively addressing this question is vital to understanding of the mechanisms by which miRNA genes control gene expression. One possibility is that pri-miRNAs and/or pre-miRNAs may have functional roles in target gene regulation before they are further processed. Supporting this idea, our findings demonstrate that pri-miRNAs and pre-miRNAs contain not only the mature miRNA sequences that can pair with cognate target sites, but also the loop nucleotides that are important for the activity and functional specificity of miRNA genes. Moreover, pre-miRNA-like stem-loop structures have been shown to be a common module for intermolecular RNA:RNA interactions [26], [27]. Thus, based on these findings (Figs 2– 6), we speculate that pre-miRNA loops may play roles in target gene regulation as a functional motif of the pri-miRNA and pre-miRNA RNAs. This model can explain why both pre-miRNA loop and “seed” nucleotides play key roles with respect to the function of corresponding wild-type and mutant mir-181a-1/c genes.
Materials and Methods
Retroviral constructs for miRNA gene expression
A double-copy retroviral vector with a human H1 polymerase III expression cassette was used to express mir-181a-1, mir-181c, and their mutant genes. Briefly, a ∼270-nt gene segment containing a ∼22-nt mature miRNA and ∼125-nt of genomic sequences flanking both sides of the miRNA was amplified from genomic DNA and placed in the U3 region of the 3′ LTR under the control of the human H1 pol III promoter [8], [9]. A GFP reporter driven by an independent murine 3-phosphoglycerate kinase promoter (PPGK) was used as a marker for infection. mir-181a-1 and mir-181c mutant constructs were generated using an overlapping PCR strategy to introduce mutations in the stem and loop regions of the miRNA genes. All mutant constructs were validated by DNA sequencing (see Materials and Methods S1 for the wild-type and mutant gene sequences). For mutations in the miRNA stem regions, compensatory mutations were also introduced to the miR* strands to preserve the integrity of the stem and loop structures (Fig. S1). High-titer retroviral supernatant was generated by co-transfecting the miRNA expression vector and pCLeco packaging construct into BOSC 23 cells (293T-based viral packaging cell line).
OP9-DL1 stromal co-culture assay for in vitro T cell differentiation
Six-week-old male C57BL/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME). The mice were given a single intravenous dose of 5-fluorouracil (5-FU; 150 mg/kg body weight; SIGMA, St. Louis, Missouri) four days before culture initiation. All mice were maintained at the Stanford University School of Medicine animal facility and treated in accordance with the Stanford University Laboratory Animal Care guidelines. Thymocytes were isolated from the 5-FU primed-mice, infected with miRNA expression vectors by spinoculation, and seeded at 1×105 infected cells/well into 24-well tissue culture plates containing a monolayer of OP9-DL1 stromal cells. For each viral construct, 12 independent culture replicates were seeded. The cells were cultured in Minimum Essential Medium (MEM) Alpha Medium supplemented with 20% FCS, 10 mM Hepes, 1 mM Sodium pyruvate, 5 ng/ml IL-7, and 27.5 ng/ml Flk2/Flt3L for 24 hours and then the medium was changed to remove non-adherent thymocytes. The cultures were fed with fresh medium on day 6. After about 8–10 days of culturing, cells were harvested and stained for surface marker CD4, CD8, and CD45. The percentage of DP cells yielded from culture was quantified by flow cytometry. Both adherent and non-adherent cells were collected. Adherent cells were dissociated by treatment with collagenase type IV (0.8 mg/ml; Worthington, Lakewood, NJ) followed by forceful pipetting. The cells were then immunolabeled with PE-conjugated anti-CD4 antibody (clone RM4-5; BD Pharmingen, San Diego, CA) and PE-Cy5-conjugated anti-CD8a antibody (clone 53-6.7; BD Pharmingen) and analyzed on a FACSCalibur (BD Biosciences, San Jose, CA) for the expression of CD4 and CD8 cell surface antigens. GFP positive thymocytes were distinguishable from GFP positive stromal cells by forward and side scatter gates and the intensity of green fluorescence. When the infection-rate was low, anti-CD45 antibody staining was used to gate out contaminating GFP+ OP9-DL1 cells. The appropriate dilution for each antibody was determined prior to use.
