Abstract
We report nanofabrication of protein dot and line patterns using a nanofountain atomic force microscopy probe (NFP). Biomolecules are continuously fed in solution through an integrated microfluidic system, and deposited directly onto a substrate. Deposition is controlled by application of an electric potential of appropriate sign and magnitude between the probe reservoir and substrate. Submicron dot and line molecular patterns were generated with resolution that depended on the magnitude of the applied voltage, dwell time, and writing speed. By using an energetic argument and a Kelvin condensation model, the quasi-equilibrium liquid–air interface at the probe tip was determined. The analysis revealed the origin of the need for electric fields in achieving protein transport to the substrate and confirmed experimental observations suggesting that pattern resolution is controlled by tip sharpness and not overall probe aperture. As such, the NFP combines the high-resolution of dip-pen nanolithography with the efficient continuous liquid feeding of micropipettes while allowing scalability to 1- and 2D probe arrays for high throughput.
Keywords: arrays, atomic force microscopy, nanolithography, patterning, nanofabrication
Nanodeposition techniques have many applications in biology and the life sciences. The ability to spatially orient and immobilize biomolecules on a substrate is valuable to the development of genomic and proteomic profiles of cells (1), drug screening, and biosensing (2, 3), which require precise high-density arrays of biological material. By reducing array spot size, less sample volume is needed because statistically, every analyte molecule will sample the entire capture surface at a greater rate (4). To fabricate nanoscale structures, biomolecules conjugated to nanowires can be patterned to direct their assembly (5, 6). Moreover, nanoscale studies of protein and cell functions, which are often mediated by ligand–receptor binding, require a device capable of precisely delivering proteins and observing the response (7). For example, immobilized ligand templates for cell binding can be created by patterning specific adhesive and inert sites. Here, protein clustering in cell focal adhesion occurs at 5- to 200-nm length scales; hence, delivery of material at this scale allows many relevant physiological single cell studies (8).
Functional biomolecule arrays have been realized in a number of ways. Dip-pen nanolithography (DPN) was used to deposit dry proteins with sub-100-nm resolution (9, 10). Here, commercial atomic force microscopy (AFM) probes were chemically modified to achieve sufficient coverage of proteins dried on the probe tip. Proteins then diffused from the tip to the substrate as it transcribed a pattern. The tips required recoating once exhausted of proteins. DPN was also used to pattern chemical templates upon which proteins were later assembled from solution (11, 12). Similarly, substrates were locally charged (13) or ionized (14) by applying an electrical bias to the probe, followed by protein assembly on the modified regions. Electrically biased probes have also been used to locally oxidize the substrate, creating binding sites for proteins diffusing from the probe tip (15). For direct protein deposition from buffer solution, electric fields have generally been applied to pipette-type devices to assist transport by electrophoretic or electro-osmotic flow with somewhat coarser 450-nm to 2-μm resolution (16–18). Electrospray systems using relatively strong electric fields have been applied to produce submicron features either through creation of a well-defined electrohydrodynamic jet from a nanopipette (19, 20) or by spraying through a dielectric mask (21) or onto a surface with localized fields to direct assembly (22).
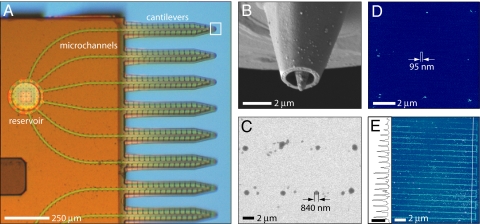
The nanofountain probe (NFP) (23–25) offers a unique combination of patterning resolution, efficiency, and generality in its ability to directly pattern a wide variety of molecular species in solution. Examples are the direct deposition of gold nanoparticles (26) and DNA (27) with resolution up to 100 nm. Direct deposition in solution (e.g., buffer) is especially significant when patterning proteins, whose function can be highly sensitive to their environment. The NFP incorporates 4 fluid reservoirs, each supplying 6 cantilevered delivery probes via enclosed microchannels (Fig. 1A), such that direct parallel molecular patterning of liquid solutions can be performed uninterrupted for long periods. The probe tips consist of an aperture formed from a volcano-like shell surrounding a pyramidal core tip with a radius of ≈200 nm (Fig. 1B). The third-generation NFP chips are batch-fabricated (23–25) on 4-in silicon wafers for use in a commercial AFM.
Fig. 1.
