Abstract
Photoactive yellow protein (PYP) is a small bacterial photoreceptor that undergoes a light-activated reaction cycle. PYP is also the prototypical PAS (Per-Arnt-Sim) domain. PAS domains, found in diverse multi-domain proteins from bacteria to humans, mediate protein-protein interactions and function as sensors and signal transducers. Here, we investigate conformational and dynamic changes in solution in wild-type PYP upon formation of the long-lived putative signaling intermediate I2 with enhanced hydrogen/deuterium exchange mass spectrometry (DXMS). The DXMS results showed that the central β-sheet remains stable but specific external protein segments become strongly deprotected. Light-induced disruption of the dark-state hydrogen-bonding network in I2 produces increased flexibility and opening of PAS core helices α3 and α4, releases the β4-β5 hairpin, and propagates conformational changes to the central β-sheet. Surprisingly, the first ~10 N-terminal residues, which are essential for fast dark state recovery from I2, become more protected. By combining the DXMS results with our crystallographic structures, which reveal detailed changes near the chromophore, but limited protein conformational change, we propose a mechanism for I2 state formation. This mechanism integrates the results from diverse biophysical studies of PYP, and links an allosteric T- to R-state conformational transition to three pathways for signal propagation within the PYP fold. Based upon the observed changes in PYP plus commonalities shared among PAS domain proteins, we further propose that PAS domains share this conformational mechanism, which explains the versatile signal transduction properties of the structurally conserved PYP/PAS module by framework-encoded allostery.
Keywords: photoactive yellow protein (PYP), hydrogen/deuterium exchange mass spectrometry (DXMS), PAS domain, light receptor, signal transduction
Introduction
Life depends on the ability of uni- and multicellular organisms to appropriately sense and respond to their environment. Typically, the signal that triggers a cell to respond is either chemical, such as a small molecule binding to a cell surface or cytoplasmic receptor, or physical like touch or light. These signals induce changes in conformation and/or charge of the receptor protein that either activate or inhibit its biological function. Understanding the molecular changes involved in the translation of environmental stimuli into biological responses in molecular to atomic detail is one of the major questions in biophysics today.
An experimentally accessible and prototypic sensing and signaling protein for these processes is the photoactive yellow protein (PYP), the bacterial photoreceptor that exemplifies the PAS (Per-Arnt-Sim) domain superfamily of sensors and signal transducers. PYP is a 125 amino acid cytosolic blue light receptor from the purple bacterium Halorhodospira halophila and presumably serves as the sensor for negative phototaxis (for reviews see ref. 1 and 2). PYP adopts an α/β fold (Figure 1) with a central six-stranded antiparallel β-sheet flanked by α-helices on both sides3,4 and covalently binds a negatively charged p-hydroxycinnamic acid chromophore with Cys69 via a thioester linkage5,6. The chromophore is buried within the major hydrophobic core on one side of the central β-sheet, and tethered to the protein by a hydrogen-bonding network (Figure 1). Upon absorption of blue light (λmax = 446 nm), the chromophore undergoes trans-cis isomerization around its vinyl double bond in about 1 picosecond and thereby triggers a photocycle, in which all subsequent steps are thermally driven. In this photocycle, the absorption maximum is first red-shifted on the pico- to nanosecond time scale (early intermediates I0, I0 ‡ and I1; I1 is also called pR or PYPL) and then blue-shifted into the UV within 200 microseconds (long-lived intermediate I2, also called pB or PYPM) before spontaneously returning to the ground or dark state (P, also called pG) within hundreds of milliseconds7–9. The I2 intermediate accumulates since it precedes the slowest step in the photocycle and is therefore believed to represent the biologically important signaling state.
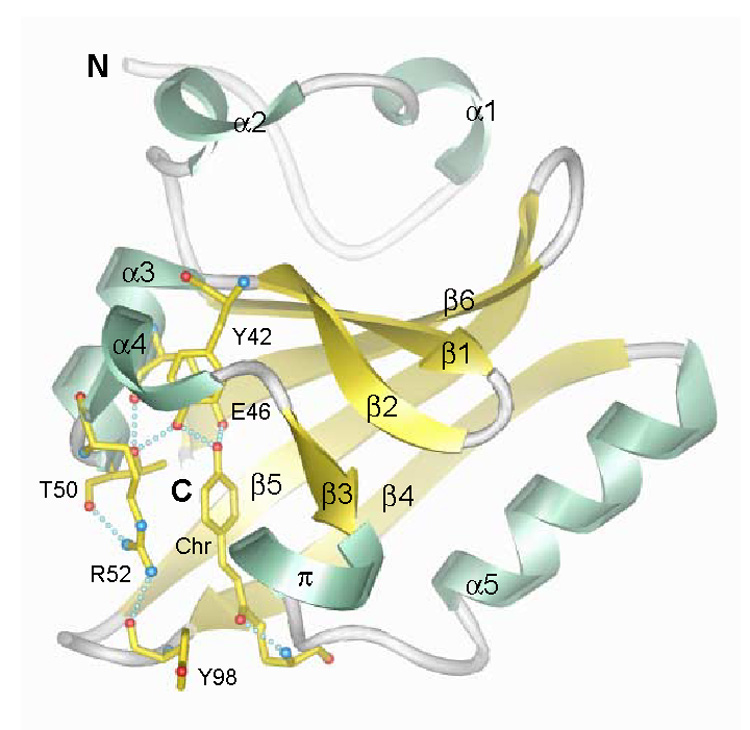
Fig. 1.
PYP fold and active site hydrogen bonding network. α-helices 1–5 and π-helix, green; β-strands 1–6, yellow; connecting loops, gray. The chromophore (Chr) and active site residues which participate in a hydrogen bonding network (dotted lines) are shown with yellow bonds (oxygen, red; nitrogen, blue; sulfur, yellow). The chromophore forms hydrogen bonds with its thioester oxygen to the backbone amide group and with its phenolate oxygen to the side chains of Tyr42 and Glu46. The side chain of Thr50 is in hydrogen bonding distance to the hydroxyl group of Tyr42 and the main-chain carbonyl oxygen of Glu46. Arg52 forms hydrogen bonds to the backbone carbonyl oxygens of Thr50 and Tyr98 by its guanidinium group and shields the chromophore from solvent. N-and C-termini are labeled (coordinates: 2PHY.pdb).
The PYP fold represents the structural prototype of the PAS domain10. PAS domains occur in hundreds of more complex multi-domain proteins from bacteria to humans, in which they mediate protein-protein interaction and function as sensors and signal transducers11. PAS domains can bind a wide variety of ligands or cofactors, such as FMN, FAD, or heme, which act as sensors of light, redox potential, or oxygen. The signals originating at these ligands are transmitted to different effector domains, such as histidine kinase domains in two-component signal transduction systems. The PAS/PYP fold can be divided into four segments10: (i) the N-terminal cap (residues 1–28), including α-helices 1 and 2, (ii) the PAS core (residues 29–69), including β-strands 1–3, intervening α-helices 3 and 4, and a short π-helix, (iii) the helical connector (residues 70–87), consisting predominantly of the long helix α5, and (iv) the β-scaffold (residues 88–125), containing β-strands 4–6.
High-resolution crystal structures for the PYP dark state3,12,13, intermediate I2, and cryotrapped I1-precursors14–16, as well as a nanosecond to second “molecular movie” by time-resolved Laue crystallography17,18 provided the following overall picture for structural rearrangements during the photocycle. In the tightly packed protein interior, rapid chromophore isomerization (~1 ps) is achieved by flipping the chromophore thioester carbonyl oxygen instead of its aromatic ring, and most protein dark state structural features are preserved15. During I2 formation, the hydrogen bonds from Glu46 and Tyr42 to the chromophore's phenolate oxygen (Figure 1) break, the phenolic ring of the chromophore rotates out of its binding pocket, and becomes protonated and exposed to solvent. To accomodate the chromophore movement, Arg52 swings out, breaking its side-chain hydrogen bonds with the main-chain carbonyl groups from Thr50 and Tyr98. Following spontaneous cis-trans re-isomerization, driven by physical strain in the non-planar cis-conformation, the protein rearranges to return to the minimum energy dark state conformation.
In PYP crystallographic structures, light-induced conformational changes are localized near the chromophore binding site. In solution, additional structural and dynamic changes during I2 formation have been observed with kinetic and thermodynamic measurements19–21; surface plasmon resonance22, NMR23,24, FTIR25,26, and CD spectroscopy27–30; small angle X-ray scattering28,31,32; transient dye binding27,33,34; and computational studies35,36. However, the many results from X-ray crystallography and other biophysical techniques have prompted interpretations for the I2 structure at opposite ends of the order-disorder spectrum.