Box-plots summarize the distribution of relative miRNA activity in DP cell development. The ends of the boxes define the 25th and 75th percentiles, a line indicates the median, and bars define the 5th and 95th percentiles. Individual outliers are also shown. The activities of mir-181a-1, mir-181c, and mutant genes in DP cell development were normalized so that the empty vector (negative control) had a median activity of “0” and mir-181a-1 expressing vector (positive control) had a median activity of “1.” The percentage of DP cells yielded from the co-culture assay varied between experiments, possibly due to the heterogeneous nature of the thymic progenitor cells and intrinsic variation between the batches of mice used. Therefore, such normalization is required to reset the baseline and allows the independent repeats to be compared. Mann-Whitney Rank Sum Tests were performed to determine whether the activities of mutants were statistically different from the control vector or the mir-181a-1 vector.
Cell culture and transfection
Adherent BOSC 23 cells were grown in DMEM, 10% FBS, 1% of pen/strep antibiotics, supplemented with glutamine. For northern and primer extension assays, BOSC 23 cells were plated at a density of 3.75×105 cells/well in a 6-well plate at 24 hours before transfection. Cells were transfected with 2 µg constructs expressing mir-181a-1, mir-181c, mutant genes, and control vector using Fugene transfection reagents (Roche).
Quantitative Northern blot analyses
Quantitative Northern blot analyses were used to determine the level of mature miRNA expression and processing of the pre-miRNA and mature miRNA. Total RNA was prepared from BOSC 23 cells transfected with constructs expressing mir-181a-1 or mir-181c, and their mutant genes, and loaded onto 15% PAGE gel (10 µg/sample). Since all of the miRNA expressing vectors contain an independent GFP reporter, the percentage of GFP positive cells were determined by FACS analyses and used to control for variations in transfection efficiency. Various amounts of synthetic mature miRNA were loaded onto the same gel to generate standard curves. Specific probes that perfectly matched mature miRNAs were used in hybridization to determine the expression of both mature and pre-miRNA species (see Materials and Methods S1 for probes sequences). Band intensity was determined by phosphoimager quantification. Blots were also probed with U6 probes to normalize loading. Exact copies/pg of total RNA were determined by comparing to the corresponding standard curve. Representative blots of three or more independent quantitative Northern blot analyses were shown. Standard curves and average results of three or more independent quantitative Northern blot analyses were summarized and plotted.
Primer Extension Analyses
Primer extension was used to map the 5′ ends of the mature miRNAs produced from the mir-181a-1/c mutant genes. Total RNA was prepared from BOSC 23 cells 48 hours after transfection with constructs expressing mir-181a-1, mir-181c, and their mutant genes. P32 labeled primer was mixed with appropriate RNA samples (10 µg total RNA) in the reaction buffer (1×RT reaction buffer with 0.25 mM of each dNTP), heated at 55°C for 20 minutes, and slowly cooled to 16°C to allow for annealing. The primer extension reaction was initiated by adding reverse transcriptase and then carried out at 16°C for 20 minutes, 42°C for 2 hours, and 85°C for 5 minutes. Samples were then resolved on a 15% denaturing PAGE gel. Synthetic miR-181a or miR-181c oligos in single nucleotide increments (15 nt–22/23 nt) were labeled and loaded onto the gel as size ladder. The primer extension results were visualized by overnight exposure to a phosphoimager screen (see Materials and Methods S1 for probes and DNA ladder sequences).