NFP chip and example protein patterns. (A) Optical image of a quarter section of the NFP chip. (B) SEM image of a probe aperture at the end of a cantilever. (C) SEM image of a 2 × 4 array of anti-BSA IgG dots patterned on a BSA substrate (46% RH). (D) Tapping-mode AFM image of a 3 × 3 array of biotin-BSA dots patterned on MHA (50% RH, height scale is 25 nm). (E) Tapping-mode AFM image and height profile of parallel lines of biotin-BSA patterned on MHA at a translation rate of 80 μm/s (50% RH, height scale bar in profile is 20 nm).
In this article, we report deposition of protein molecules [positively charged IgG and negatively charged biotinylated BSA (biotin-BSA)] controlled through application of an electric field between the NFP and substrate. Initial demonstrations of basic protein deposition are presented, followed by a study of the effects of patterning parameters (e.g., applied bias and dwell time). As we show later through modeling, feature size with the NFP is controlled by the sharpness of the core tip rather than the aperture diameter as in pipette devices. Implications of this finding on fluid transport and ultimate resolution are discussed.
Results and Discussion
In all electric field-driven patterning experiments, the substrate was grounded while the relative sign and magnitude of the voltage applied to the NFP reservoir was controlled by using a voltage source. The applied bias was held constant while the NFP transcribed the desired pattern as guided by the AFM in contact mode. Unless otherwise specified, the substrate consisted of a self-assembled monolayer of 16-mercaptohexadecanoic acid (MHA) on gold-coated silicon. Patterning was conducted at 35–60% relative humidity (RH), although it is reasonable to assume that deposition would also be possible beyond this range, as in the case of DNA patterning via NFP (27).
Examples of IgG and biotin-BSA patterns generated with the described field-driven technique are shown in Fig. 1 c–e. Dot arrays and line features of IgG were initially patterned on MHA substrates, because IgG is known to electrostatically bind to carboxyl-terminated thiols such as MHA (11). Although deposition was possible, extraneous aggregates of protein often formed outside the designed feature locations. We hypothesize that high protein mobility on the substrate, due to weak electrostatic interactions and complex electric field contours, allowed some of the biomolecules to migrate away from the main feature. To reduce mobility, stronger antibody–antigen binding in the form of anti-BSA IgG deposition on BSA-coated substrates was tested in place of electrostatic immobilization. Fig. 1C shows an array of anti-BSA IgG dots patterned on a BSA substrate with 8-s dwell times at 5-V bias. These IgG dots were up to 66% smaller than those patterned under similar conditions on MHA, likely because of reduced surface mobility with specific antibody–antigen binding. Rodolfa et al. (18) also observed reduced feature sizes in pipette deposition when exploiting biotin–streptavidin binding in place of electrostatic interactions. Carboxyl-terminated thiols (e.g., MHA) have also proven effective in immobilizing negatively charged BSA (28, 29). Fig. 1D shows an array of biotin-BSA dots patterned on MHA at −1.5-V bias. Fig. 1E shows a set of parallel lines of biotin-BSA patterned on MHA with a translation rate of 80 μm/s at −2-V bias. After these initial demonstrations of protein deposition under electric field, a more detailed study of the underlying transport mechanisms and the effects of patterning parameters (e.g., dwell time and applied bias) was conducted to better understand and optimize the process.
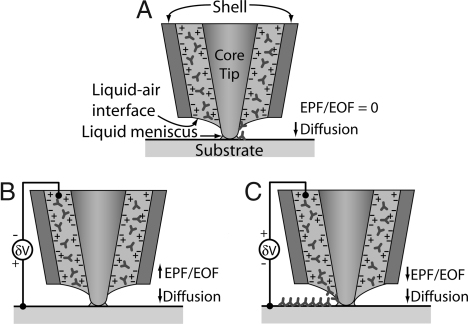
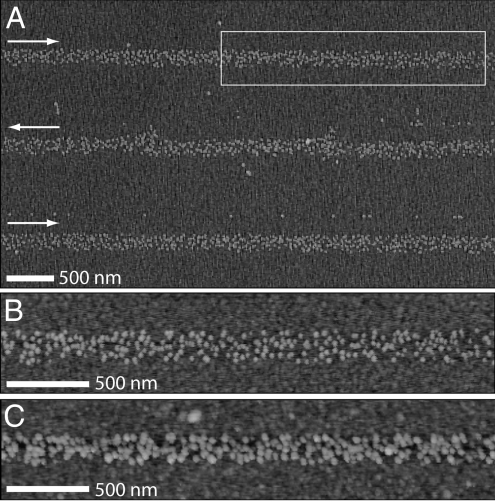
Transport of biomolecules through the liquid column between the probe tip and the substrate can occur because of different mechanisms. Without application of an electric field (Fig. 2A), protein deposition was sporadic and not very repeatable. For example, several lines of IgG were deposited on an MHA substrate at relatively low speeds of 0.8 μm/s. However, these lines were discontinuous and nonuniform in width. Furthermore, discrete dot patterns could not be made with dwell times ranging from 2 to 180 s. When the NFP reservoir was negatively biased relative to the substrate (Fig. 2B), IgG deposition was not observed. By contrast, controlled IgG deposition was possible when the NFP was positively biased (Fig. 2C). These observations suggest that the dominant protein transport mechanism through the NFP is not diffusion but rather electrophoretic (EPF) or electro-osmotic flow (EOF).