Structural characterizations of the I2 intermediate can be facilitated by greatly extending its lifetime through acidification, N-terminal truncations, or site-directed mutations (e.g. of E46 and M100), although these modifications may also alter the native structure. Recently, the solution structure of the I2 intermediate for a PYP variant lacking the first 25 N-terminal amino acid residues (Δ25) was determined by NMR spectroscopy37. This study, which provides no information for the protein's N-terminus, suggested disordering of segments encompassing residues 42–58 (helices α3 and α4), 63–78 (first four residues of the connector helix α5 and flanking loops), and 96–103 (loop joining β4 to β5), and concluded that the main event of I2 intermediate formation is an unfolding of all helices in the protein except part of connector helix α5. The I2 intermediate in solution is proposed to exist in an equilibrium between a well-ordered crystal-like and a partially unfolded state which exchange on a millisecond time scale24,37. On the other hand, recent time-resolved Laue crystallographic studies for the PYP E46Q mutant38,39 and the PYP wild-type18 mapped a pathway of small conformational changes from the chromophore binding site, via helix α3, to the N-terminus of the protein. The authors suggested that these crystal structures represent precursors to more extensive conformational changes in solution, that are inhibited in the E46Q mutant26,33,40, and by crystal lattice forces. Thus, the detailed localization and extent of the light-activated protein conformational and dynamic changes, as well as the pathway for signal propagation within the structure of the PYP signaling state, remain elusive.
To improve understanding of PYP signaling, we determined for full-length, wild-type PYP in solution at neutral pH the specific sites that undergo changes in conformation or dynamics in the biologically important I2 intermediate, relative to the dark state, by applying the technique of enhanced hydrogen/deuterium (H/D) exchange mass spectrometry (DXMS)41. The technique consists of two distinct steps. In the initial labeling step, deuterium exchange is performed under physiological conditions for various exchange times. In the second step, the location of the deuterium label on individual exchangeable protein backbone amide groups is determined. This is done by denaturating the protein under conditions that dramatically slow the deuterium exchange rate of all amide protons and lock the attached deuterium label in place. The protein is then proteolyzed, and the peptide fragments are analyzed by HPLC separation and subsequent electrospray LCQ ion trap and time of flight (TOF) mass spectrometry. The proteolysis conditions, using pepsin and additional fungal proteases, are fine tuned to generate a maximal number of overlapping fragments. The DXMS methods used here provide complete protein coverage, high resolution, and high-throughput efficiency41–55. Consequently, our enhanced DXMS analyses for full-length, wild-type PYP in the dark state and the long-lived I2 intermediate allow us to localize conformational and dynamic changes occuring upon I2 formation in solution.
The DXMS studies identified increased flexibility and opening of PAS core helices α3 and α4, and release of the β-hairpin connecting strands β4 and β5, together with specific increased regularization of the β–sheet and increased protection of the N-terminus. The results are interpreted as a tense (T) to relaxed (R) state transition upon I2 formation: the tension imposed on the dark state by the active-site hydrogen-bonding network is released, yielding a more open, flexible and relaxed protein fold. We propose that the allosteric T- to R-state conformational transitions and signal propagation observed in PYP provide a probable general mechanism and pathway for signal transduction directed by the conserved structure of the PYP/PAS module.
Results
PYP proteolytic fragmentation and deuterium on-exchange
To achieve high-resolution localization of deuterium on the backbone amide groups of individual amino acid residues within PYP requires the generation of many, densely overlapping peptides. Prior to deuterium on-exchange experiments, we therefore determined the optimal proteolysis conditions on undeuterated PYP under exchange-quenched conditions (0.5% formic acid) by varying the GuHCl concentrations. The largest number of peptides was produced at 1.0 M GuHCl and 250 µl/min flow rate over a pepsin and subsequent fungal protease XIII column (66 µl bed volume each). The resulting peptides cover the whole PYP sequence without any gaps.
Deuterium/hydrogen exchange experiments were performed for the dark state and the I2 intermediate of PYP, generated under continuous illumination, once the optimal quench-compatible fragmentation conditions were established. PYP was incubated at 4 °C in 5 mM Hepes, pD (read) 7.0 containing 87.5% mole-fraction deuterated water for 10 time points varying from 30 s to 30000 s (8 h 20 min). Aliquots were exchange-quenched by adjusting to 0.5% formic acid and 1.0 M GuHCl at 4 °C, followed by immediate cooling to −80 °C. We produced 158 high-quality peptides for the PYP dark state and identified 57 identical peptides for the I2 state. The deuterium content of each of the 57 peptides from each sample was calculated from the LCMS data for all on-exchange times with the DXMS software. The peptide map is shown in Figure 2.
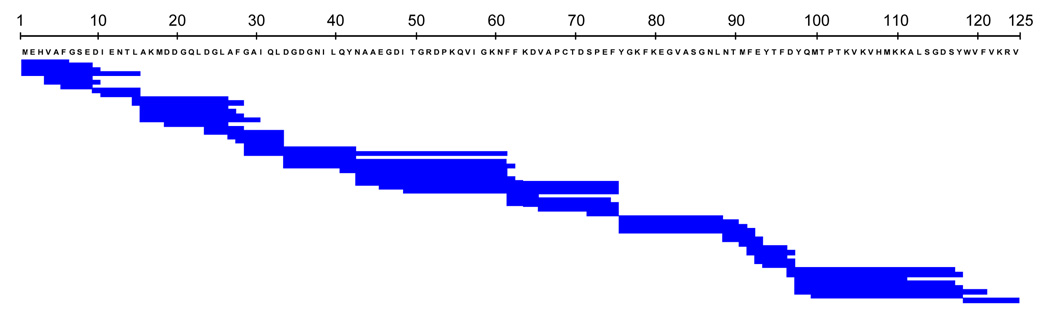
Fig. 2.
Peptide coverage map of PYP. 57 identical peptides were obtained for the dark state and the I2 intermediate of PYP by tandem online column-immobilized pepsin and fungal protease XIII digestion. The peptides cover all 125 amino acids of PYP shown in single letter code.
Construction of high-resolution deuterium exchange maps
For high-resolution analysis of deuterium incorporation, we determined the smallest protein sequence segments that are resolved by overlapping peptides, and calculated the mass gain for each such sequence segment at each time point measured in the experiment as previously described51. Figure 3 shows the result of this analysis for the PYP dark state and the I2 intermediate. With the fragmentation map of 57 peptides there are 46 such sequence segments with sizes ranging from 1 to 11 amides in length. 24 segments or 19.2% of the sequence are single amides.
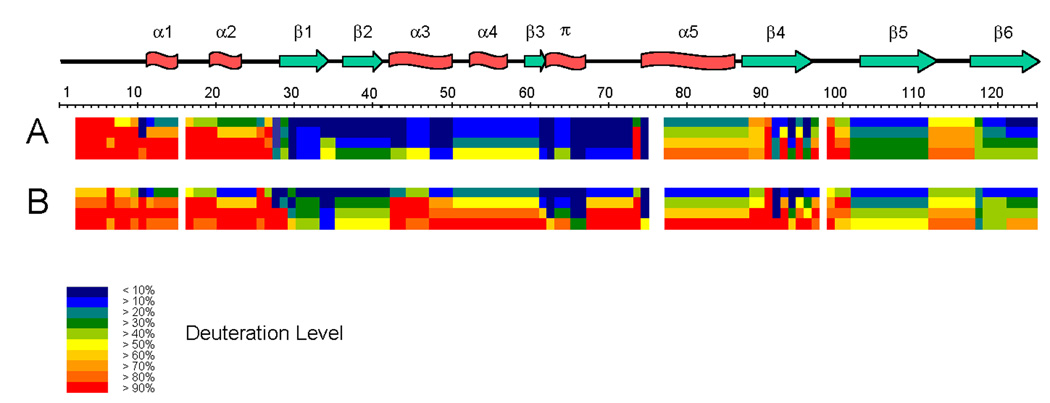
Fig. 3.
Amide H/D exchange for the PYP dark state (A) and the I2 intermediate (B). Each block represents protein sequence segments defined by overlapping peptides and consists of 4 time points (from top: 30 s, 300 s, 3000 s, 30000 s). The deuteration levels of each such segment at each time point are shown by different colors from blue (< 10% deuteration) through red (> 90% deuteration), as indicated in the figure. Residues 1–2, 16, 76–77, and 98 are not covered by any sequence segment. The secondary structure elements are shown on top of the protein sequence; α stands for α-helix, π for π-helix, β for β-sheet.