miRNA qPCR analyses
In the OP9-DL1 culture assay, mixed DN (CD4− CD8−) thymic progenitor cells differentiate into DP cells though multiple stages that are characterized by unique cell surface markers and complex molecular events. The DP cell population in the OP9-DL1 culture represents a relatively homogeneous population and can be isolated by FACS-sorting in reasonable quantity for miRNA qPCR analyses. In brief, GFP positive DP cells from OP9-DL1 co-culture assay were isolated by FACS-sorting (>94% pure). Synthetic miR-223 was spiked into sorted cells at the ratio of 100 pmol of miR-223 per 100,000 cells before RNA purification. Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA). We assumed that the ratio of spiked miR-223 to a miRNA of interest would not change during RNA purification. cDNA was then synthesized using miRNA-specific looped primers (Applied Biosystems, Foster city, CA) and amplified with miRNA-specific forward primers, a TaqMan probe, and reverse primers (Applied Biosystems). PCR amplification was performed in triplicate in an ABI-7000 sequence detection system (Applied Biosystems) at 95°C for 10 minutes followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. To determine the exact copy number of a miRNA in sorted DP cells, we carried out absolute miRNA quantification with miRNA qPCR assay. Exact copies of test and spiked miRNAs in the defined amount of total RNA input were determined by using standard curves for mature miR-181a, miR-181c, and spiked miR-223. miR-181a or miR-181c expression was normalized using miR-15b as an endogenous loading control. Representative results of three miRNA qPCR analyses of independently-sorted, virally infected DP cells were shown. All reactions were carried out according to the manufacturer's instructions.
Supporting Information
(0.03 MB DOC)
Schematics and nucleotide sequences depict mature mir-181a-1 mutants. Compensatory mutations are introduced to maintain the integrity of the pre-miRNA secondary structure.
(0.95 MB TIF)
Expression and processing of wild-type mir-181a-1 and the M1 stem mutant gene. (A) Nucleotide sequences of the wild-type miR-181a and the M1 mutant. (B) Northern blot analyses of mature miRNA expression from the wild-type miR-181a and the M1 mutant. Total RNA was prepared from BOSC cells transfected with constructs expressing mir-181a-1, or the M1 mutant genes. Relative transfection efficiencies were determined by qPCR analyses of GFP mRNA levels produced from the transfected miRNA constructs, then used to normalize RNA loadings in Northern blot analyses. A shorter probe that perfectly matches to both mature miR-181a and the M1 mutant forms is used in hybridization to determine the expression of mature miR-181a and its mutant forms. Relative expression levels of the mature miRNAs determined by phosphoimager quantification is indicated.
(1.46 MB TIF)
Members of the mir-181 gene family. (A) Alignment of the mature miR-181 miRNAs. (B) Schematics and nucleotide sequences depicting the pre-miRNAs of the human and mouse mir-181 gene family members.
(0.98 MB TIF)
Developmental regulation of miR-181c expression in various purified thymocyte populations determined by miRNA qPCR.
(0.23 MB TIF)
The effects of mutations in the mature miRNA region of the mir-181a-1 genes on DP cell development (SI Fig. 1). (A) Box-plots summarize the percent of DP cells generated from DN progenitor cells infected with mir-181a-1, or mature miRNA mutant genes (gated on GFP positive). A representative OP9-DL1 stromal co-culture assay (12 independent replicates for each construct) is shown. The ends of the boxes define the 25th and 75th percentiles, a line indicates the median, and bars define the 5th and 95th percentiles. (B) Statistical summary. Mann-Whitney Rank Sum Tests were performed on this representative data set to determine whether the activity of mir-181a-1, mir-181c, or their chimeric mutants is statistically different from the control vector or the mir-181a-1 vector.
(2.34 MB TIF)
The effects of the chimeric mir-181a-1 and mir-181c genes on DP cell development (Fig. 2). (A) Box-plots summarize the percent of DP cells generated from DN progenitor cells infected with mir-181a-1, mir-181c, or their chimeric mutants (GFP positive). A representative OP9-DL1 stromal co-culture assay (12 independent replicates for each construct) is shown. The ends of the boxes define the 25th and 75th percentiles, a line indicates the median, and bars define the 5th and 95th percentiles. (B) Statistical summary. Mann-Whitney Rank Sum Tests were performed on this representative data set to determine whether the activity of mir-181a-1, mir-181c, or their chimeric mutants is statistically different from the control vector or the mir-181a-1 vector.