Fig. 2.
Schematic of NFP tip and possible protein-transport mechanisms. Proteins are shown as Y-shaped IgG and are assumed to be positively charged. The NFP walls are negatively charged (45). (A) With zero applied field, the molecular transport is through diffusion. (B) NFP is at negative potential relative to substrate; little or no deposition is expected. (C) NFP is at positive potential relative to the substrate; deposition of positively charged molecules is expected. Note the “+” at the NFP walls indicates positive countercharge ions in the double layer. The estimated Debye length (≈3 nm) for our solution is much less than the microchannel dimensions (≈780 nm). Therefore, the NFP will not display ion permselective transport.
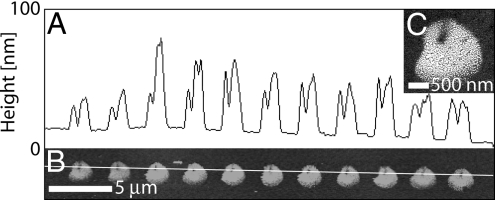
The repeatability for a given set of patterning parameters was tested. Fig. 3 shows a row of dots of biotin-BSA deposited on MHA with constant dwell time at −2.5-V bias. The dots have an average diameter of 1.9 ± 0.1 μm (n = 11). Their morphology is also consistent and reflects that of the NFP tip, with the area where the tip was in contact with the substrate exhibiting a depression. As shown in Fig. 3C, the dot features were composed of small, densely packed aggregates of protein similar to those formed by microcontact printing (3). In general, dot features larger than ≈500 nm tended to have this aggregate composition, whereas smaller features were a solid structure (Fig. 4 A and B).
Fig. 3.
Assessing uniformity of patterned features. (A and B) Height profile (A) and tapping mode (B) AFM image of a row of dots of biotin-BSA deposited with 5-s dwell times (50% RH). (C) SEM image of an individual spot.
Fig. 4.
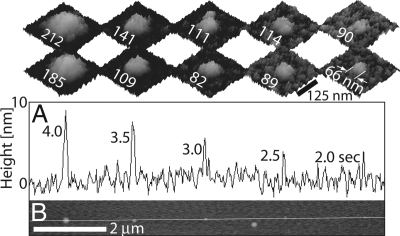
Effects of dwell time. (A) Height profile and (B) tapping mode AFM image of a row of 5 biotin-BSA dots deposited with decreasing dwell times (57% RH). The Insets above A are tapping-mode AFM images showing the diameter of the corresponding dots before (lower row) and after (upper row) immersion in streptavidin and then in biotin-IgG solutions (height scale is 15 nm).
The dependence of biotin-BSA feature size on applied bias and dwell time was investigated with voltages ranging from 0 to −5 V. At voltages from 0 to −1.5 V, no deposition was observed for dwell times as high as 20 s on MHA, suggesting that a minimum threshold voltage is required for deposition. At −1.5 V (near the required threshold), the dot diameter and height did not depend strongly on dwell time. For example, the dots in Fig. 1D were patterned with dwell times (dot diameters) of 3 s (117 ± 19 nm), 4 s (118 ± 5 nm), and 5 s (151 ± 12 nm) in the first, second, and third rows, respectively. The liquid meniscus between the NFP tip and substrate takes time to reach an equilibrium width (30). Thus, the relatively small bias and short dwell times may allow only smaller menisci that are less stable and have larger fluctuations [fluctuations may even be greater than their average width (31)]. Within a −1.5- to −2.5-V range, biotin-BSA deposition was controlled. Fig. 4 shows a row of 5 biotin-BSA spots deposited at −2.0-V bias with decreasing dwell times. The height of the features decreases monotonically with dwell time and approaches that of a monolayer of protein [BSA is 4 × 4 × 14 nm (32)]. The diameter of the dots also decreases with dwell time. Unlike the larger dots shown in Fig. 3, these comprised a single, solid feature (see lower row of Insets in Fig. 4). Beyond −3 V, biotin-BSA deposition became uncontrolled, with solution tending to flood a large area around the tip.
