Abstract
Protein p53 is a transcription factor crucial for cell cycle and genome integrity. It is able to induce both cell arrest when DNA is damaged and the expression of DNA repair machinery. When the damage is irreversible, it triggers apoptosis. Indeed, the protein, which is a homotetramer, is mutated in most human cancers. For instance, the inherited mutation p53-R337H results in destabilization of the tetramer and, consequently, leads to an organism prone to tumor setup. We describe herein a rational designed molecule capable of holding together the four monomers of the mutated p53-R337H protein, recovering the tetramer integrity as in the wild-type structure. Two ligand molecules, based on a conical calix[4]arene with four cationic guanidiniomethyl groups at the wider edge (upper rim) and hydrophobic loops at the narrower edge (lower rim), fit nicely and cooperatively into the hydrophobic clefts between two of the monomers at each side of the protein and keep the tetrameric structure, like molecular templates, by both ion-pair and hydrophobic interactions. We found a good agreement between the structure of the complex and the nature of the interactions involved by a combination of theory (molecular dynamics) and experiments (circular dichroism, differential scanning calorimetry and 1H saturation transfer difference NMR).
Keywords: molecular recognition, multivalent calix[4]arene ligands, oligoprotein stabilization, protein–protein interactions
Protein–protein interactions play key roles in biological processes (1). Inside the cells, proteins rarely act alone, but interact instead with each other, assembled in complexes either homooligomeric (2) or formed by several different components. The design of ligands with the capacity to modulate protein–protein interactions is a current pursuit in drug discovery (3). Most of the effort has concentrated so far on molecules able to disrupt specific protein interactions (4) or to inhibit unspecific aggregation (5). However, little precedents refer to molecules that could induce oligomerization (6) or allow the stabilization (7) or recovery of proteins that have lost their ability to oligomerize. Several diseases are caused by modified or mutated proteins that do not interact in a functional positive way. Amyloid aggregates in Alzheimer's disease are a clear example of the negative consequences resulting from “inappropriate” protein–protein interactions (8). On the contrary, it has been shown that mutations compromising the ability to form the tetrameric functional structure in the tumor suppressor protein p53 may end in cancer development (9). For instance, the most frequent inherited mutation, p53-R337H, is associated to adrenocortical carcinoma in children of southern Brazil, as well as to a wide spectrum of other cancer forms (10). Mutated p53-R337H cannot efficiently assemble the active tetramer and, consequently, leads to an organism prone to tumor setup (11).
The system constituted by the p53 tetramerization domain (p53TD) and its mutants with defective association properties represents a challenging model for the design, synthesis, and evaluation of molecules that could stabilize and recover the tetrameric structure.
Protein p53, often called the “genome guardian,” is a key transcription factor that induces cell arrest when DNA is damaged and triggers the expression of DNA repair machinery or apoptosis when the damage is irreversible (12). The protein folds into several domains, the most relevant ones being the DNA-binding domain (p53DBD), at the core of the protein, and the tetramerization domain (p53TD), close to the C terminus (13). Because tumor-inducing mutations are mainly located at the p53DBD, this domain has gathered a major interest in anticancer research. However, the activity of the protein strongly depends on its tetrameric integrity (14). Hence, molecules able to stabilize the tetrameric structure of mutated proteins with compromised tetramerization abilities could be valuable therapeutical tools.
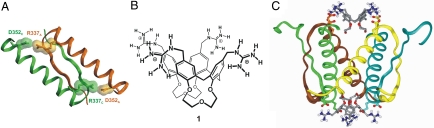
The structure of p53TD was elucidated both by x-ray crystallography (15) and NMR (16). Two identical chains of 31 residues folded into single α-helix–β-hairpin secondary structures associate to form intertwined dimers, which in turn stack into tetramers. Hydrogen bonding, hydrophobic interactions, and salt bridges stabilize the dimer-to-dimer association. One of these interactions, namely the ion-paired bridge Arg-337–Asp-352 between two monomers is lost in mutant p53-R337H (Fig. 1A), and the tetramer is destabilized, because the histidine is not fully protonated at physiological conditions (17, 18).
Fig. 1.
Protein p53TD and docking of ligand 1. (A) Primary p53 dimer showing the two major R337–D352 interactions that stabilize the tetramer by establishing strong hydrogen-bonded ion pairs between different chains. (B) Chemical formula of the tetraguanidiniomethylcalix[4]arene ligand 1. (C) Tetramerization domain with two molecules of ligand 1 showing the interactions of the ligand guanidinium groups and glutamates E336 and E339, as well as the fitting of the lower rim of the ligand into the hydrophobic pocket of the protein.