The H/D exchange map for the PYP dark state (Figure 3A) shows that contiguous segments with similar H/D exchange rates demarcate the four regions defining the PYP PAS module10. For the N-terminal 27 residues of PYP, including helices α1 and α2, all main-chain amides exchange rapidly (>90% at 3000 or 30000 s). This region corresponds with the solvent-exposed PYP/PAS N-terminal cap (residues 1–28). Residues 28–73, including the 3 N-terminal β-strands, intervening turns, intervening α-helices α3 and α4, and the π-helix that overlaps with β-strand 3, have very low H/D exchange rates (exchanging ~10% at 30 s, and increasing at most to >50% at 30000 s). This region of very low H/D exchange maps to the conserved PAS core (residues 29–69) and the beginning of the helical connector (residues 70–87). Amides in the long sequence segment (residues 78–88) in the helical connector show intermediate H/D exchange rates (from >20% at 30 s to –80% at 30000 s), and correspond to the long helix α5 (residues 75–86). The remaining C-terminal H/D exchange segments (residues 88–125), covering the three longer β-strands and two intervening loops, correspond to the β-scaffold (residues 87–125) of the PYP/PAS module, and exhibit exchange rates reflecting their secondary structural elements. The two C-terminal β-strands show slow H/D exchange, comparable to that of the PAS core. In strand β4, which lies at the solvent-exposed edge of the β-sheet, single-residue resolution highlights a pattern of alternating fast (residues 91, 93, 95) and slow (residues 92, 94, 96) H/D exchange, showing protection provided by the main-chain β-sheet hydrogen bonds from the amide groups of residues 92, 94, and 96 to the corresponding backbone carbonyls in strand β5.
Changes in H/D exchange upon light-activation of PYP
Light-activation to produce the I2 intermediate of PYP significantly alters the H/D exchange (compare Figure 3A with 3B). For I2, overall deuterium incorporation is ~17% higher than for the dark state, yet PYP remains stable as shown by measurable protection from deuterium incorporation. The central strands of the β-sheet (strands β1, β2, β5, and β6) remain the most protected, the N-terminal cap the least protected, and the connector helix α5 shows little change. The largest increases in deuteration upon I2 formation occur in sequence segments encompassing a continuous stretch around the chromophore binding site (residues 43–73), especially in helices α3 and α4 and strand β3 of the PAS core, and in the linker residues preceding connector helix α5. The main-chain amides of these residues are significantly less protected from H/D exchange in I2 than in the dark state.
To localize light-activated deuteration changes precisely, we determined for each protein sequence segment the average deuterium incorporation over 4 time points (30 s, 300 s, 3000 s, and 30000 s), distributed at evenly spaced intervals on a logarithmic scale (10-fold increments), calculated the differences between the I2 and the dark state (Figure 4), and mapped them onto the PYP dark state structure (Figure 5).
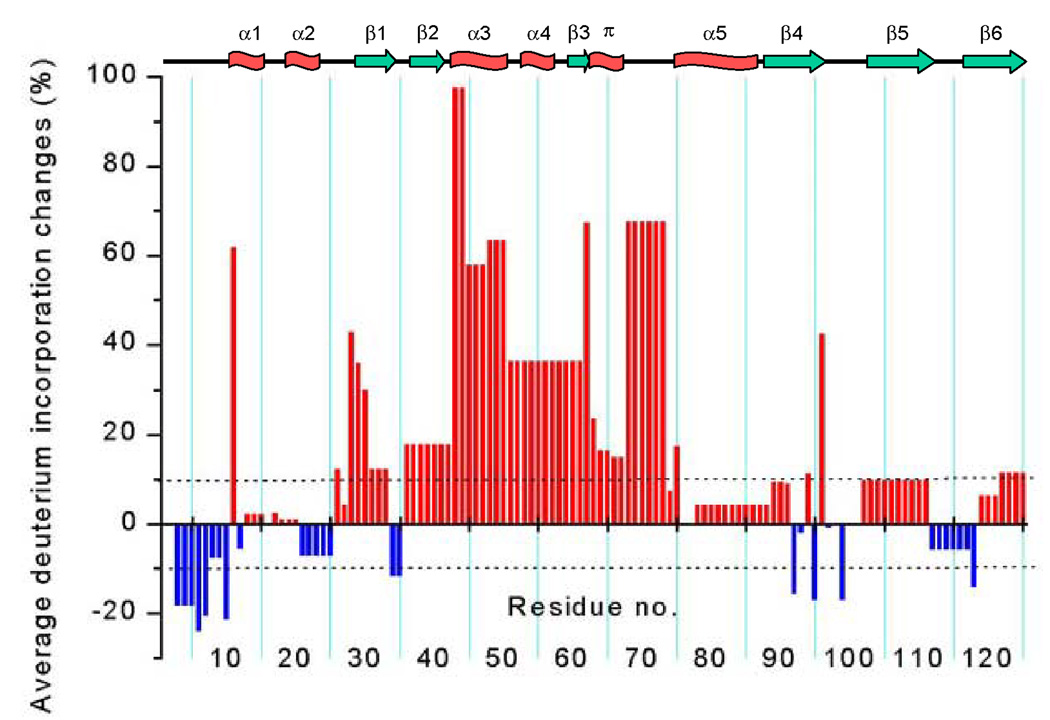
Fig. 4.
Average deuterium incorporation changes upon formation of the I2 intermediate. A comparison of the light-activated I2 state and the dark state of PYP is shown with a positive value (red bar) representing increased deuteration upon I2 formation and a negative value (blue bar) decreased deuteration upon I2 formation. Changes greater than 10% (dashed lines) are considered to be significant. The secondary structure elements are shown on top of the figure; α stands for α-helix, π for π-helix, β for β-sheet.
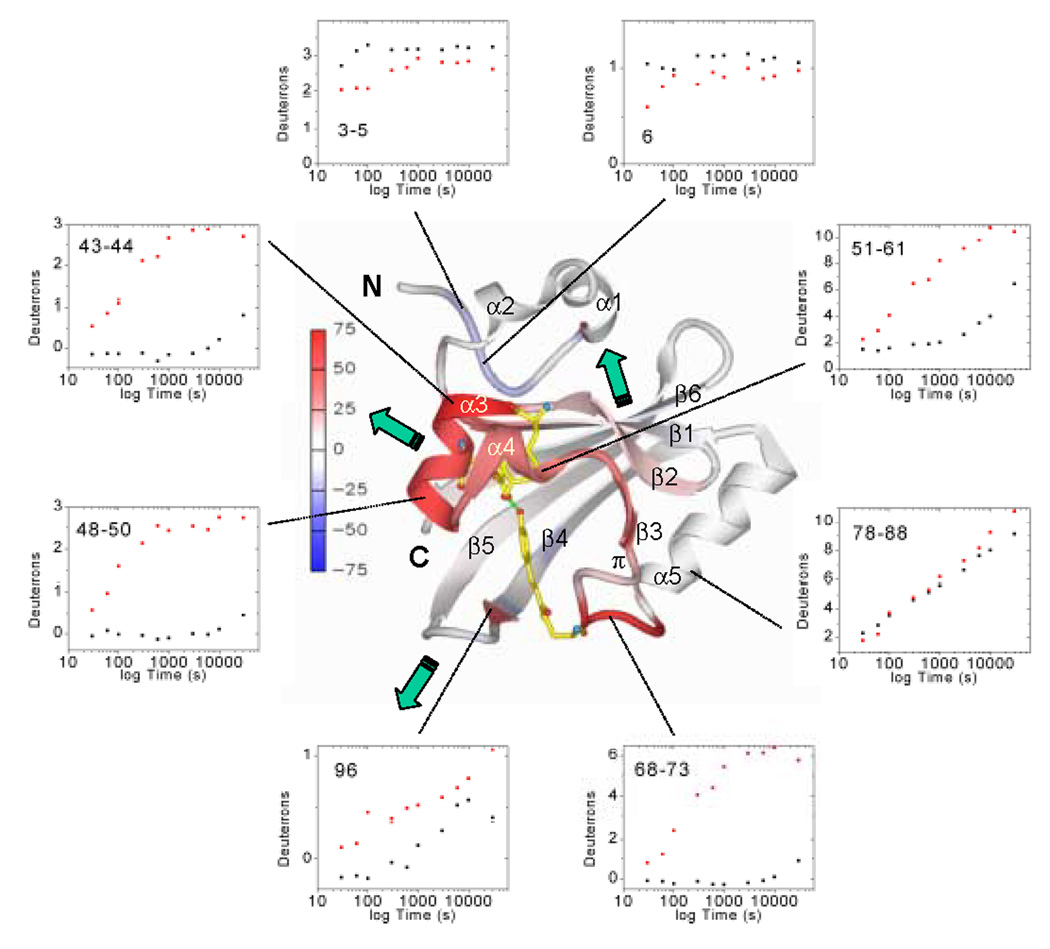
Fig. 5.
Average deuteration changes upon I2 state formation mapped on PYP structure, H/D exchange kinetics, and signal transduction pathways. Average deuterium incorporation changes upon I2 formation from Figure 4 are mapped on the PYP dark state ribbon diagram colored from −75 to +75% change according to the color bar. α-helices 1–5, the π-helix, β-strands 1–6, and N-and C-termini are labeled. The chromophore and active site residues Tyr42 and Glu46 which form hydrogen bonds (green dotted lines) to the phenolate oxygen of the chromophore in the dark state are shown for better orientation as well. Green arrows indicate the three signal transduction pathways (through the PAS core helices, the β-hairpin, or the β-sheet interface) as discussed in the text. H/D exchange kinetics (black dots, dark state; red dots, I2 intermediate) are shown as examples for the data quality and the time course of H/D exchange in representative PYP sequence segments.