To ensure that the patterned features were composed of protein rather than residual salt from the buffer solution, all substrates were rinsed first in pure buffer then in NanoPure water. Before rinsing, the height of the deposited features varied from 9 to >80 nm, depending on their lateral size. This was due either to protein agglomeration on the substrate or salts accumulating from the buffer solution. After rinsing, the height decreased significantly. For smaller dots, the height was consistent with that of a monolayer of protein (Fig. 4A). Initially, samples were rinsed and imaged multiple times to ensure that the feature size did not continue to decrease with further rinsing. The fact that the features remained intact and stable through repeated rinsing suggests that the patterned features were, in fact, composed of protein rather than salt from the buffer, because the latter would have been dissolved in rinsing.
A major advantage of the NFP is its ability to preserve the liquid state of the solution during deposition. Not only does this greatly simplify deposition of nanoparticle sols, it is significant for biomolecule patterning as they may be deposited in their natural hydrated state. In previous work, the biological activity of NFP-patterned single-stranded DNA was confirmed by hybridization with complementary DNA-functionalized gold nanoparticles (27). In the present study, the activity of the deposited biotin-BSA proteins was investigated by observing an increase in biotin-BSA feature size due to molecular recognition after immersion in streptavidin then biotin-IgG solutions. Biotin-BSA was first patterned on MHA (Fig. 4, lower row of Insets). Then the substrates were immersed in a streptavidin solution to provide streptavidin at the biotin on the surface of the features. Finally, the substrates were immersed in biotin-IgG solution. Interestingly, the diameter of the patterned dots increased an average of 29 ± 4 nm (n = 7) after drying (Fig. 4, upper row of Insets), whereas negligible height increase was observed. This increase in diameter is consistent with a monolayer of streptavidin and biotin-IgG surrounding the features (streptavidin is 5.4 × 5.8 × 4.8 nm (33) and IgG is Y-shaped 14.5 × 8.5 × 4.0 nm (34), giving a predicted diameter increase of 17.6–40.6 nm depending on the orientation of the biotin on the IgG). We hypothesize that the IgG lays flat upon drying of the substrate, which is likely a favorable conformation because of the electrostatic attraction between the positively charged IgG and negatively charged MHA [see supporting information (SI) Text for further discussion and Figs. S1 and S2]. Further tests of preserved biological activity will be the focus of future work in which the electric field-assisted protein-deposition technique is applied to cell-adhesion studies.
Parallel lines were patterned by scanning the NFP at a constant rate (Figs. 1E and 5). For example, biotin-BSA lines of uniform width were rapidly patterned at a rate of 80 μm/s by using an applied bias of −1.5 V or lower at 55% RH. Lines deposited at −1.5-V bias (Fig. 5) were ≈170 nm in width and composed of small, densely packed aggregates similar to those observed in larger dot features (Fig. 3C) and in lines patterned by microcontact printing (3). As above, patterned substrates were immersed in streptavidin then biotin-IgG to investigate their biological activity. Comparing Fig. 5 B and C, there is a clear increase in the diameter of the aggregates upon immersion, which is consistent with that observed for dot features (Fig. 4). Deposition at rates >80 μm/s was not tested, although it is 3 orders of magnitude faster than that demonstrated by using direct-write DPN of IgG [250-nm line widths written at 0.08 μm/s (10)]. We believe that even higher deposition rates and, thus, high-throughput patterning will be possible by further controlling the applied potential, humidity, and temperature.
Fig. 5.
Tapping-mode AFM images of protein lines. (A) Biotin-BSA lines patterned at 80 μm/s. Arrows indicate the translation direction. (B and C) Images of the boxed region in A before (B) and after (C) incubation in streptavidin and biotin-IgG. (All height scales are 25 nm).