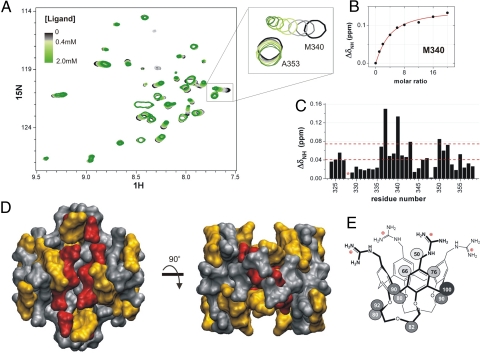
The surface of the α-helices contains a number of anionic residues at regular i,i + 3 or i,i + 4 positions, namely, glutamates E336, E339, E343, E346, and E349 and aspartate D352. An earlier report from our laboratories demonstrated that linear synthetic ligands carrying four positively charged guanidinium moieties and designed to wrap around the monomers are able to bind to these anionic patches at each strand (19). Taking advantage of the spatial arrangement of E336 and E339 residues in strands of two different monomers, whose free carboxylate side chains form an almost perfect square just above the hydrophobic pocket located within the strands, we reasoned that a hydrophobic, conically shaped molecule endowed with four cationic residues at suitable distances to the above-mentioned anionic residues would efficiently complement the shape of the protein tetramer, thus stabilizing the whole ensemble. This is the case of 1, featuring (Fig. 1 B and C): (i) a conical calix[4]arene platform with four guanidiniomethyl residues at the upper rim to interact with glutamates E336 and E339 via ion-pairing and hydrogen bonding, and (ii) a hydrophobic surface (the calixarene and the lower-rim loops) to fit into the hydrophobic pocket. Moreover, each guanidine motif is linked to the calixarene platform by means of a methylene spacer, to enable the correct alignment of the group for an optimal chelation with a minimal distortion of the side chains of the protein. The simultaneous binding of multiple functional groups of 1 to two strands of p53-TD nicely illustrates the concept of multivalency, widely studied in carbohydrate interactions (20–22) that results in strong binding from the sum of multiple interactions of moderate strength (23). Moreover, the loops bridging vicinal phenol rings keep the calixarene into an almost perfect conical shape and prevent it from collapsing into pinched or winged conformations (24). Also, the guanidinium moieties hang on top of the platform, easily orienting their hydrogen donors to the carboxylate groups of the protein at their native conformations (preorganization).
Results and Discussion
Molecular Dynamics.
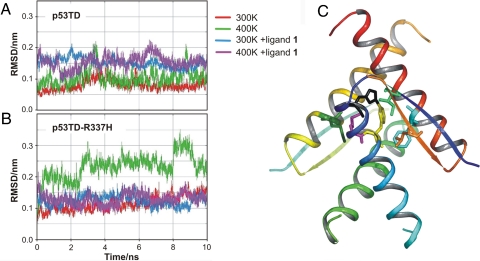
To validate the above-described rational, although intuitive design, protein and protein-ligand stabilities were assessed by analyzing the trajectories obtained in 10-ns molecular dynamics simulations in a box of water at 300 and 400 K, aimed at simulating thermal denaturing conditions. Additional details are provided in Methods. Although a recent 15-ns trajectory study reports on the changes caused by the R337H mutation in the hydrogen bonding network at different pH values (17), we wished to have a deeper insight into the contribution of both the hydrogen bonds and the hydrophobic contacts along the trajectory. As a measure of the structural changes of the protein backbone over time, hence of structural stability, we studied the root-mean-square deviation (rmsd) of the backbone referred to the x-ray crystal structure. In the absence of ligand (Fig. 2A), the structure of wild-type p53TD was highly stable at either 300 or 400 K over the whole dynamics. However, for mutated p53TD-R337H (Fig. 2B) striking differences were observed. At 300 K, the rmsd grows gradually along the trajectory, never recovering its initial value. At thermal denaturing conditions (400 K), the differences between wild and mutated proteins were evident from the very early steps of the trajectory. Ligand 1, docked on the protein, remained stable at the same site it was initially positioned, interacting with the protein through hydrogen bonds and hydrophobic interactions. Most remarkably, whereas ligand binding did not significantly influence the rmsd of wild-type p53TD, it strongly stabilized mutant p53-R337H, as the decrease in the rmsd value and the attenuation of the fluctuations clearly show (Fig. 2B). This fully agrees with the design principles stated above.
Fig. 2.
Molecular dynamics. (A) Wild-type p53TD rmsd of the backbone atoms relative to the starting crystal architecture at 300 and 400 K, in the absence and the presence of the calixarene ligand 1. (B) Mutant protein p53-R337H rmsd of the backbone atoms at 300 K (blue trace) and 400 K (magenta trace). (C) Hydrophobic patch. Only residues in chain A and other chains showing contacts with it are highlighted: Leu-330D (orange), Ile-332D (bright green), His-337A (black), Met-340A (magenta), Met-340C (dark green), Phe-341A (yellow), and Phe-341D (cyan). In wild-type p53, most hydrophobic contacts remained stable at 400 K within 2- to 3Å or 2- to 4Å van der Waals distances, whereas some interactions were notably weakened in mutant p53-R337H: His-337A-Phe-341A, Ile-332A-Phe-341A, Met-340A-Met-340C, and Ile-332A-His-337A, only Leu-330D-Phe-341A turning shorter. All distances remained at their wild-type values in the presence of ligand 1.