The biggest increases in H/D exchange for I2 are focused on the chromophore binding site, where significant (>10%) changes extend from strand β2 to the connector helix (residues 36–73 and 75, in foreground of Figure 5). The most significant decreases in H/D exchange are found in the PYP N-terminus: segments 3–5, 6, 7 and 10, which are >90% deuterated in the dark state, become more protected in I2 (top of Figure 5). Significant differences from the dark state (Figure 4 and Figure 5) also occur at the junction of the N-terminal cap and strand β1 (segments 26, 28, 29, 30 and 31–33) which exhibit more deuteration (less protection), and in the hairpin joining β1 to β2 (segment 34–35), which exhibits less deuteration (more protection). The strong alternating protection pattern for edge strand β4 in the dark state becomes less distinct in I2 (Figure 3), as residues 92, 95, and 99 become significantly less deuterated and intervening hydrogen-bonded amides at 94 and 96 become more deuterated (Figure 4). At the C-terminal end of PYP, segments 118 (decrease) and 122–125 (increase) slightly change deuteration. The N-terminal cap helices α1 through α2 (residues 11–15 and 19–23) show little change between the dark state and I2 intermediate.
H/D exchange kinetics
We also examined the H/D exchange kinetics in both states for individual sequence segments at all 10 time points (Figure 5). For the dark state (Figure 3A), the most rapid rates of H/D exchange occur in the most flexible and solvent-exposed N-terminus (for example, Figure 5, sequence segments 3–5 and 6); then in segments forming the remainder of the N-terminal cap, encompassing solvent-exposed, but less flexible, α-helices 1 and 2. The connector helix α5 (Figure 5, segment 78–88) and most of the C-terminal β-scaffold (residues 87–125) are characterized by a steady, nearly linear time course for H/D exchange (for example, Figure 5, segment 96). For dark-state H/D exchange kinetics, helix α4 and short edge strand β3 exhibit a more delayed transition (Figure 5, segment 51–61). The dark-state PAS core region predominantly exhibits little and delayed H/D exchange (for example, Figure 5, segments 43–44, 48–50 and 68–73) characteristic of the least flexible or most buried parts of a folded protein.
Upon I2 formation, the rates of H/D exchange (Figure 5) increase dramatically for all sequence segments encompassing helices α3 and α4 and strand β3 of the PAS core (residues 43–73). First, segments 43–44 and 45–47 achieve >90% deuteration within 5 minutes (300 s) of H/D exchange, followed by segment 68–73 at 50 minutes (3000 s), and then segments 48–50, 51–61. Within this region (residues 43–73), only the π-helix segments after β3 (63, 64–65 and 66–67) show slower exchange rates (Figure 3). In contrast, H/D exchange rates decrease at the N-terminus especially at the beginning of the time course (for example segments 3–5 and 6 in Figure 5).
Discussion
Conformational changes and DXMS results
Our DXMS results and analyses show that PYP undergoes significant dynamic and conformational changes upon I2 intermediate formation in solution (at neutral pH and under continuous illumination). Overall, the protein is more open and flexible, as indicated by a ~17% increase in deuterium incorporation in the I2 state, relative to the dark state (Figure 3). This protein relaxation is consistent with the disruption of the active-site hydrogen-bonding network and loss of the chromophore ring from the major hydrophobic core, originally identified from the X-ray crystal structure of the I2 intermediate14. For I2 formation in solution, the DXMS results identify significant increases in deuterium incorporation for ~46% of the backbone amides of PYP, and significant decreases for ~10% (Figure 4). Most of the increased deuteration occurs in residues 43–73, encompassing α-helices 3 and 4, β-strand 3 and the overlapping π-helix, and the chromophore attachment site (Cys69) of the PAS core. In contrast, most of the decreased deuteration occurs near the N-terminus (residues 3–7 and 10). Thus, both the significant increases and decreases in deuteration between the dark and I2 states map to the periphery of the PYP molecule (Figure 5).
By identifying the specific PYP residues that undergo changes in flexibility, hydrogen-bonding or surface exposure upon I2 formation, these DXMS analyses help to explain and unify observations of conformational change and partial disorder in the I2 state in solution by other experimental techniques. (i) The dependence of I2 formation and decay kinetics on solvent hydrophobicity19, increased binding of PYP to lipid bilayers in the I2 state22, and transient binding of dyes27,33,34 indicated that buried hydrophobic sites are exposed in I2. (ii) Curved Arrhenius plots associated with I2 formation and decay were analyzed with a thermodynamic model incorporating heat capacity changes and interpreted as partial (~40 residues) protein unfolding and refolding, respectively20,21. (iii) Exchange broadening and 58% loss of NMR cross peaks indicated structural and dynamic disorder in the I2 state23,24. (iv) H/D exchange experiments using FTIR spectroscopy and electrospray ionization mass spectrometry showed that 23% of the protected amide backbone protons become solvent-exposed in I2 after 30 min on-exchange21. (v) A free energy loss of ~3.5 kcal/mol for PYP during I2 formation was measured with urea denaturation experiments56. (vi) Considerable changes in the protein backbone during I2 formation were detected by time-resolved FTIR spectroscopy25,26. (vii) The 19–27% reduction in 222 nm ellipticity for I2 in wild-type PYP27,30 was attributed to loss in α-helical content. (viii) Small angle X-ray scattering (SAXS) experiments showed a small increase in the radius of gyration (Rg) for the I2 intermediate, ranging from 0.4–1.1 Å depending on the experimental conditions28,31,32. Despite the limitations of each of these biophysical and biochemical techniques, and the seemingly significant variations in both the PYP proteins (wild-type vs. site-directed mutants vs. N-terminally truncated protein constructs) and the conditions used for the experiments, a common theme emerges, as described below.
Integrating structural changes in PYP upon I2 state formation
The DXMS results reveal the conformational and dynamic changes that propagate through the protein upon light-activated I2 formation in solution. These changes begin with those identified by comparing the crystallographic structures for PYP in the dark state and the long-lived I2 intermediate proposed to be the signaling state. In the dark state structure3, an active-site hydrogen-bonding network (Figure 1) interconnects the buried phenolate ring of the trans chromophore with Tyr42, Glu46, Thr50 and Arg52 (residues in and flanking α3). The guanidinium group of Arg52 protects the chromophore from solvent and is anchored by hydrogen bonds to the main-chain carbonyl groups of Thr 50 and Tyr98 (in the β4-β5 hairpin connecting strands 4 and 5). During I2 formation, the cis chromophore moves from the hydrophobic protein interior to the solvent-exposed protein surface, disrupting this dark state hydrogen-bonding network14. In the DXMS experiment, the strongest deuteration changes are found in the continuous stretch of residues 43–73 (Figure 4 and Figure 5). This stretch spans from α-helix 3 (residues 43–50), which contributes most residues to the hydrogen bonding network, through α-helix 4 (residues 53–57), short β-strand 3 (residues 60–62) and the overlapping π-helix (residues 62–67), to chromophore-bearing Cys69. The large increases in deuteration for I2 in this part of the protein therefore directly reflect the chromophore movements and the ensuing complete loss of the dark-state hydrogen-bonding network.
Changes in the H/D exchange kinetics between the I2 and dark states can provide insights into the conformational and dynamical changes that characterize this transformation. For the continuous stretch of residues (43–73) around the chromophore binding site, increased H/D exchange rates (segments 43–44, 48–50, 51–61, 68–73 as examples in Figure 5) describe the relaxation of these PAS core residues upon I2 formation. The overall pattern of H/D exchange rates in this region is higher (less protection) at the ends (α3 and the chromophore attachment site), and decreasing (more protection) toward the π-helix (Figure 3). This pattern suggests a breathing mode, in which the π-helix (residues 62–67) remains attached to the β-sheet, while preceding helices α3 (residues 43–50) and α4 (residues 53–57) become less hydrogen bonded and more solvent-exposed. Overall, the PAS core region from 43–73 is not highly connected to the rest of the PYP dark state structure and, as described below, the connections are weakened upon I2 formation.
The light-activated conformational and dynamic changes at the chromophore binding site subsequently propagate through the PYP protein to the β4–β5 hairpin, the β–sheet, and the N-terminus. By combining our DXMS results with crystal structures, which resolve the chromophore movements but are limited in protein conformational change, we can map the following signal transduction pathways for PYP.