To gain insight into the geometry of the liquid meniscus at the NFP tip, a study of the equilibrium liquid–air interface at the tip was conducted. Here, the meniscus shape and width not only determine the lower bound of the NFP resolution but also whether deposition occurs at all. In considering the mechanics of protein deposition, the effects of probe tip geometry, liquid–tip and liquid–substrate contact angles, and RH were assessed. In the absence of external forces, the equilibrium shape of a given volume of liquid can be determined by minimizing the total surface energy, which is given by,
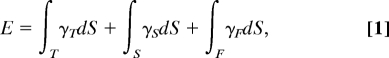 |
where γ denotes the surface tension, and subscripts T, S, and F refer to the liquid–tip, liquid–substrate, and liquid–air interfaces, respectively. Numerical computations to investigate this energy landscape were performed by using Surface Evolver (35) to determine the equilibrium configuration of the liquid surface as shaped by surface tension and other energies.
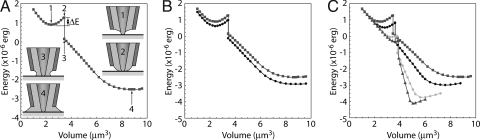
For the NFP tip shown in Fig. 1B, we modeled the flow of liquid by determining the equilibrium shape of the liquid surface for a series of prescribed volumes starting from an arbitrary reference volume. In this way, we obtained a series of snapshots of a quasi-equilibrium flow of liquid to the probe tip because of capillary force. The low-concentration protein solution was modeled as water (36) and the contact angles of the liquid–tip and liquid–substrate taken as variables. Fig. 6A shows the computed total energy E as a function of prescribed volume when both contact angles are 30°. A gap of 2 nm between the bottom of the tip and the substrate was used in the calculations. The volume was computed from an arbitrary reference height and therefore should not be taken as the absolute volume of fluid in the NFP.
Fig. 6.
Modeling the liquid surface energy landscape. (A) Total surface energy of the liquid as a function of increasing volume. Insets show the evolution of the liquid as a function of increasing volume. (B) Surface energies for liquid–tip contact angles of 30° (squares) and 10° (circles), for the probe tip geometry of Fig. 1. (C) Surface energies for core tip protrusion lengths of 0.62 μm (squares), 0.5 μm (circles), 0.3 μm (diamonds), and 0.2 μm (triangles). The liquid–tip contact angle is 30°.
The plot in Fig. 6A indicates two minimum-energy states, with an energy well depth ΔE that must be overcome for the transition to the global minimum. The corresponding sequence of liquid configurations with increasing volume is shown as Insets in Fig. 6A. As the liquid flows along the probe aperture, the liquid–air interface stretches across the aperture from the shell to the protruding tip. The two minimum-energy states labeled 1 and 4 show distinctly different configurations. In state 1 there is no liquid bridge from the tip to the substrate. Further increase in liquid volume causes the liquid to contact the substrate (state 2) and form a continuous column of liquid between the probe and substrate (state 3). In state 4, the global minimum-energy state, the column of liquid has spread to the outer shell. This state is similar to that of pipette-type devices (16–18), and the function of the sharp tip in preserving resolution is lost. The energy well ΔE that separates states 1 and 3 is 3.71 × 10−7 erg, much too large to be overcome by thermal fluctuations at room temperature, which are on the order of kBT = 4.1 × 10−14 erg. The height of the liquid at the tip in state 1 is 0.315 μm above the substrate, and therefore under these conditions, there is no fluid connecting the probe to the substrate.
In actuality, there will always be a small liquid meniscus between the tip and substrate because of capillary condensation if the tip is close enough to the substrate. Capillary condensation, which corresponds to condensation of liquid bridges across small gaps, occurs because of the pressure difference across the interface of a curved surface (31, 37–39). It can be described by the Kelvin equation
where v is the molecular volume, T is the temperature, p is the equilibrium vapor pressure, p0 is the saturated vapor pressure, r1 and r2 are the two principal radii of the curved liquid surface, and rK is the Kelvin radius. Thus, the relative vapor pressure p/p0 plays an important role, and the water meniscus is a direct consequence of the RH.