Analysis of the dimer-to-dimer interactions revealed that hydrogen bond distances for the Arg-337–Glu-349 ion pair were quite constant and short in wild-type p53TD, whereas the corresponding His-337–Glu-349 pair was never hydrogen bonded along the trajectory. Other analyzed distances, such as Arg-333–Asp-352, or Arg/His-337–Asp-345, fully supported a weaker dimer-to-dimer hydrogen bonding network in the mutated protein. However, relevant hydrophobic contacts that are essential for tetramer integrity (Fig. 2C) were strongly affected by the mutation. For instance (naming as A and C the two monomers forming one primary dimer, and B and D those forming the other one), His-337A–Phe-341A, Ile-332A–Phe-341A, Met-340A–Met-340C, and Ile-332A–His-337A, were notably weakened, whereas Leu-330D–Phe-341A turned shorter. However, all distances remained at their wild-type values in the presence of ligand 1. Therefore, the replacement of Arg-337 by His in the mutated protein caused not only missing three hydrogen bonding contacts (with Glu-349, Asp-352, and Asn-345), but also deeply disturbed the hydrophobic surface responsible of the stability of the structure. His-337 of chain A is not ion-paired anymore to chain D (as His-337 of chain C is paired to chain B), but orients instead away from the protein core. Consequently, the hydrophobic interaction with Ile-332A is lost (distance increases to 7–8 Å). The structural disruption further spreads to keep apart other residues, whereas some others, closer to the protein nucleus, become shorter [see details in supporting information (SI) Figs. S1 and S2].
Addition of ligand 1 to the mutated protein does not produce significant shortening in the hydrogen bonding distances, but the structure keeps fairly fixed along the whole trajectory, as in wild-type protein, because of hydrophobic contacts with the bridges at the lower rim of the calixarene, which prevent, from the very beginning, the cascade of events leading to disassembly.
Synthesis.
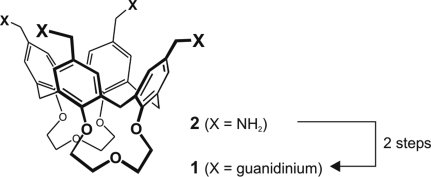
We readily synthesized ligand 1 by guanidilation of the O-protected, doubly bridged tetraaminomethylcalix[4]arene 2 (25) with N,N′-bis-(tert-butoxycarbonyl)guanidine N″-triflate (26) (Fig. 3). The resulting bis-BOC-protected derivative was transformed into 1 by a standard acidic treatment. For full details on the synthesis see SI Text and Fig. S3.
Fig. 3.
Synthetic scheme. Preparation of 1 from tetraamine 2 (see text and SI for details).
Stability Effects.
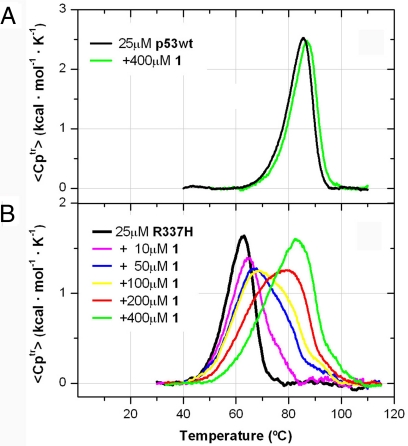
Experimentally, the thermal effects caused by ligand 1 on the stability of both wild-type and mutated proteins were evaluated by circular dichroism (see Fig. S4) and differential scanning calorimetry (DSC). The presence of 400 μM of calixarene 1 hardly shifted a couple of degrees the DSC unfolding curve of the highly stable wild-type p53TD (27) (from 85.5°C to 86.9°C, Fig. 4A), which indicated that, at the melting temperature, the calixarene ligand displayed rather low affinity for the stable protein (28, 29). Conversely, ligand 1 induced an outstanding thermal stabilization in mutant p53TD-R337H (Fig. 4B), which beyond a shift toward higher melting temperatures (from 62°C to 82°C at 400 μM 1) resulted also in a substantial increase in the unfolding enthalpy (area of the heat absorption peak). Thermal changes detected on the mutant protein not only indicated a tight protein–ligand interaction (27–30) but also suggested that the tetramer was the actual stabilized structure. In full agreement with the molecular dynamic simulations described above, DSC experiments suggested that the tight binding of calixarene ligand 1 did stabilize the tetramerization of the mutated protein.
Fig. 4.
Thermal stabilization induced by ligand 1. The presence of ligand 1 shifted the unfolding endotherms for p53TD (A) and p53TD-R337H (B) toward higher temperatures.
Additional evidence for robustness of the final assembly came from electrospray mass spectrometry (ESI-MS) measurements, in which the tetrameric protein, noncovalently bound to two synthetic ligands, was detected (Fig. S5).
Thermal stabilization induced by ligand 1 was not unique for mutant p53TD-R337H. Indeed, p53TD-G334V, another natural mutant associated to cancer (31, 32) of weak tetrameric integrity (33), was also structurally and thermally stabilized by the calixarene (Fig. S4 and Fig. S6).
Structural Characterization.
Structural aspects were examined by NMR, on both the ligand and the protein. These experiments not only determined that p53TD and ligand 1 interacted specifically, but also mapped the interacting molecules.