(1.92 MB TIF)
The effects of the pre-miR-181a-1 loop mutants on DP cell development (Fig. 3). (A) Box-plots summarize the percent of DP cells generated from DN progenitor cells infected with mir-181a-1, or pre-miR-181a-1 loop mutant genes (GFP positive). A representative OP9-DL1 stromal co-culture assay (12 independent replicates for each construct) is shown. The ends of the boxes define the 25th and 75th percentiles, a line indicates the median, and bars define the 5th and 95th percentiles. (B) Statistical summary. Mann-Whitney Rank Sum Tests are performed on this representative data set to determine whether the activity of mir-181a-1 and pre-miRNA loop mutant genes is statistically different from the empty vector (negative control) or the mir-181a-1 expressing vector (positive control).
(1.07 MB TIF)
Mature and pre-miRNA expression levels from the chimeric mir-181a-1 and mir-181c genes (Fig. 5A, B). Total RNA was prepared from BOSC cells transfected with constructs expressing mir-181a-1, mir-181c, and the chimeric mir-181a-1 and mir-181c genes. Since all miRNA vectors contain an independent GFP reporter, percentage cells that are GFP positive were determined by FACS analyses and used to control for variations in transfection efficiency. Quantitative Northern blot analyses were carried out to determine the expression of mir-181a-1, mir-181c, and the chimeric mir-181a-1 and mir-181c genes. Specific probes that perfectly match to mature miR-181a or miR-181c were used in hybridization to determine the expression of mature and pre-miRNA forms. Band intensities were determined by phosphoimager quantification and normalized to the levels of wild-type controls accordingly. (A) Standard Curves for miR-181a and miR-181c. (B) The copies of mature miR-181a and miR-181c in BOSC 23 cells transfected with mir-181a-1/c mutants determined by quantitative Northern blot analyses. Average results of four independent experiments were plotted ( Table S4 for statistics). (C) Relative levels of pre-miR-181a and pre-miR-181c in BOSC 23 cells transfected with mir-181a-1/c mutants determined by Northern blot analyses. Average results of four independent experiments were plotted.
(2.36 MB TIF)
Mature and pre-miRNA expression levels from the pre-miR-181a-1 loop mutant genes (Fig. 5E). Total RNA was prepared from BOSC cells transfected with constructs expressing the mir-181a-1 loop mutant genes. Since all miRNA vectors contain an independent GFP reporter, percentage cells that are GFP positive were determined by FACS analyses and used to control for variations in transfection efficiency. Quantitative Northern blot analyses were carried out to determine the expression of the pre-mir-181a-1 loop mutant genes. A probe that perfectly matches to the mature miR-181a was used in hybridization to determine the expression of mature and pre-miRNA forms. Band intensity was determined by phosphoimager quantification and normalized to the levels of wild-type controls accordingly. (A) Standard Curves for miR-181a. (B) The copies of mature miR-181a in BOSC 23 cells transfected with mir-181a-1 loop mutants determined by quantitative Northern blot analyses. Average results of four independent experiments were plotted ( Table S7 for statistics). (C) Relative levels of pre-miR-181a in BOSC 23 cells transfected with mir-181a-1 loop mutants determined by Northern blot analyses. Average results of four independent experiments were plotted.
(0.92 MB TIF)
Summary of the statistical analyses on the activity of mir-181a-1 genes with mutations in the mature miRNA region. The activities of the wild-type mir-181a-1 and its variants with mutations in the mature miRNA regions in promoting DP cell development are normalized so that the empty vector (negative control) has a median activity of “0” and the wild-type mir-181a-1 vector (positive control) has a median activity of “1.” Normalized data from 3–5 independent T cell assays (each with 12 independent replicates, total 36–60 replicates) are pooled and graphed in the distribution box plots. Mann-Whitney Rank Sum Tests are performed to determine whether the activity of mir-181a-1 and mature miRNA mutant genes is statistically different from the empty vector (negative control) or the mir-181a-1 expressing vector (positive control).