The largest meniscus due to capillary condensation occurs at 100% RH (p/p0 = 1). The computed liquid menisci that form because of capillary condensation at 100% RH are shown in Fig. 2A. The heights of the menisci from the substrate are 0.20 and 0.22 μm for liquid–tip contact angles of 30° and 10°, respectively. These heights are still too small to reach the bulk liquid in state 1. At 90% RH, the meniscus is dramatically smaller, with a height from the substrate of 2.5 nm and a diameter on the substrate of 26 nm for a liquid–tip contact angle of 30°. These dimensions are comparable with the lattice gas Monte Carlo simulation results of Jang et al. (31), who reported meniscus height and width values of ≈3.2 and 20 nm for an elliptic tip with a tip–substrate distance of 2 nm at 90% RH.
Therefore, at equilibrium, there is still a substantial portion of the tip that is not covered with liquid. This provides a plausible explanation of the need for electric fields and EPF/EOF effects to pattern proteins, where passive diffusion does not seem to take place in the presence of a discontinuity in the liquid at the tip. The experimental results confirm the meniscus stability (resolution) and the effect of electric fields on transport. In fact, the biotin-BSA spots ranged from ≈70 nm to 2 μm in diameter, which falls within the predicted meniscus diameter on the substrate through condensation (≈20 nm) and the NFP aperture (2.2 μm).
The mechanism of protein patterning by NFP therefore seems to be distinctly different from the double molecular layer model proposed by Cho et al. (40) for DPN processes. In DPN, the molecules exist on the AFM tip in the solid phase, and an adsorbed layer of water promotes formation of a thin mobile layer of molecules on top of the solid layer. In NFP patterning, protein solutions are deposited entirely in liquid form. Although an adsorbed water layer will form on the exposed parts of the hydrophilic tip in addition to the condensed meniscus, the proteins are likely too large and immobile to easily diffuse across the thin adsorbed layer of water to reach the liquid meniscus and therefore the substrate. An additional driving force in the form of an electric field is required in order for the proteins to gain sufficient mobility.
We note here an additional possible transport mechanism in NFP patterning where the field may alter the meniscus and thereby reduce or eliminate the gap between liquid bodies on the tip. Electrowetting describes electric field-induced changes in surface energy of the liquid–air and liquid–solid interfaces (41) that alter the contact angles of the liquid and, in turn, result in a different equilibrium position of the meniscus. This effect has been exploited in the past, for example, to alter capillary flow in a channel (42). Inclusion of this effect in the modeling is beyond the scope of this work; however, it will be explored in future studies. In more extreme cases, liquid may be ejected from the probe by an electrospray mechanism as used for protein mass spectrometry (43), patterning (19–22), and some forms of inkjet printing (44). This effect is driven by the accumulation of repulsive charges within a solution, causing an effective outward motion of the fluid from the capillary. Although the electric fields required for this to occur are relatively large, the locally-concentrated fields occurring within the NFP tip may result in a similar process. Future studies will be needed to explore this possibility.
The presence and dimensions of the discontinuity of bulk liquid at the NFP tip can be controlled by varying the tip geometry and buffer chemistry and the ambient temperature and RH. This allows us a measure of control over the stability of equilibrium state 1 (Fig. 6A) and, by extension, the threshold electric potential required for deposition to occur. This is demonstrated in Fig. 6 B and C, where the effects of contact angles and core tip protrusion distances on the total energy are plotted. Smaller liquid–tip contact angles θT result primarily in lower total surface energy (Fig. 6B). The depth of the energy well, ΔE, does not change dramatically. However, the bulk liquid reaches down the tip to a height from the substrate of 0.253 μm for θT = 10°, versus 0.315 μm for θT = 30°. Including the liquid meniscus heights calculated at 100% RH, the width of the gap between the upper liquid body and the meniscus is reduced from 115 to ≈30 nm by the smaller contact angle. This suggests that the use of a better wetting buffer would lower the electric field required for patterning.
The core tip protrusion length affects the energy well depth more significantly, and for a protrusion length of 0.2 μm, ΔE is reduced to 5.29 × 10−8 erg from 3.71 × 10−7 erg at 0.62 μm protrusion. In addition, at a protrusion length of 0.2 μm, the liquid body reaches down the tip to a height of 0.107 μm above the substrate, and therefore, at sufficient RH, the entire tip will be covered by bulk liquid. Thus, for certain tip geometry and buffer chemistry, passive protein patterning becomes possible under high-humidity conditions. The above results explain the relatively large voltages required to pattern IgG compared with biotin-BSA, because NFPs with greater tip protrusion were used in those experiments. For this reason, we do not make comparisons in the magnitudes patterning parameters between different protein solutions. Instead we emphasize the experimental trends within a given protein solution and NFP chip. These results provide a framework for directing further experimental investigation toward a better understanding of the NFP patterning mechanisms.