1H Saturation Transfer Difference (1H-STD) experiments (34) were used to resolve the ligand binding mode (see details in Methods). Normalized values for the different protons are directly labeled on formula 1 (Fig. 5E and Fig. S7), 100% meaning the highest saturation, that is, the proton closest to the protein. STD results completely agreed with the design rationale for ligand 1 (Fig. 1C), with the hydrophobic lower rim of the calixarene “inserted” like a cork into the protein cleft (hence, experiencing higher saturation), whereas the upper rim remained outside (lower saturation). Different STD effect for protons occupying similar positions (i.e., the two types of aromatic protons, for instance) suggested a favored orientation of the ligand when interacting with the protein.
Fig. 5.
Protein p53TD mapping. (A) 1H-15N-HSQC overlaid spectra for the titration of p53TD with ligand 1. Only the central region is shown. Color code: in black, the free protein (125 μM tetramer); in the gray scale, up to 500 μM ligand; in the green scale, from 500 μM to 2.0 mM ligand. (B) Perturbation of Met-340 resonance adjusted to a simplified 1:1 binding model (red line) (see Methods for details). (C) Chemical shift mapping at the end of the titration (*, not available data). Dashed red lines drawn across indicate the cutoff limits used to categorize the perturbation degrees for mapping the 3D protein structure represented in D: in red, residues shifted >0.075 ppm (the mean shift plus one standard deviation); in orange, those shifted >0.04 ppm (the mean shift). (E) STD epitope mapping of ligand 1. Saturation normalized percentages are displayed for each proton of the molecule; 100% corresponds to the 1H with the highest saturation degree, thus, the one closer to the protein.
Regarding the protein, a thorough titration of 15N-p53TD with ligand 1, followed by 1H-15N-heteronuclear sequential quantum correlation (HSQC) proved which parts of the protein structure were directly involved in the interaction. Only some resonances experienced an easily observable perturbation on addition of the ligand, whereas others remained basically unaltered (Fig. 5A), as should correspond to a specific molecular recognition event. Moderate changes in the HSQC spectra suggested no significant alteration of the protein structure. Representation of the most strongly shifted residues at the end of the titration (Fig. 5C) led to the mapping of the protein binding site (Fig. 5D), which was in perfect agreement with the design model, because all of the solvent exposed residues constituting the protein cleft were sensitive to the presence of ligand 1.
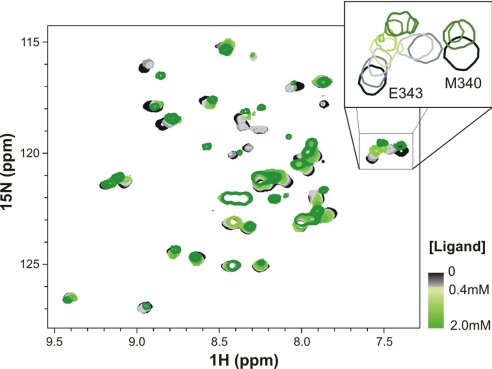
When a similar titration was carried out for mutant p53TD-R337H (Fig. 6 and Fig. S8), a totally different evolution for the HSQC spectra occurred, indicating that interaction of ligand 1 with p53TD-R337H took place through a different and more complex mechanism (see later). Likely, the histidine-mutated residue, as a part of the hydrophobic pocket, might cause significant distortions affecting the binding of the ligand with respect to the wild-type protein mode.
Fig. 6.
1H-15N-HSQC overlaid spectra for the titration of p53TD-R337H with ligand 1. Two molecules of ligand sequentially bind to the protein, as the two clear stages in which resonances evolve during the titration suggest (SI Text). Initially resonances decreased in intensity or totally disappeared; newly emerged resonances in the second stage could not be readily reidentified, so we were not able to map the protein at the end of the titration. For color code, see Fig. 5.
Insights into the Macromolecular Interaction Mechanism.
NMR data not only provided structural information about the ligand–protein system, but also further understanding about the binding mechanism and its thermodynamics by analyzing changes in NMR line shape, shift, and intensity on titration of the protein with the ligand (35).
The interaction between the wild-type p53TD and the calixarene underwent a fast chemical exchange (in the NMR timescale), as proved by the progressive shifts of the protein resonances when the ligand was added (Fig. 5A). Moreover, the unidirectional shift in the 2D spectrum movement was only feasible if both sites on the protein were equivalent and independent. The adjustment of the clear chemical shifts for Met-340 resonance (Fig. 5B) to a 1:2 protein–ligand binding model (Fig. 1C) resulted in an apparent dissociation constant of ≈280 ± 40 μM.
The interaction of p53TD with ligand 1 was entropy-driven (isothermal titration calorimetry, unpublished results), which correlated well with both hydrophobic interactions and charge–charge matching (36).
The contribution of the guanidinium groups to the binding was assessed by comparison with the ammonium derivative 2 (as the HCl salt, Fig. S9). Although 2 also bound to the protein, it displayed considerably lower affinity (37). These results underscored the importance of both hydrophobic and electrostatic contributions, as well as hydrogen bonding in the protein recognition event.