Signaling pathway 1: PAS core α-helices
The DXMS results, in combination with the crystallographic structures, show a pathway for destabilization of the PAS core, starting from α-helix 3. In the dark state structure of PYP3, Asn43 is an N-cap57,58 forming stabilizing hydrogen bonds with the N-terminus of helix α3. Overall, Asn43 participates in 6 dark-state hydrogen bonds: Asn43 N to Ala30 O, Asn43 OD1 to Ala30 N and Glu46 N, Asn43 ND2 to Leu23 O and Phe28 O, and Asn43 O to Gly47 N. The main-chain amides participating in α3 hydrogen bonds, as well as in other hydrogen bonds internal to α3 (Glu46 to Thr50), exhibit some of the highest increases in deuteration for PYP during I2 formation (Figure 4 and Figure 5). Thus α3 undergoes increased flexibility or unfolding during I2 formation. These changes extend the distortions of α3 helical hydrogen bonds in the I2 crystal structure, as residues Tyr42, Ala45, Glu46, Gly47, Ile48, Thr50, and Gly51 moved inward to partially fill the cavity previously left by the exit of the chromophore ring14.
The increased flexibility and opening of helix α3 propagate to adjacent secondary structural elements β2, α4, and β3, as seen from deuteration increases encompassing residues 36–62 (Figure 4). CD spectroscopy has detected a 19–40% decrease in α-helical structure27–30 upon I2 formation. Because helix α1 in PYP's N-terminus and connector helix α5 are almost unaffected in the DXMS experiment (Figure 4–Figure 5) and helix α2 does not form in solution4, the unfolding reactions detected with CD spectroscopy are mainly attributed to helices 3 and 4, in agreement with the NMR results on the PYP Δ25 truncation variant37. Unfolding of helical segments is inhibited in PYP crystals, due to the intermolecular contacts between protein molecules within the crystal lattice, and perhaps to the reduced water content and the low pH (~5) at which the crystals grow.
DXMS analyses also show the release of the central PAS core from its connections to the rest of the PYP structure. Large increases in deuterium exchange at the main-chain amides of Ala30 (hydrogen-bonded to Asn43 OD1 in the dark state), Asn43 (hydrogen-bonded to Ala30 O in the dark state), and Phe62 (hydrogen-bonded to Gly37 O in the dark state) highlight the release of Asn43, α3 and β3 from their tethers to the N-terminal side of the β-sheet. Similarly, large increases in deuterium exchange at α3 amides of Ala44 (hydrogen-bonded to Asp24 OD1 in the dark state) and Ala45 (in water bridged hydrogen bonds to Asp24 and Leu26 main-chain carbonyl groups) shows the release of α3 from the N-terminal cap.
Signaling pathway 2: β-hairpin
In the β4-β5 hairpin, the connecting loop (residues 97–102) and the C-terminal end of edge strand β4 (residues 89–96) are in close proximity to the chromophore, and thus are directly affected by its movements. Residues 92, 94–96, and 99 show significant changes in deuterium incorporation upon I2 formation (Figure 4–Figure 5). These deuteration changes reflect the disruption of the dark state hydrogen bond between the Tyr98 backbone amide and the Arg52 guanidinium group (Figure 1), and consequent conformational rearrangement of this β-sheet connection. Although Tyr98 is not covered by any sequence segment in the DXMS experiment (Figure 3), its direct neighbour Gln99 shows a significant decrease in deuterium incorporation (Figure 4).
The deuteration changes in the β4–β5 hairpin of PYP may also be caused by the rearrangement of residues, including Tyr42, Phe62, Phe63, Phe75, Phe92, Tyr94, Phe96 and Tyr118, in the major hydrophobic core that surrounds the cavity left behind by the chromophore after it moves to the protein surface in I2 1,14. All of these aromatic residues show significantly altered deuterium incorporation (Figure 4). The transient exposure of hydrophobic sites in I2 was inferred from several experiments19,22,27,33,34, and the fluorescent dye Nile Red binds to the major hydrophobic core in PYP in the I2 state34. This indicates that the major hydrophobic core, which is part of the chromophore binding pocket, becomes deprotected in I2, in nice agreement with the observation that residues 43–73, 92, 94–96, and 99 around the chromophore binding site strongly change deuteration.
Signaling pathway 3: β-sheet interface
Disruptions of Asn43 hydrogen bonds to residues Phe28 and Ala30 in strand β1 (residues 29–34) relaxes distortions in the N-terminal half of the β-sheet. From strand β1 the signal spreads further to segments 122–125 and 118, respectively, which are located in C-terminal β-strand 6 (residues 117–125), as shown by small, but significant, deuteration changes in segments 118 and 122–125 (Figure 4–Figure 5). Gly29 makes antiparallel main-chain β-sheet hydrogen bonds to Val122 and Leu33 to Tyr118, joining the central N-terminal and C-terminal strands of the antiparallel β-sheet. Rearrangements in the C-terminal end of PYP were also proposed from fluorescence spectroscopy on Trp119, the only Trp in PYP59, but may primarily reflect the influence of the changed dipole moment of the isomerized chromophore on Trp fluorescence60. Decreased H/D exchange at the amides of Asp34, Gly35, and Tyr118 suggests that the release of the Asn38 side chain and Asp36 main chain carbonyl from their hydrogen bonds with π-helix N-cap Asn61, as well as the release of Gly37 O by Phe62 N, allows regularization of the β–sheet hydrogen bonds from Tyr118 N to Leu33 O, Asp34 N to Asn38 O, and Gly35 N to Asp116 O. Thus, the β–sheet relaxes, becomes less distorted and more regular in the I2 intermediate.
Remarkably, on the opposite side of the β-sheet from the chromophore, a significant decrease in deuteration is found for the first 7 N-terminal PYP residues and residue 10 (Figure 4–Figure 5), indicating that these residues become more protected upon I2 formation. The N-terminal cap (helical lariat) of PYP (residues 1–28) is closed by a main-chain hydrogen bond from Glu2 to Gly25, but forms few hydrogen bonds with the remainder of the protein and is therefore relatively flexible in the dark state3. In good agreement with the DXMS results, N-terminal truncation experiments have shown that the first 6 residues are most important within the N-terminal cap30,61. Truncation of residues 1–6 prolongs the lifetime of I2 140 fold and results in a significant change of the amide I infrared band, indicative of protein backbone motions. Additional truncation of residues 7–15 and 16–23 leads to smaller shifts of the amide I band and increases the I2 lifetime an additional 16 and 32 fold, respectively. The DXMS results suggest that the secondary structure elements in the N-terminal cap remain stable, in contrast to previous assumptions39,62. The N-terminus has an important function in stabilizing helices 3–4 (residues 43–50 and 53–57) in the PYP core, as the protection factors derived from H/D exchange experiments are substantially smaller in the Δ25 truncation variant than in wild-type PYP37. The decrease of the recovery kinetics from I2 by 2–3 orders of magnitude in N-terminal deletion mutants61,62 indicates that the N-terminus, besides possibly interacting with a PYP binding partner protein, is important for tuning the lifetime of the I2 state and catalyzing PYP recovery.
Allosteric T-state to R-state switch for PYP/PAS module
Based upon the above results and analyses, we propose that PYP and likely other PAS domains undergo an allosteric (“other shape”) transition, switching between two conformations for signaling. By analogy with the nomenclature developed for oligomeric allostery63–65, we define tense (T) and relaxed (R) states within the single domain of PYP. In the T-state (the dark state for PYP), the β-sheet is distorted and under tension imposed by restraints (the PYP activesite hydrogen-bonding network) which lock the PAS core helices (PYP α3 and α4) to the underlying β-sheet. The conformational switch is triggered by release of these restraints (light-induced conformational changes in the PYP chromophore disrupt the hydrogen-bonding network). In the resulting R-state, the PAS core helices and the β4–β5 hairpin are released, relaxing the distortions in the β-sheet. Thus, the versatile PYP/PAS module can propagate a conformational signal in three directions (Figure 5): through the PAS core helices, the β-hairpin or the β-sheet interface (with the N-terminal cap in PYP).
General implications
By applying enhanced hydrogen/deuterium exchange mass spectrometry (DXMS), we have localized the light-activated changes in PYP protein conformation and dynamics that accompany formation of the proposed signaling state I2. The H/D exchange took place in the wild-type protein in solution at neutral pH and under continuous illumination, so the results were neither perturbed by a crystalline environment, extreme pH, or other unnatural conditions, nor affected by mutations or truncations of the protein to artifically extend the lifetime of the photocycle intermediate. The observed ~17% overall increase in the deuteration of wild-type PYP upon light–activation, indicated a more open, flexible and relaxed protein fold for the proposed signaling intermediate I2 than for the dark state.