Conclusions
Rapid direct deposition of IgG and BSA proteins from solution into submicron dot and line arrays was demonstrated by using a nanofountain probe with an applied electric field. The biomolecules were transported in buffer solution through an integrated microfluidic system to an apertured probe tip. Patterning resolution was found to depend on the magnitude of the applied voltage. Resolution also depended on the type of biomolecule–substrate interaction, with antibody–antigen binding exhibiting more effective immobilization than electrostatic interactions. This technique, which combines strengths of high-resolution DPN and continuously fed micropipettes, is readily scaled to 1- and 2D probe arrays for high throughput.
By means of an energetic argument, the quasi-equilibrium liquid–air interface at the probe tip was determined. This, together with a Kelvin condensation model, provided insight into the possible mechanisms for biomolecular transport from the NFP to the substrate. Likewise, the need for electric fields to achieve transport of charged proteins emerged from the analysis. Much modeling work remains to quantify the effect of all of the variables involved in the transport, deposition, and binding kinetics of biomolecules in fountain probe patterning mode. However, the quasi-equilibrium analysis provides a first step in rationalizing the experimental observations and assessing the effect of thermal fluctuations on meniscus stability and consequently pattern resolution.
Materials and Methods
Materials.
MHA (90%), polyclonal anti-BSA IgG from rabbit, biotin-labeled BSA, streptavidin from Streptomyces avidinii, biotin-labeled anti-mouse IgG from goat, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), PBS, Tween-20, and NanoPure water were purchased from Sigma–Aldrich. Alexa Fluor 488-labeled BSA was purchased from Invitrogen.
Protein Solution Preparation.
Solutions of polyclonal anti-BSA IgG were prepared to a final concentration of 15 μg/ml in 10 mM Hepes buffer (pH 7.4) with 0.02% Tween-20 nonionic surfactant. The solutions were filtered through a 0.2-μm pore syringe filter to remove large aggregates. Alexa Fluor 488-labeled BSA was prepared to a final concentration of 35 μg/ml in 10 mM Hepes buffer. These values are typical of protein concentrations used in DPN studies (11, 12). Solutions of biotin-BSA, streptavidin, and biotin-IgG were prepared to a final concentration of 50 μg/ml in 10 mM PBS (pH 7.4). The solutions were filtered through a 0.2-μm pore syringe filter. To test biological activity, biotin-BSA-patterned substrates were rinsed in buffer, followed by NanoPure water. After imaging by tapping-mode AFM, they were immersed in streptavidin solution at room temperature for 30 min, rinsed in buffer then NanoPure water, immersed in biotin-IgG for 30 min, and rinsed again.
Substrates.
Substrates were prepared by evaporating >80-nm gold onto a silicon wafer, with a titanium or chromium adhesion layer. The wafer was then cut into pieces of desired dimensions. MHA monolayers were formed on the gold by immersing the substrates in a 1.5 mM ethanol solution for 1–2 h, followed by copious rinsing with ethanol. The RMS roughness of the MHA-coated substrates was 1.3 nm as measured by tapping-mode AFM. For patterning of anti-BSA IgG on BSA, gold substrates were directly incubated in a solution of Alexa Fluor 488-labeled BSA at room temperature for 1 h. The substrates were then rinsed in a 10 mM Hepes buffer solution with 0.5% Tween-20 to remove protein multilayers before being rinsed in deionized water and allowed to dry. The substrates were then fixed to an insulating glass slide.
Patterning and Characterization.
A layer of gold ≈30 nm thick was sputtered (Denton Vacuum) onto the reservoir side of the NFP (back side of the cantilevers) to act as an electrode, whereas the volcano tips were left nonconductive. Patterning experiments were conducted in contact mode at room temperature (≈25°C) by using a Veeco DI 3100 AFM (Veeco Instruments) equipped with an nPoint 100-μm closed-loop two-axes scanner (nPoint.) and NanoScript software (Veeco Instruments). A Keithley 4200 Semiconductor Characterization System (Keithley Instruments) was used to apply voltage and measure current between the gold-coated NFP reservoir and substrate. Humidity was controlled with a commercial humidifier (Kenmore).
Unless otherwise noted, substrates were rinsed in buffer and then in NanoPure water and dried with compressed air before imaging. Tapping-mode AFM images were taken on a Veeco DI 3100 AFM (Veeco Instruments). SEM images were taken by using a LEO Gemini 1525 SEM.