For mutant p53TD-R337H the interaction with the calixarene 1 took place by a more complex mechanism, as suggested the unconventional HSQC evolution (Fig. 6 and S8). At the lowest ligand ratios, most of the resonances became broad and decreased in intensity up to complete disappearance. Remarkably, residues within the α-helix also experienced a shift during this first stage, whereas those in the β-strand remained nearly at their positions (Fig. S8). Further addition of ligand made the signals that had disappeared to emerge back, although not always near the former position. Wherever the new resonances appeared, some of them shifted again, but in a different direction (or even backward).
These complex behaviors could be interpreted considering two sequential events, each of them probably corresponding to the binding of a molecule of calixarene ligand to the tetramer. The fact that any clear intermediate was not detected suggested that the second binding should be of larger affinity than the first one, that is, the two molecules of calixarene bind sequentially in a cooperative manner (38). Indeed, assuming this mechanism, the dissociation constants for both binding events were estimated to be KD1 ≈130 μM and KD2 ≈65 μM (Fig. S8), clearly lower than that for the wild-type p53TD. These values would also agree with the behavior deduced from the DSC endotherms.
The sequential binding, the different behavior of several resonances, the slower kinetics, and the slight perturbation of residues out of the binding pocket further suggested that the protein underwent some—likely minor—structural rearrangements on ligand binding.
Moreover, for the control amino-calixarene ligand 2 only changes on p53TD-R337H corresponding to the first stage could be detected although to a lesser extent (Fig. S9). This would prove that the absence of the guanidinium groups decreased the affinity so much that under the working conditions just the first binding event was probably detected. Should this be the case, it would clearly emphasize the essential role of the guanidinium groups in the interaction process.
The lesser packed tetrameric structure of p53TD-R337H and the direct alteration of the binding site by the mutation H337 were probably the reasons for the different binding mechanism. Furthermore, protein conformational plasticity would enable to adopt a better optimized binding site and hence to increase affinity by establishing tighter interactions with the ligand.
In conclusion, the experimental results here presented fully validate the rational design. Calorimetric experiments have clearly shown how the presence of ligand 1 can strongly enhance the thermal stability of p53TD-R337H mutant. In addition, the noncovalent complex of the tetrameric protein and two ligands was unequivocally confirmed by mass spectrometry, and characterization by NMR has assessed the structural binding model on both the protein and the ligand. Further information provided by NMR data helped us to better understand the mechanisms of the protein–ligand recognition events. Although for the stable and well packed wild-type p53TD, the two binding sites are likely equivalent and independent, for the mutated protein, which forms a less stable tetramer, the binding of the two ligand molecules appears to be sequential and it can even imply some structural rearrangements, possible only because of the plasticity of the mutant protein. In this case, binding of the second ligand appears to be slightly cooperative.
Thus, both the experimental evidence and molecular dynamics simulations reveal how hydrophobic interactions combine with ion-pairing and hydrogen bonding to contribute significantly to the protein–ligand recognition event. Our results encourage strategies in drug discovery, based on the use of small molecules acting as templates for the structural recovery of the supramolecular integrity of damaged protein assemblies.
Methods
Protein Expression and Purification.
Recombinant p53TD (residues 311–367) was expressed in E. coli BL21(DE3) cultures by using autoinducing media (39) and purified as described by Mateu et al. (40) The clone for p53TD-R337H was obtained by site-directed mutagenesis (Stratagene) of p53TD and expressed and purified as the wild-type protein. For the uniformly 15N-labeled proteins, expression was performed in BL21(DE3)pLys competent cells in defined autoindicing media containing 15N-ammonium chloride (41).
Differential Scanning Calorimetry.
DSC thermograms were obtained in a VP-DSC microcalorimeter (Microcal), scanning from 10°C to 120°C, at a rate of 30°C·h−1 and a constant pressure of 2 atm. Samples containing 25 μM (tetramer) protein and 10, 50, 100, 200, and 400 μM ligand 1 were prepared in water (thoroughly adjusting pH at 7.0) and degassed under vacuum at 18°C for 30 min before filling the calorimeter cell; the reference cell was filled with the corresponding degassed sample containing only ligand 1. The sample cell was systematically washed between samples and water blanks were recorded. Experimental data were processed and analyzed with Microcal Origin 7.0 software. After subtraction of the buffer baseline to the raw thermogram, the excess heat capacity function for the unfolding transition, 〈CPtr〉, was obtained by subtracting a progressive baseline traced between the native and the unfolded states.
1H Saturation Transfer Difference NMR.