(0.05 MB DOC)
Summary of the statistical analyses on the activity of the mir-181a-1 and mir-181c mutant genes. The activities of mir-181a-1, mir-181c, and mutant genes in promoting DP cell development are normalized so that the empty vector (negative control) has a median activity of “0” and the mir-181a-1 expressing vector (positive control) has a median activity of “1.” Normalized data from 3–7 independent T cell assays (each with 12 independent replicates, total 36–84 replicates) are pooled and graphed in the distribution box plots. Mann-Whitney Rank Sum Tests are performed on the pooled data set to determine whether the activity of mir-181a-1 and mir-181c mutant genes is statistically different from the empty vector (negative control) or the mir-181a-1 expressing vector (positive control).
(0.04 MB DOC)
Summary of the statistical analyses on the activity of the mir-181a-1 genes and mutants with nucleotides in the pre-miRNA loop region altered. The activity of mir-181a-1 and its pre-miRNA loop mutant genes in promoting DP cell development are normalized so that the empty vector (negative control) has a median activity of “0” and the mir-181a-1 expressing vector (positive control) has a median activity of “1.” Normalized data from at least 6 independent T cell assays (each with 12 independent replicates, total 72 replicates) are pooled and graphed in the distribution box plots. Mann-Whitney Rank Sum Tests are performed on the pooled data set to determine whether the activity of mir-181a-1 and pre-miRNA loop mutant genes is statistically different from the empty vector (negative control) or the mir-181a-1 expressing vector (positive control).
(0.03 MB DOC)
Summary of the statistical analyses on the mature miR-181a and miR-181c levels in transfected BOSC cells. The copy numbers of mature miR-181a and miR-181c expressed in BOSC cells transfected with constructs expressing mir-181a-1, mir-181c, and the chimeric mir-181a-1 and mir-181c genes were determined by quantitative Northern blot. Statistical significance was determined by analyzing the results of four independent quantitative Northern blot analyses using an unpaired two-tailed student's t test.
(0.03 MB DOC)
Summary of the statistical analyses on the mature miR-181a levels in infected DP T cells. The copy numbers of mature miR-181a expressed in the DP thymocytes transduced with viral vectors expressing mir-181a-1, mir-181c, “pre-chimeric”, and “loop-chimeric” miRNAs were determined by miRNA qPCR analyses. Mature miR-181a copy numbers in DP cells were determined using standard curve miRNA qPCR quantification and normalized using miR-15b as an endogenous control. Representative results of three miRNA qPCR analyses of independently sorted infected DP cells were shown. Statistical significance was determined by an unpaired two-tailed sutdent's t test.
(0.04 MB DOC)
Summary of the statistical analyses on the mature miR-181c levels in infected DP T cells. The copy numbers of mature miR-181c expressed in the DP thymocytes transduced with viral vectors expressing mir-181a-1, mir-181c, “pre-chimeric”, and “loop-chimeric” miRNAs were determined by miRNA qPCR analyses. Mature miR-181c copy numbers in DP cells were determined using standard curve miRNA qPCR quantification and normalized using miR-15b as an endogenous control. Representative results of three miRNA qPCR analyses of independently sorted infected DP cells were shown. Statistical significance was determined by an unpaired two-tailed student's t test.
(0.04 MB DOC)
Summary of the statistical analyses on the mature miR-181a in transfected BOSC cells. The copy numbers of mature miR-181a expressed in BOSC cells transfected with constructs expressing mir-181a-1 loop mutants were determined by quantitative Northern blot. Statistical significance was determined by analyzing the results of four independent quantitative Northern blot analyses using an unpaired two-tailed student's t test.
(0.03 MB DOC)
Summary of the statistical analyses on the mature miR-181a levels in infected DP T cells. The copy numbers of mature miR-181a expressed in the DP thymocytes transduced with viral vectors expressing mir-181a-1 loop mutants were determined by miRNA qPCR analyses. Mature miR-181a copy numbers in DP cells were determined using standard curve miRNA qPCR quantification and normalized using miR-15b as an endogenous control. Representative results of three miRNA qPCR analyses of independently sorted infected DP cells were shown. Statistical significance was determined by an unpaired two-tailed student's t test and summarized in the table.