Light-induced movements of the PYP chromophore upon formation of the I2 signaling intermediate disrupt the active-site hydrogen-bonding network, thus destabilizing the protein. The most significant increases and decreases in deuteration between the dark and I2 states map to the periphery of the PYP molecule (Figure 5). More specifically, our DXMS analysis, in combination with structural results, reveals propagation of allosteric signals to open PAS core helices α3 and α4, release the β4–β5 hairpin, and produce conformational and dynamic changes in the central β-sheet and the N-terminal cap. In good agreement with these results, signaling through the β4–β5 hairpin and the α3–α4 helices was suggested from analysis of the “I2-like” PYP domain structure of Ppr66, a bacterial photoreceptor consisting of N-terminal PYP, central phytochrome, and C-terminal His kinase domains67. All three signal transduction pathways identified in this work may also provide versatility for signalling in other PAS domains. The key role for the N-cap of α3 in unlocking the structurally conserved PAS core helices and propagating the signal is underscored by functional conservation among the PAS domains10,39,57. In the clock protein PERIOD, the dimerization interface between the PAS A and PAS B domains is mediated by the interdomain insertion of a Trp side chain into the hydrophobic core68. This Trp interacts with residues structurally equivalent to those in the PYP signal transduction pathways shown by DXMS. In the LOV (light, oxygen or voltage) domain branch of the PAS domain family, a conserved pathway of structural connectivity from the flavin cofactor to a surface salt bridge (linking α4 with the β4–β5 hairpin) is proposed to modulate LOV-partner interactions69. In the LOV domain of the plant blue-light receptor phototropin, light-induced signal propagation from the FMN chromophore through the β-sheet interface is coupled to unfolding of a C-terminal helix that lies in the same three-dimensional location as the PYP N-terminal cap, on the far side of the β-sheet from the chromophore70. Similarly, the β-sheet interface of the hypoxia-inducible factor HIF-α PAS B domain forms the heterodimerization interface with the PAS domain of its partner the aryl hydrocarbon nuclear translocator ARNT71. Thus, accumulating evidence from PAS family members suggests that they share the three signal transduction pathways that we propose for PYP.
Taken together, our results and analyses show that, upon I2 formation, wild-type PYP undergoes a tense (T) to relaxed (R) state transition: the tension imposed on the dark state by the active-site hydrogen-bonding network is released, yielding a more open, flexible and relaxed protein fold. The high resolution of our DXMS studies revealed that signal propagation from chromophore movement, through disruption of the hydrogen-bonding network, then follows three pathways directed by the PYP/PAS fold: through 1) the PAS core helices, 2) the β-hairpin or 3) the β-sheet interface (with the N-terminal cap in PYP) (Figure 5). Furthermore, we propose that the allosteric T- to R-state conformational transition observed in PYP represents a probable unified mechanism and general pathway for signal transduction directed by the conserved structure and framework-encoded allostery of the PYP/PAS module.
Materials and Methods
Protein expression and purification
Protein expression, chromophore attachment, and protein purification were performed as described previously72. In this procedure, the PYP gene was inserted in a pET-20b vector (Novagen) which carries an N-terminal pelB signal sequence for export of apo-PYP into the growth medium. The chromophore was subsequently attached by addition of carbonyl-diimidazole-activated p-hydroxycinnamic acid to the cell-free growth medium. Holo-PYP with a final absorbance ratio A280/A446 of ~0.5 was purified by FPLC using hydrophobic interaction, anion exchange, and gel filtration columns.
Deuterium exchange mass spectrometry (DXMS)
Establishment of optimal proteolysis conditions
2.5 µl PYP (20 mg/ml in 5 mM Hepes, pH 7.0) were diluted with 17.5 µl 5 mM Hepes, pH 7.0, and mixed with 30 µl quench solution (0.8 % formic acid and 0.08 M, 0.8 M, 1.6 M, 3.2 M, 6.4 M guanidine hydrochloride, respectively) on ice, transferred to ice-cooled autosampler vials, and frozen on dry ice one minute after addition of quench solution. Vials with frozen samples were stored at −80 °C until transferred to the dry ice-containing sample basin of the autosampler module of the DXMS apparatus as previously described43,47–49. Temperature control was maintained by storing valves, tubing, columns, and autosampler within a refrigerator maintained at 5 °C, with columns also immersed in melting ice. The samples were held at dry ice temperature until individually melted at 5 °C. 45 µl sample were pumped through two protease columns (0.05% trifluoracetic acid (TFA), 250 µl/min, 16 s protease exposure) containing immobilized porcine pepsin (coupled to 20AL support from PerSeptive Biosystems at 30 mg/ml; 66 µl column bed volume) and fungal protease type XIII from Aspergillus saitoi (20 mg/ml; 66 µl column bed volume). Protease-generated fragments were collected on a C18 HPLC column, eluted by a linear acetonitrile gradient (5–45 % solvent B in 30 min, 50 µl/ min; solvent A: 0.05 % TFA, solvent B: 80 % acetonitrile, 20 % water, 0.01 % TFA), and injected directly into the mass spectrometer with data acquisition in either MS1 profile mode or data-dependent MS2 mode. Column effluent was analyzed on both an LCQ electrospray ion trap type mass spectrometer (Thermo Finnigan Inc.) operated with capillary temperature at 200 °C and an electrospray Q-TOF mass spectrometer (Micromass), as previously described43,47–49. The SEQUEST software program (Thermo Finnigan Inc.) was used to identify the likely sequence of the parent peptide ions and these tentative identifications were tested with specialized DXMS data reduction software developed in collaboration with Sierra Analytics (Modesto, CA). Fragmentation maps for all denaturant concentrations were generated. 1 M guanidine hydrochloride (final concentration) worked best and was used in all subsequent experiments.
On-exchange protein deuteration
Deuterium exchange was initiated by the addition of 2.5 µl PYP (20 mg/ml in 5 mM Hepes, pH 7.0) to 17.5 µl 5 mM Hepes in D2O, pD (read) 7.0. Samples were incubated in the cold room on ice for 30, 60, 100, 300, 600, 1000, 3000, 6000, 10000, and 30000 s, then mixed with 30 µl quench solution (0.8 % formic acid, 1.6 M guanidine hydrochloride, 16.6 % glycerol), transferred to ice-cooled autosampler vials, directly frozen on dry ice, and stored at −80 °C prior to DXMS analysis. The quench solution brings the PYP solution down to pH 2.5, denatures the protein, and quenches the deuterium exchange. During deuterium exchange, the samples were either kept in the dark (dark state sample set) or illuminated with >400 nm light from three optical fibers using two 150 W cold light fiber optic illuminators for accumulation of the I2 intermediate (light-activated sample set). The I2 intermediate accumulates since it precedes the slowest kinetic step in the photocycle (recovery to the dark state). Non-deuterated and fully deuterated samples were prepared for comparison by addition of 2.5 µl PYP (20 mg/ml in 5 mM Hepes, pH 7.0) to either 17.5 µl 5 mM Hepes, pH 7.0 on ice (non-deuterated sample) or to 17.5 µl 0.5 % formic acid in D2O over night at room temperature (fully deuterated sample). Quench was performed with 30 µl quench solution as described above. Data on the deuterated sample sets were acquired in a single automated 8-hour run with Q-TOF analysis only. Subsequent data reduction was performed with the DXMS data reduction software as described above.
Sub-localization of deuterium
The smallest protein sequence segments that were resolved by overlapping peptides in the final set of 57 peptides and their deuteration levels were determined by subtracting deuteration levels of overlapping peptides as previously described51. For example, the deuteration level for residue 10 of PYP can be obtained by subtracting the deuterium incorporation of peptide 3–9 from that of peptide 3–10. The calculations have to take into account that the first two N-terminal amide protons of each peptide exchange within the dead time of the experiment73 and cannot be used. Therefore, residues 1–2, 16, 76–77, and 98 are not covered by any sequence segment (Figure 3). If a sequence segment was determined by more than 2 peptides the average deuterium incorporation was calculated.