1H-STD experiments were recorded at 288 K (42) on a Bruker Digital Advance 600 MHz spectrometer equipped with a triple resonance probe and Z-pulsed field gradient. A sample containing 1 mM 1 and 12.5 μM (tetramer) p53TD was lyophilized several times and solved in “100%” D2O to minimize the residual HDO resonance, which could not be suppressed because most of ligand 1 resonances would also be undesirably suppressed. On-resonance irradiation was set to 0.72 ppm and the off-resonance to ≈30 ppm. Selective presaturation of the protein was achieved by a train of Gausian-shaped pulses of a 50-ms length each. A spin-lock pulse of 20 ms was used to eliminate background protein resonances. 1H spectra with varying saturation times were all recorded with the same sample and the same parameters, only changing the saturation time from 0.2 s to 3 s (in random order). Build-up curves were obtained by plotting the STD amplificator factor (34) against the saturation time; experimental data were fitted to the monoexponential equation STD = STDmax [1 − exp(−ksat × tsat)] (43). The slope of the curve at time 0 was taken as the measure of the proximity to the protein. Values for the different protons were normalized by assigning 100% to the closest one.
15N-1H-HSQC NMR Spectroscopy.
Samples containing 125 μM (tetramer) 15N-labeled protein in water (10% D2O) at pH 7.04 were used for the HSQC titration. To avoid dilution, ligands 1 and 2 were added lyophilized. A total of nine ligand-to-protein ratios where recorded: 0, 1, 2, 4, 6, 8, 12, 16, and 24. Titrations were repeated with a different batch of protein and ligand to ensure reproducibility. Sensitivity enhancement 1H-15N-HSQC spectra were recorded at 298 K on a Bruker Digital Advance 600 MHz spectrometer equipped with cryoprobe. All spectra were processed with the package NMRPipe/NMRView (44) and visualized and analyzed with the NMRViewJ software (One Moon Scientific, Inc.).
Dissociation Constant Estimation.
Perturbation of the protein resonances were experimentally measured as . Chemical shift perturbations were expressed as Δδ = fB × ΔδB, where fB is the fraction of bound protein, and ΔδB is the final perturbation. For the interaction between the wild-type p53TD and calixarenes 1 and 2 (in which the two binding sites of p53TD were equivalent and independent: P4 + 2L ⇆ P4L2), the system was simplified considering that only one ligand interacted with one dimer of protein (“P2 + L ⇆ P2L”), and therefore
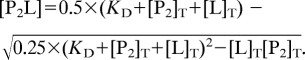 |
Dissociation constants were determined by nonlinear least-squares employing Origin 7.0 software. The simplification was not conceptually wrong, but the virtual concentration of dimer is twice that of tetramer and the resulting dissociation constant would overestimate the real one, although it provided a sufficiently accurate idea of the affinity.
Dissociation Constant Estimation for p53TD-R337H and Ligand 1.
The concentrations for the free, the single-bound, and the double-bound protein were estimated according to the intensity and the shift of the former resonances and the intensity of the newly emerged ones. Those values were mathematically adjusted to the corresponding equations KD1 and KD2 for a sequential binding (see Fig. S8).
Molecular Dynamics.
Simulations were carried out with p53TD/p53TD-R337H in the presence and in the absence of ligand 1 at standard conditions (300 K) and at denaturing conditions (400 K). The starting structure of the wild-type p53TD was taken from the x-ray structure [Protein Data Bank (PDB) ID code 1aie (45)], which is formed by four identical chains of 31 residues each. The initial structure for the p53TD-R337H mutant was generated by a single-residue replacement in the wild-type structure, thus arginine in position 337 was replaced by histidine. The protonation status of every amino acid residue was the corresponding situation at a medium of pH 7. The histidines in the mutated protein were deprotonated, simulating mildly basic conditions. The protein–ligand complexes were composed of one protein and two equal calixarenes, each of them docked by hand in the specific way to have their lower rims embedded in the hydrophobic pockets of the protein between the four chains forming the tetramer. At the same time the guanidinium groups in the upper rim had the appropriate length and orientation to interact with glutamate 336 and glutamate 339 of two different chains situated in an “A-C” or an “B-D” position. Thus, the calixarenes remain almost encapsulated and protected from the solvent being surrounded by parts of the protein that have notable affinity for corresponding parts of the ligand. The complexes were constructed with Maestro (Schrodinger Inc.) molecular modeling environment. Each of the protein or protein–ligand structures was embedded in a rectangular box of water molecules. The volume of the box was such that there was a distance of 9 Å from every wall to the nearest part of the complex. Water molecules were represented applying the SPC (46, 47) model. Counterions were added to ensure the overall electric neutrality of the system. Periodic boundary conditions were used. Amber94 (48) force-field parameters were applied to describe the bonding, van der Waals, and electrostatic interactions. All molecular dynamics simulations were carried out by using the GROMACS simulation package (49). The database files of GROMACS were modified to include suitable parameters for the calixarenes. The atomic charges were calculated by means of the restrained electrostatic potential fit (RESP) (50) from a quantum mechanics optimization with Gaussian03 (51) at the HF/6–31G(d) level. The geometry of the hydrated systems was initially optimized by using a steepest-descent algorithm, and then the system was subjected to a three-stage equilibration protocol: 100 ps of constant temperature; 100 ps of constant pressure, allowing volume changes; 100 ps of constant temperature. After this, a 10-ns-long simulation was submitted, collecting data every 2 ps. The same protocol was used to carry out molecular dynamics simulations at 300 and 400 K. Long-range electrostatic interactions were taken into account by the Particle-mesh Ewald (PME) method (52). Van der Waals interactions were truncated at a cutoff distance of 10 Å. Newton equations were integrated by using a time step of 2 fs. The simulations were performed in the NVT ensemble by using Berendsen coupling method (53) to keep temperature constant, with a coupling time of 0.1 ps. The LINCS algorithm (54) was chosen to satisfy the constraints to the equilibrium for all bond lengths. GROMACS analysis tools were used to analyze the trajectories.