(0.03 MB DOC)
Acknowledgments
We would like to thank the members of the Chen lab, and Drs. Helen Blau, Thomas Cech, Andrew Fire, Karla Kirkegaard, and Peter Sarnow, for their helpful discussions and comments on our manuscript. We are very grateful to Dr. J.C. Zúñiga-Pflücker for the OP9-GFP and OP9-DL1 cells.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the NIH R01-HL081612 grant and the Baxter and Terman faculty awards (to C.-Z. C.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bushati N, Cohen SM. microRNA Functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Bettinger JC, Pasquinelli AE, et al. The 21 nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 6.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA Targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 9.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, et al. miR-181a Is an Intrinsic Modulator of T Cell Sensitivity and Selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grad Y, Aach J, Hayes GD, Reinhart BJ, Church GM, et al. Computational and experimental identification of C. elegans microRNAs. Mol Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 15.Steiner FA, Hoogstrate SW, Okihara KL, Thijssen KL, Ketting RF, et al. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat Struct Mol Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- 16.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs Are Sorted into Functionally Distinct Argonaute Complexes after Production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 18.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 23.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 25.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984;38:861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- 27.Eguchi Y, Itoh T, Tomizawa J. Antisense RNA. Annu Rev Biochem. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(0.03 MB DOC)
Schematics and nucleotide sequences depict mature mir-181a-1 mutants. Compensatory mutations are introduced to maintain the integrity of the pre-miRNA secondary structure.
(0.95 MB TIF)
Expression and processing of wild-type mir-181a-1 and the M1 stem mutant gene. (A) Nucleotide sequences of the wild-type miR-181a and the M1 mutant. (B) Northern blot analyses of mature miRNA expression from the wild-type miR-181a and the M1 mutant. Total RNA was prepared from BOSC cells transfected with constructs expressing mir-181a-1, or the M1 mutant genes. Relative transfection efficiencies were determined by qPCR analyses of GFP mRNA levels produced from the transfected miRNA constructs, then used to normalize RNA loadings in Northern blot analyses. A shorter probe that perfectly matches to both mature miR-181a and the M1 mutant forms is used in hybridization to determine the expression of mature miR-181a and its mutant forms. Relative expression levels of the mature miRNAs determined by phosphoimager quantification is indicated.
(1.46 MB TIF)
Members of the mir-181 gene family. (A) Alignment of the mature miR-181 miRNAs. (B) Schematics and nucleotide sequences depicting the pre-miRNAs of the human and mouse mir-181 gene family members.
(0.98 MB TIF)
Developmental regulation of miR-181c expression in various purified thymocyte populations determined by miRNA qPCR.
(0.23 MB TIF)
The effects of mutations in the mature miRNA region of the mir-181a-1 genes on DP cell development (SI Fig. 1). (A) Box-plots summarize the percent of DP cells generated from DN progenitor cells infected with mir-181a-1, or mature miRNA mutant genes (gated on GFP positive). A representative OP9-DL1 stromal co-culture assay (12 independent replicates for each construct) is shown. The ends of the boxes define the 25th and 75th percentiles, a line indicates the median, and bars define the 5th and 95th percentiles. (B) Statistical summary. Mann-Whitney Rank Sum Tests were performed on this representative data set to determine whether the activity of mir-181a-1, mir-181c, or their chimeric mutants is statistically different from the control vector or the mir-181a-1 vector.
(2.34 MB TIF)
The effects of the chimeric mir-181a-1 and mir-181c genes on DP cell development (Fig. 2). (A) Box-plots summarize the percent of DP cells generated from DN progenitor cells infected with mir-181a-1, mir-181c, or their chimeric mutants (GFP positive). A representative OP9-DL1 stromal co-culture assay (12 independent replicates for each construct) is shown. The ends of the boxes define the 25th and 75th percentiles, a line indicates the median, and bars define the 5th and 95th percentiles. (B) Statistical summary. Mann-Whitney Rank Sum Tests were performed on this representative data set to determine whether the activity of mir-181a-1, mir-181c, or their chimeric mutants is statistically different from the control vector or the mir-181a-1 vector.