Acknowledgements
We thank Ilona L. Canestrelli and Tammy T. Woo for purification of PYP protein, Michael E. Pique for help with preparing Figure 5, and Krissi Hewitt for introduction to the Sequest and DXMS programs. This work was supported by NIH grants GM37684 to E.D.G. and CA118595 and CA099835 to V.L.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cusanovich MA, Meyer TE. Photoactive yellow protein: a prototypic PAS domain sensory protein and development of a common signaling mechanism. Biochemistry. 2003;42:4759–4770. doi: 10.1021/bi020690e. [DOI] [PubMed] [Google Scholar]
- 2.Hellingwerf KJ, Hendriks J, Gensch T. Photoactive yellow protein, a new type of photoreceptor protein: will this “yellow lab” bring us where we want to go? J. Phys. Chem. A. 2003;107:1082–1094. [Google Scholar]
- 3.Borgstahl GEO, Williams DR, Getzoff ED. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 4.Düx P, Rubinstenn G, Vuister GW, Boelens R, Mulder FAA, Hård K, Hoff WD, Kroon AR, Crielaard W, Hellingwerf KJ, Kaptein R. Solution structure and backbone dynamics of the photoactive yellow protein. Biochemistry. 1998;37:12689–12699. doi: 10.1021/bi9806652. [DOI] [PubMed] [Google Scholar]
- 5.Baca M, Borgstahl GEO, Boissinot M, Burke PM, Williams DR, Slater KA, Getzoff ED. Complete chemical structure of photoactive yellow protein: novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemistry. Biochemistry. 1994;33:14369–14377. doi: 10.1021/bi00252a001. [DOI] [PubMed] [Google Scholar]
- 6.Hoff WD, Düx P, Hård K, Devreese B, Nugteren-Roodzant IM, Crielaard W, Boelens R, Kaptein R, van Beeumen J, Hellingwerf KJ. Thiol ester-linked p-coumaric acid as a new photoactive prosthetic group in a protein with rhodopsin-like photochemistry. Biochemistry. 1994;33:13959–13962. doi: 10.1021/bi00251a001. [DOI] [PubMed] [Google Scholar]
- 7.Meyer TE, Yakali E, Cusanovich MA, Tollin G. Properties of a water-soluble, yellow protein isolated from a halophilic phototrophic bacterium that has photochemical activity analogous to sensory rhodopsin. Biochemistry. 1987;26:418–423. doi: 10.1021/bi00376a012. [DOI] [PubMed] [Google Scholar]
- 8.Hoff WD, van Stokkum IH, van Ramesdonk HJ, van Brederode ME, Brouwer AM, Fitch JC, Meyer TE, van Grondelle R, Hellingwerf KJ. Measurement and global analysis of the absorbance changes in the photocycle of the photoactive yellow protein from Ectothiorhodospira halophila. Biophys. J. 1994;67:1691–1705. doi: 10.1016/S0006-3495(94)80643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devanathan S, Pacheco A, Ujj L, Cusanovich M, Tollin G, Lin S, Woodbury N. Femtosecond spectroscopic observations of initial intermediates in the photocycle of the photoactive yellow protein from Ectothiorhodospira halophila. Biophys. J. 1999;77:1017–1023. doi: 10.1016/S0006-3495(99)76952-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellequer J-L, Wager-Smith KA, Kay SA, Getzoff ED. Photoactive yellow protein: a structural prototoype for the three-dimensional fold of the PAS domain superfamiliy. Proc. Natl. Acad. Sci. USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Getzoff ED, Gutwin KN, Genick UK. Anticipatory active-site motions and chromophore distortion prime photoreceptor PYP for light activation. Nat. Struct. Biol. 2003;10:663–668. doi: 10.1038/nsb958. [DOI] [PubMed] [Google Scholar]
- 13.Anderson S, Crosson S, Moffat K. Short hydrogen bonds in photoactive yellow protein. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1008–1016. doi: 10.1107/S090744490400616X. [DOI] [PubMed] [Google Scholar]
- 14.Genick UK, Borgstahl GEO, Ng K, Ren Z, Pradervand C, Burke PM, Srajer V, Teng T-Y, Schildkamp W, McRee DE, Moffat K, Getzoff ED. Structure of a photocycle intermediate by millisecond time-resolved crystallography. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- 15.Genick UK, Soltis SM, Kuhn P, Canestrelli IL, Getzoff ED. Structure at 0.85 Å resolution of an early protein photocycle intermediate. Nature. 1998;392:206–209. doi: 10.1038/32462. [DOI] [PubMed] [Google Scholar]
- 16.Anderson S, Srajer V, Moffat K. Structural heterogeneity of cryotrapped intermediates in the bacterial blue light photoreceptor, photoactive yellow protein. Photochem. Photobiol. 2004;80:7–14. doi: 10.1562/2004-03-15-RA-115.1. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt M, Pahl R, Srajer V, Anderson S, Ren Z, Ihee H, Rajagopal S, Moffat K. Protein kinetics: structures of intermediates and reaction mechanism from time-resolved x-ray data. Proc. Natl. Acad. Sci. USA. 2004;101:4799–4804. doi: 10.1073/pnas.0305983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihee H, Rajagopal S, Srajer V, Pahl R, Anderson S, Schmidt M, Schotte F, Anfinrud PA, Wulff M, Moffat K. Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds. Proc. Natl. Acad. Sci. USA. 2005;102:7145–7150. doi: 10.1073/pnas.0409035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer TE, Tollin G, Hazzard JH, Cusanovich MA. Photoactive yellow protein from the purple phototrophic bacterium, Ectothiorhodospira halophila. Qunatum yield of photobleaching and effects of temperature, alcohols, glycerol, and sucrose on kinetics of photobleaching and recovery. Biophys. J. 1989;56:559–564. doi: 10.1016/S0006-3495(89)82703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Brederode ME, Hoff WD, van Stokkum IHM, Groot ML, Hellingwerf KJ. Protein folding thermodynamics applied to the photocycle of the photoactive yellow protein. Biophys. J. 1996;71:365–380. doi: 10.1016/S0006-3495(96)79234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoff WD, Xie A, van Stokkum IHM, Tang XJ, Gural J, Kroon AR, Hellingwerf KJ. Global conformational changes upon receptor stimulation in photoactive yellow protein. Biochemistry. 1999;38:1009–1017. doi: 10.1021/bi980504y. [DOI] [PubMed] [Google Scholar]
- 22.Salamon Z, Meyer TE, Tollin G. Photobleaching of the photoactive yellow protein from Ectothiorhodospira halophila promotes binding to lipid bilayers - evidence from surface plasmon resonance spectroscopy. Biophys. J. 1995;68:648–654. doi: 10.1016/S0006-3495(95)80225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinstenn G, Vuister GW, Mulder FAA, Düx PE, Boelens R, Hellingwerf KJ, Kaptein R. Structural and dynamic changes of photoactive yellow protein during its photocyclce in solution. Nat. Struct. Biol. 1998;5:568–570. doi: 10.1038/823. [DOI] [PubMed] [Google Scholar]
- 24.Craven CJ, Derix NM, Hendriks J, Boelens R, Hellingwerf KJ, Kaptein R. Probing the nature of the blue-shifted intermediate of photoactive yellow protein in solution by NMR: hydrogen-deuterium exchange data and pH studies. Biochemistry. 2000;39:14392–14399. doi: 10.1021/bi001628p. [DOI] [PubMed] [Google Scholar]
- 25.Brudler R, Rammelsberg R, Woo TT, Getzoff ED, Gerwert K. Structure of the I1 early intermediate of photoactive yellow protein by FTIR spectroscopy. Nat. Struct. Biol. 2001;8:265–270. doi: 10.1038/85021. [DOI] [PubMed] [Google Scholar]
- 26.Xie A, Kelemen L, Hendriks J, White BJ, Hellingwerf KJ, Hoff WD. Formation of a new buried charge drives a large-amplitude protein quake in photoreceptor activation. Biochemistry. 2001;40:1510–1517. doi: 10.1021/bi002449a. [DOI] [PubMed] [Google Scholar]
- 27.Lee BC, Croonquist PA, Sosnick TR, Hoff WD. PAS domain receptor photoactive yellow protein is converted to a molten globule state upon activation. J. Biol. Chem. 2001;276:20821–20823. doi: 10.1074/jbc.C100106200. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki J, Kumauchi M, Hamada N, Oka T, Tokunaga F. Light-induced unfolding of photoactive yellow protein mutant M100L. Biochemistry. 2002;41:1915–1922. doi: 10.1021/bi011721t. [DOI] [PubMed] [Google Scholar]
- 29.Chen E, Gensch T, Gross AB, Hendriks J, Hellingwerf KJ, Kliger DS. Dynamics of protein and chromophore structural changes in the photocycle of photoactive yellow protein monitored by time-resolved optical rotatory dispersion. Biochemistry. 2003;42:2062–2071. doi: 10.1021/bi020577o. [DOI] [PubMed] [Google Scholar]
- 30.Harigai M, Imamoto Y, Kamikubo H, Yamazaki Y, Kataoka M. Role of an N-terminal loop in the secondary structural change of photoactive yellow protein. Biochemistry. 2003;42:13893–13900. doi: 10.1021/bi034814e. [DOI] [PubMed] [Google Scholar]
- 31.Imamoto Y, Kamikubo H, Harigai M, Shimizu N, Kataoka M. Light-induced global conformational change of photoactive yellow protein in solution. Biochemistry. 2002;41:13595–13601. doi: 10.1021/bi0264768. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu N, Kamikubo H, Mihara K, Imamoto Y, Kataoka M. Effect of organic anions on the photoreaction of photoactive yellow protein. J. Biochem. 2002;132:257–263. doi: 10.1093/oxfordjournals.jbchem.a003219. [DOI] [PubMed] [Google Scholar]
- 33.Borucki B, Devanathan S, Otto H, Cusanovich MA, Tollin G, Heyn MP. Kinetics of proton uptake and dye binding by photoactive yellow protein in wild type and in the E46Q and E46A mutants. Biochemistry. 2002;41:10026–10037. doi: 10.1021/bi0256227. [DOI] [PubMed] [Google Scholar]