Supplementary Material
Acknowledgments.
S.G. thanks B. Claasen for her help on the STD experiments design and T. Tarragó for biological support. Some of the simulations were run at the Barcelona Supercomputing Center–Mare Nostrum. This work was supported by the Spanish Ministry of Science and Education (MEC) Projects BIO2005-00295, CTQ2005-06909-C02-02/BQU, and CTQ2005-08989-C02/BQU, Consolider Ingenio 2010 Grant CSD2006-0003, and the Institute of Chemical Research of Catalonia (ICIQ) Foundation. S.G., V.M., and E.S. received predoctoral fellowships from Generalitat de Catalunya–Agència de Gestió d'Ajuts Universitaris i de Recerca, MEC, and the ICIQ Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805658105/DCSupplemental.
References
- 1.Jeong H, Mason SP, Barabasi AL, Oltvai Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 2.Ali MH, Imperiali B. Protein oligomerization: How and why. Bioorg Med Chem. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 3.Fry DC, Vassilev LT. Targeting protein-protein interactions for cancer therapy. J Mol Med. 2005;83:955–963. doi: 10.1007/s00109-005-0705-x. [DOI] [PubMed] [Google Scholar]
- 4.Yin H, Hamilton AD. Strategies for targeting protein-protein interactions with synthetic agents. Angew Chem Int Ed. 2005;44:4130–4163. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- 5.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 6.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. Influencing receptor-ligand binding mechanisms with multivalent ligand architecture. J Am Chem Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 7.Block P, Weskamp N, Wolf A, Klebe G. Strategies to search and design stabilizers of protein-protein interactions: A feasible study. Proteins. 2007;68:170–186. doi: 10.1002/prot.21296. [DOI] [PubMed] [Google Scholar]
- 8.Ryan DP, Matthews JM. Evolving strategies for enzyme engineering. Curr Opin Struct Biol. 2005;15:441–446. doi: 10.1016/j.sbi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 10.Achatz MI. The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer Lett. 2007;245:96–102. doi: 10.1016/j.canlet.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 11.Figueiredo BC, et al. Penetrance of adrenocortical tumours associated with the germline TP53 R337H mutation. J Med Genet. 2006;43:91–96. doi: 10.1136/jmg.2004.030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 13.Joerger AC, Fersht AR. Structure–function–rescue: The diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 14.Chène P. The role of tetramerization in p53 function. Oncogene. 2001;20:2611–2617. doi: 10.1038/sj.onc.1204373. [DOI] [PubMed] [Google Scholar]
- 15.Jeffrey PD, Gorina S, Pavletich NP. Crystal structure of the tetramerization domain of the p53 tumor suppressor. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 16.Lee W. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 17.Mateu MG, Sánchez Del Pino MM, Fersht AR. Mechanism of folding and assembly of a small tetrameric protein domain from tumor suppressor p53. Nat Struct Biol. 1999;6:191–198. doi: 10.1038/5880. [DOI] [PubMed] [Google Scholar]
- 18.DiGiammarino EL, et al. A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nat Struct Biol. 2002;9:12–16. doi: 10.1038/nsb730. [DOI] [PubMed] [Google Scholar]
- 19.Salvatella X, et al. A tetraguanidinium ligand binds to the surface of the tetramerization domain of protein p53. Angew Chem Int Ed. 2004;43:196–198. doi: 10.1002/anie.200352115. [DOI] [PubMed] [Google Scholar]
- 20.Bovin NV, Gabius HJ. Polymer-immobilized carbohydrate ligands: Versatile chemical tools for biochemistry and medical sciences. Chem Soc Rev. 1995:413–421. [Google Scholar]
- 21.Kanai M, Mortell KH, Kiessling LL. Varying the size of multivalent ligands: the dependence of Concavaline A binding on neoglycopolymer length. J Am Chem Soc. 1997;119:9931–9932. [Google Scholar]
- 22.Dam TK, Brewer CF. Thermodynamic studies of lectin-carbohydrate interactions by isothermal titration calorimetry. Chem Rev. 2002;102:387–429. doi: 10.1021/cr000401x. [DOI] [PubMed] [Google Scholar]
- 23.Mammen M, Choi S-K, Whitesides GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Arduini A, et al. Calix[4]arenes blocked in a rigid cone conformation by selective functionalization at the lower rim. J Org Chem. 1995;60:1454–1457. [Google Scholar]
- 25.Casnati A, Pirondini L, Pelizzi N, Ungaro R. New tetrafunctionalized cone calix[4]arenes as neutral hosts for anions. Supramol Chem. 2000;12:53–65. [Google Scholar]
- 26.Feichtinger K, Heather LS, Baker TJ, Matthews K, Goodman M. Triurethane-protected guanidines and triflyldiurethane-protected guanidines: New reagents for guanidinylation reactions. J Org Chem. 1998;63:8432–8439. [Google Scholar]