(1.92 MB TIF)
The effects of the pre-miR-181a-1 loop mutants on DP cell development (Fig. 3). (A) Box-plots summarize the percent of DP cells generated from DN progenitor cells infected with mir-181a-1, or pre-miR-181a-1 loop mutant genes (GFP positive). A representative OP9-DL1 stromal co-culture assay (12 independent replicates for each construct) is shown. The ends of the boxes define the 25th and 75th percentiles, a line indicates the median, and bars define the 5th and 95th percentiles. (B) Statistical summary. Mann-Whitney Rank Sum Tests are performed on this representative data set to determine whether the activity of mir-181a-1 and pre-miRNA loop mutant genes is statistically different from the empty vector (negative control) or the mir-181a-1 expressing vector (positive control).
(1.07 MB TIF)
Mature and pre-miRNA expression levels from the chimeric mir-181a-1 and mir-181c genes (Fig. 5A, B). Total RNA was prepared from BOSC cells transfected with constructs expressing mir-181a-1, mir-181c, and the chimeric mir-181a-1 and mir-181c genes. Since all miRNA vectors contain an independent GFP reporter, percentage cells that are GFP positive were determined by FACS analyses and used to control for variations in transfection efficiency. Quantitative Northern blot analyses were carried out to determine the expression of mir-181a-1, mir-181c, and the chimeric mir-181a-1 and mir-181c genes. Specific probes that perfectly match to mature miR-181a or miR-181c were used in hybridization to determine the expression of mature and pre-miRNA forms. Band intensities were determined by phosphoimager quantification and normalized to the levels of wild-type controls accordingly. (A) Standard Curves for miR-181a and miR-181c. (B) The copies of mature miR-181a and miR-181c in BOSC 23 cells transfected with mir-181a-1/c mutants determined by quantitative Northern blot analyses. Average results of four independent experiments were plotted ( Table S4 for statistics). (C) Relative levels of pre-miR-181a and pre-miR-181c in BOSC 23 cells transfected with mir-181a-1/c mutants determined by Northern blot analyses. Average results of four independent experiments were plotted.
(2.36 MB TIF)
Mature and pre-miRNA expression levels from the pre-miR-181a-1 loop mutant genes (Fig. 5E). Total RNA was prepared from BOSC cells transfected with constructs expressing the mir-181a-1 loop mutant genes. Since all miRNA vectors contain an independent GFP reporter, percentage cells that are GFP positive were determined by FACS analyses and used to control for variations in transfection efficiency. Quantitative Northern blot analyses were carried out to determine the expression of the pre-mir-181a-1 loop mutant genes. A probe that perfectly matches to the mature miR-181a was used in hybridization to determine the expression of mature and pre-miRNA forms. Band intensity was determined by phosphoimager quantification and normalized to the levels of wild-type controls accordingly. (A) Standard Curves for miR-181a. (B) The copies of mature miR-181a in BOSC 23 cells transfected with mir-181a-1 loop mutants determined by quantitative Northern blot analyses. Average results of four independent experiments were plotted ( Table S7 for statistics). (C) Relative levels of pre-miR-181a in BOSC 23 cells transfected with mir-181a-1 loop mutants determined by Northern blot analyses. Average results of four independent experiments were plotted.
(0.92 MB TIF)
Summary of the statistical analyses on the activity of mir-181a-1 genes with mutations in the mature miRNA region. The activities of the wild-type mir-181a-1 and its variants with mutations in the mature miRNA regions in promoting DP cell development are normalized so that the empty vector (negative control) has a median activity of “0” and the wild-type mir-181a-1 vector (positive control) has a median activity of “1.” Normalized data from 3–5 independent T cell assays (each with 12 independent replicates, total 36–60 replicates) are pooled and graphed in the distribution box plots. Mann-Whitney Rank Sum Tests are performed to determine whether the activity of mir-181a-1 and mature miRNA mutant genes is statistically different from the empty vector (negative control) or the mir-181a-1 expressing vector (positive control).