- 34.Hendriks J, Gensch T, Hviid L, van der Horst MA, Hellingwerf KJ, van Thor JJ. Transient exposure of hydrophobic surface in the photoactive yellow protein monitored with nile red. Biophys. J. 2002;82:1632–1643. doi: 10.1016/S0006-3495(02)75514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itoh K, Sasai M. Dynamical transition and proteinquake in photoactive yellow protein. Proc. Natl. Acad. Sci. USA. 2004;101:14736–14741. doi: 10.1073/pnas.0402978101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vreede J, Crielaard W, Hellingwerf KJ, Bolhuis PG. Predicting the signaling state of photoactive yellow protein. Biophys. J. 2005;88:3525–3535. doi: 10.1529/biophysj.104.055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernard C, Houben K, Derix NM, Marks D, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NAJ. The solution structure of a transient photoreceptor intermediate: Δ25 photoactive yellow protein. Structure. 2005;13:953–962. doi: 10.1016/j.str.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Anderson S, Srajer V, Pahl R, Rajagopal S, Schotte F, Anfinrud P, Wulff M, Moffat K. Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein. Structure. 2004;12:1039–1045. doi: 10.1016/j.str.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Rajagopal S, Anderson S, Srajer V, Schmidt M, Pahl R, Moffat K. A structural pathway for signaling in the E46Q mutant of photoactive yellow protein. Structure. 2005;13:55–63. doi: 10.1016/j.str.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Derix NM, Wechselberger RW, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NAJ. Lack of negative charge in the E46Q mutant of photoactive yellow protein prevents partial unfolding of the blue-shifted intermediate. Biochemistry. 2003;42:14501–14506. doi: 10.1021/bi034877x. [DOI] [PubMed] [Google Scholar]
- 41.Woods VL, Jr, Hamuro Y. High resolution, high-throughput amide deuterium exchange- mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J. Cell. Biochem. Suppl. 2001;37:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- 42.Hamuro Y, Burns LL, Canaves JM, Hoffman RC, Taylor SS, Woods VL., Jr Domain organization of D-AKAP2 revealed by deuterium exchange-mass spectrometry (DXMS) J. Mol. Biol. 2002;321:703–714. doi: 10.1016/s0022-2836(02)00419-9. [DOI] [PubMed] [Google Scholar]
- 43.Hamuro Y, Wong L, Shaffer J, Kim JS, Stranz DD, Jennings PA, Woods VL, Jr, Adams JA. Phosphorylation-driven motions in the COOH-terminal Src kinase, CSK, revealed through enhanced hydrogen-deuterium exchange and mass spectrometry (DXMS) J. Mol. Biol. 2002;323:871–881. doi: 10.1016/s0022-2836(02)01003-3. [DOI] [PubMed] [Google Scholar]
- 44.Englander JJ, Del Mar C, Li W, Englander SW, Kim JS, Stranz DD, Hamuro Y, Woods VL., Jr Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2003;100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamuro Y, Zawadzki KM, Kim JS, Stranz DD, Taylor SS, Woods VL., Jr Dynamics of cAPK type II beta activation revealed by enhanced amide H/2H exchange mass spectrometry (DXMS) J. Mol. Biol. 2003;327:1065–1076. doi: 10.1016/s0022-2836(03)00234-1. [DOI] [PubMed] [Google Scholar]
- 46.Zawadzki KM, Hamuro Y, Kim JS, Garrod S, Stranz DD, Taylor SS, Woods VL., Jr Dissecting interdomain communication within cAPK regulatory subunit type IIbeta using enhanced amide hydrogen/deuterium exchange mass spectrometry (DXMS) Protein. Sci. 2003;12:1980–1990. doi: 10.1110/ps.03166903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 48.Hamuro Y, Anand GS, Kim JS, Juliano C, Stranz DD, Taylor SS, Woods VL., Jr Mapping intersubunit interactions of the regulatory subunit (RIalpha) in the type I holoenzyme of protein kinase A by amide hydrogen/deuterium exchange mass spectrometry (DXMS) J. Mol. Biol. 2004;340:1185–1196. doi: 10.1016/j.jmb.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 49.Pantazatos D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL., Jr Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc. Natl. Acad. Sci. USA. 2004;101:751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong L, Lieser S, Chie-Leon B, Miyashita O, Aubol B, Shaffer J, Onuchic JN, Jennings PA, Woods VL, Jr, Adams JA. Dynamic coupling between the SH2 domain and active site of the COOH terminal Src kinase, Csk. J. Mol. Biol. 2004;341:93–106. doi: 10.1016/j.jmb.2004.05.060. [DOI] [PubMed] [Google Scholar]
- 51.Burns-Hamuro LL, Hamuro Y, Kim JS, Sigala P, Fayos R, Stranz DD, Jennings PA, Taylor SS, Woods VL., Jr Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 2005;14:2982–2992. doi: 10.1110/ps.051687305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL., Jr Structure and properties of {alpha}-synuclein and other amyloids determined at the amino acid level. Proc. Natl. Acad. Sci. USA. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia RA, Pantazatos DP, Gessner CR, Go KV, Woods VL, Jr, Villarreal FJ. Molecular interactions between matrilysin and the matrix metalloproteinase inhibitor doxycycline investigated by deuterium exchange mass spectrometry. Mol. Pharmacol. 2005;67:1128–1136. doi: 10.1124/mol.104.006346. [DOI] [PubMed] [Google Scholar]
- 54.Wong L, Lieser SA, Miyashita O, Miller M, Tasken K, Onuchic JN, Adams JA, Woods VL, Jr, Jennings PA. Coupled motions in the SH2 and kinase domains of Csk control Src phosphorylation. J. Mol. Biol. 2005;351:131–143. doi: 10.1016/j.jmb.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Garrod SM, Deal MS, Anand GS, Woods VL, Jr, Taylor S. Allosteric network of cAMP-dependent protein kinase revealed by mutation of Tyr204 in the P+1 loop. J. Mol. Biol. 2005;346:191–201. doi: 10.1016/j.jmb.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 56.Ohishi S, Shimizu N, Mihara K, Imamoto Y, Kataoka M. Light induces destabilization of photoactive yellow protein. Biochemistry. 2001;40:2854–2859. doi: 10.1021/bi001846i. [DOI] [PubMed] [Google Scholar]
- 57.Pellequer J-L, Brudler R, Getzoff ED. Biological sensors: More than one way to sense oxygen. Curr. Biol. 1999;9:R416–R418. doi: 10.1016/s0960-9822(99)80257-7. [DOI] [PubMed] [Google Scholar]
- 58.Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 59.Gensch T, Hendriks J, Hellingwerf KJ. Tryptophan fluorescence monitors structural changes accompanying signalling state formation in the photocycle of photoactive yellow protein. Photochem. Photobiol. Sci. 2004;3:531–536. doi: 10.1039/b401600a. [DOI] [PubMed] [Google Scholar]
- 60.Otto H, Hoersch D, Meyer TE, Cusanovich MA, Heyn MP. Time-resolved single tryptophan fluorescence in photoactive yellow protein monitors changes in the chromophore structure during the photocycle via energy transfer. Biochemistry. 2005;44:16804–16816. doi: 10.1021/bi051448l. [DOI] [PubMed] [Google Scholar]
- 61.Harigai M, Yasuda S, Imamoto Y, Yoshihara F, Tokunaga F, Kataoka M. Amino acids in the N-terminal region regulate the photocycle of photoactive yellow protein. J. Biochem. 2001;130:51–56. doi: 10.1093/oxfordjournals.jbchem.a002961. [DOI] [PubMed] [Google Scholar]
- 62.van der Horst MA, van Stokkum IH, Crielaard W, Hellingwerf KJ. The role of the N-terminal domain of photoactive yellow protein in the transient partial unfolding during signaling state formation. FEBS Lett. 2001;497:26–30. doi: 10.1016/s0014-5793(01)02427-9. [DOI] [PubMed] [Google Scholar]
- 63.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 64.Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 65.Changeux J-P, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 66.Rajagopal S, Moffat K. Crystal structure of a photoactive yellow protein from a sensor histidine kinase: conformational variability and signal transduction. Proc. Natl. Acad. Sci. USA. 2003;100:1649–1654. doi: 10.1073/pnas.0336353100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang ZY, Swem LR, Rushing BG, Devanathan S, Tollin G, Bauer CE. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science. 1999;285:406–409. doi: 10.1126/science.285.5426.406. [DOI] [PubMed] [Google Scholar]
- 68.Yildiz O, Doi M, Yujnovsky I, Cardone L, Bernt A, Hennig S, Schulze S, Urbanke C, Sassone-Corsi P, Wolf E. Crystal structure and interactions of the PAS repeat region of the Drosophila clock protein PERIOD. Mol. Cell. 2005;17:69–82. doi: 10.1016/j.molcel.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 69.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 70.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 71.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix-loop-helix transcription factor hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Genick UK, Devanathan S, Meyer TE, Canestrelli IL, Williams E, Cusanovich MA, Tollin G, Getzoff ED. Active site mutants implicate key residues for control of color and light cycle kinetics of photoactive yellow protein. Biochemistry. 1997;36:8–14. doi: 10.1021/bi9622884. [DOI] [PubMed] [Google Scholar]
- 73.Bai Y, Milne JS, Mayne LC, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins: Struct. Funct. Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]