- 27.Johnson CR, Morin PE, Arrowsmith CH, Freire E. Thermodynamic analysis of the structural stability of the tetrameric oligomerization domain of p53 tumor suppressor. Biochemistry. 1995;34:5309–5316. doi: 10.1021/bi00016a002. [DOI] [PubMed] [Google Scholar]
- 28.Brandts JF, Lin LN. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry. 1990;29:6927–6940. doi: 10.1021/bi00481a024. [DOI] [PubMed] [Google Scholar]
- 29.Shrake A, Ross PD. Ligand-induced biphasic protein denaturation. J Biol Chem. 1990;265:5055–5059. [PubMed] [Google Scholar]
- 30.Fukada H, Sturtevant JM, Quiocho FA. Thermodynamics of the binding of L-arabinose and of D-galactose to the L-arabinose-binding protein of Escherichia coli. J Biol Chem. 1983;258:13193–13198. [PubMed] [Google Scholar]
- 31.D'Amico D, et al. High frequency of somatically acquired p53 mutations in small-cell lung cancer cell lines and tumors. Oncogene. 1992;7:339–346. [PubMed] [Google Scholar]
- 32.Fujita T, Kiyama M, Tomizawa Y, Kohno T, Yokota J. Comprehensive analysis of p53 gene mutation characteristics in lung carcinoma with special reference to histological subtypes. Int J Oncol. 1999;15:927–934. doi: 10.3892/ijo.15.5.927. [DOI] [PubMed] [Google Scholar]
- 33.Higashimoto Y, et al. Unfolding, aggregation, and amyloid formation by the tetramerization domain from mutant p53 associated with lung cancer. Biochemistry. 2006;45:1608–1619. doi: 10.1021/bi051192j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 35.Nageswara Rao BD. Nuclear magnetic resonance line-shape analysis and determination of exchange rates. Methods Enzymol. 1989;176:279–311. doi: 10.1016/0076-6879(89)76016-x. [DOI] [PubMed] [Google Scholar]
- 36.Velazquez-Campoy A, Leavitt SA, Freire E. Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol Biol. 2004;261:35–54. doi: 10.1385/1-59259-762-9:035. [DOI] [PubMed] [Google Scholar]
- 37.Ohara K, et al. Amine-guanidine switch: A promising approach to improve DNA binding and antiproliferative activities. J Med Chem. 2007;50:6465–6475. doi: 10.1021/jm701207m. [DOI] [PubMed] [Google Scholar]
- 38.Stevens SY, Sanker S, Kent C, Zuiderweg ER. Delineation of the allosteric mechanism of a cytidylyltransferase exhibiting negative cooperativity. Nat Struct Biol. 2001;8:947–952. doi: 10.1038/nsb1101-947. [DOI] [PubMed] [Google Scholar]
- 39.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Mateu MG, Fersht AR. Nine hydrophobic side chains are key determinants of the thermodynamic stability and oligomerization status of tumour suppressor p53 tetramerization domain. EMBO J. 1998;17:2748–2758. doi: 10.1093/emboj/17.10.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyler RC, et al. Auto-induction medium for the production of [U-15N]- and [U-13C, U-15N]-labeled proteins for NMR screening and structure determination. Protein Expr Purif. 2005;40:268–278. doi: 10.1016/j.pep.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 42.Groves P, et al. Temperature dependence of ligand-protein complex formation as reflected by saturation transfer difference NMR experiments. Magn Reson Chem. 2007;45:745–748. doi: 10.1002/mrc.2041. [DOI] [PubMed] [Google Scholar]
- 43.Mayer M, James TL. NMR-based characterization of phenothiazines as a RNA binding scaffold. J Am Chem Soc. 2004;126:4453–4460. doi: 10.1021/ja0398870. [DOI] [PubMed] [Google Scholar]
- 44.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.Mittl PRE, Chène P, Grutter MG. Crystallization and structure solution of p53 (residues 326–356) by molecular replacement using an NMR model as template. Acta Crystallogr D. 1998;54:86–89. doi: 10.1107/s0907444997006550. [DOI] [PubMed] [Google Scholar]
- 46.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular Forces. Dordrecht, The Netherlands: Reidel; 1981. pp. 331–342. [Google Scholar]
- 47.Robinson GW, Zhu SB, Singh S, Evans MW. Water in Biology, Chemistry and Physics; Experimental Overviews and Computational Methodologies. Singapore: World Scientific; 1996. [Google Scholar]
- 48.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1996;118:2309. [Google Scholar]
- 49.Van der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 50.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—The Resp model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 51.Frisch MJ, et al. Gaussian03. Wallingford, CT: Gaussian, Inc.; 2004. Revision C.02. [Google Scholar]
- 52.Darden T, York D, Pederson L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 53.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 54.Hess B, Beker H, Berendsen HJC, Fraaije JGEM. Lincs: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.