(0.05 MB DOC)
Summary of the statistical analyses on the activity of the mir-181a-1 and mir-181c mutant genes. The activities of mir-181a-1, mir-181c, and mutant genes in promoting DP cell development are normalized so that the empty vector (negative control) has a median activity of “0” and the mir-181a-1 expressing vector (positive control) has a median activity of “1.” Normalized data from 3–7 independent T cell assays (each with 12 independent replicates, total 36–84 replicates) are pooled and graphed in the distribution box plots. Mann-Whitney Rank Sum Tests are performed on the pooled data set to determine whether the activity of mir-181a-1 and mir-181c mutant genes is statistically different from the empty vector (negative control) or the mir-181a-1 expressing vector (positive control).
(0.04 MB DOC)
Summary of the statistical analyses on the activity of the mir-181a-1 genes and mutants with nucleotides in the pre-miRNA loop region altered. The activity of mir-181a-1 and its pre-miRNA loop mutant genes in promoting DP cell development are normalized so that the empty vector (negative control) has a median activity of “0” and the mir-181a-1 expressing vector (positive control) has a median activity of “1.” Normalized data from at least 6 independent T cell assays (each with 12 independent replicates, total 72 replicates) are pooled and graphed in the distribution box plots. Mann-Whitney Rank Sum Tests are performed on the pooled data set to determine whether the activity of mir-181a-1 and pre-miRNA loop mutant genes is statistically different from the empty vector (negative control) or the mir-181a-1 expressing vector (positive control).
(0.03 MB DOC)
Summary of the statistical analyses on the mature miR-181a and miR-181c levels in transfected BOSC cells. The copy numbers of mature miR-181a and miR-181c expressed in BOSC cells transfected with constructs expressing mir-181a-1, mir-181c, and the chimeric mir-181a-1 and mir-181c genes were determined by quantitative Northern blot. Statistical significance was determined by analyzing the results of four independent quantitative Northern blot analyses using an unpaired two-tailed student's t test.
(0.03 MB DOC)
Summary of the statistical analyses on the mature miR-181a levels in infected DP T cells. The copy numbers of mature miR-181a expressed in the DP thymocytes transduced with viral vectors expressing mir-181a-1, mir-181c, “pre-chimeric”, and “loop-chimeric” miRNAs were determined by miRNA qPCR analyses. Mature miR-181a copy numbers in DP cells were determined using standard curve miRNA qPCR quantification and normalized using miR-15b as an endogenous control. Representative results of three miRNA qPCR analyses of independently sorted infected DP cells were shown. Statistical significance was determined by an unpaired two-tailed sutdent's t test.
(0.04 MB DOC)
Summary of the statistical analyses on the mature miR-181c levels in infected DP T cells. The copy numbers of mature miR-181c expressed in the DP thymocytes transduced with viral vectors expressing mir-181a-1, mir-181c, “pre-chimeric”, and “loop-chimeric” miRNAs were determined by miRNA qPCR analyses. Mature miR-181c copy numbers in DP cells were determined using standard curve miRNA qPCR quantification and normalized using miR-15b as an endogenous control. Representative results of three miRNA qPCR analyses of independently sorted infected DP cells were shown. Statistical significance was determined by an unpaired two-tailed student's t test.
(0.04 MB DOC)
Summary of the statistical analyses on the mature miR-181a in transfected BOSC cells. The copy numbers of mature miR-181a expressed in BOSC cells transfected with constructs expressing mir-181a-1 loop mutants were determined by quantitative Northern blot. Statistical significance was determined by analyzing the results of four independent quantitative Northern blot analyses using an unpaired two-tailed student's t test.
(0.03 MB DOC)
Summary of the statistical analyses on the mature miR-181a levels in infected DP T cells. The copy numbers of mature miR-181a expressed in the DP thymocytes transduced with viral vectors expressing mir-181a-1 loop mutants were determined by miRNA qPCR analyses. Mature miR-181a copy numbers in DP cells were determined using standard curve miRNA qPCR quantification and normalized using miR-15b as an endogenous control. Representative results of three miRNA qPCR analyses of independently sorted infected DP cells were shown. Statistical significance was determined by an unpaired two-tailed student's t test and summarized in the table.
(0.03 MB DOC)