Supplementary Material
Acknowledgments.
We thank Prof. Igal Szleifer (Northwestern University) for discussions in interpreting protein deposition results. This work was supported by the Nanoscale Science and Engineering Initiative of the National Science Foundation (NSF) under NSF Award EEC-0647560. H.D.E. acknowledges support provided by the NSF through Nanoscale Interdisciplinary Research Team Project CMS00304472. P.K. acknowledges support provided by NSF Career Award CHE-0748676. We acknowledge use of the fabrication facilities of the Cornell University NanoScale Facility (Ithaca, NY), which is supported by NSF Grant ECS-0335765. O.Y.L. acknowledges the Northwestern University Ryan Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806651105/DCSupplemental.
References
- 1.Stears RL, Martinsky T, Schena M. Trends in microarray analysis. Nat Med. 2003;9:140–145. doi: 10.1038/nm0103-140. [DOI] [PubMed] [Google Scholar]
- 2.Henderson E, et al. NanoArrays. Microsc Microanal. 2004;10:1432–1433. [Google Scholar]
- 3.Bernard A, et al. Microcontact printing of proteins. Adv Mater. 2000;12:1067–1070. [Google Scholar]
- 4.Lynch M, et al. Functional protein nanoarrays for biomarker profiling. Proteomics. 2004;4:1695–1702. doi: 10.1002/pmic.200300701. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Tang ZY, Tan SS, Kotov NA. Biological assembly of nanocircuit prototypes from protein-modified CdTe nanowires. Nano Lett. 2005;5:243–248. doi: 10.1021/nl0482682. [DOI] [PubMed] [Google Scholar]
- 6.Salem A, et al. Directed assembly of multisegment Au/Pt/Au nanowires. Nano Lett. 2004;4:1163–1165. [Google Scholar]
- 7.Tang QL, et al. Protein delivery with nanoscale precision. Nanotechnology. 2005;16:1062–1068. [Google Scholar]
- 8.Arnold M, et al. Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 9.Lee K-B, Lim J-H, Mirkin C. Protein nanostructures formed via direct-write dip-pen nanolithography. J Am Chem Soc. 2003;125:5588–5589. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 10.Lim J-H, et al. Direct-write dip-pen nanolithography of proteins on modified silicon oxide surfaces. Angew Chem. 2003;115:2411–2414. doi: 10.1002/anie.200351256. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, et al. Biologically active protein nanoarrays generated using parallel dip-pen nanolithography. Adv Mater. 2006;18:1133–1136. [Google Scholar]
- 12.Lee K-B, et al. protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295:1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 13.Naujoks N, Stemmer A. Using local surface charges for the fabrication of protein patterns. Colloids Surf A. 2004;249:69–72. [Google Scholar]
- 14.Agarwal G, Naik R, Stone M. Immobilization of histidine-tagged proteins on nickel by electrochemical dip pen nanolithography. J Am Chem Soc. 2003;125:7408–7412. doi: 10.1021/ja029856p. [DOI] [PubMed] [Google Scholar]
- 15.Cai YG, Ocko BM. Electro pen nanolithography. J Am Chem Soc. 2005;127:16287–16291. doi: 10.1021/ja054951u. [DOI] [PubMed] [Google Scholar]
- 16.Bruckbauer A, et al. Writing with DNA and protein using a nanopipet for controlled delivery. J Am Chem Soc. 2002;124:8810–8811. doi: 10.1021/ja026816c. [DOI] [PubMed] [Google Scholar]
- 17.Bruckbauer A, et al. Multicomponent submicron features of biomolecules created by voltage controlled deposition from a nanopipet. J Am Chem Soc. 2003;125:9834–9839. [Google Scholar]
- 18.Rodolfa KT, et al. Two-component graded deposition of biomolecules with a double-barreled nanopipette. Angew Chem Int Ed. 2005;44:6854–6859. doi: 10.1002/anie.200502338. [DOI] [PubMed] [Google Scholar]
- 19.Park J-U, et al. High-resolution electrohydrodynamic jet printing. Nature. 2007;6:782–789. doi: 10.1038/nmat1974. [DOI] [PubMed] [Google Scholar]
- 20.Lee D-Y, et al. Electrohydrodynamic printing of silver nanoparticles by using a focused nanocolloid jet. Appl Phys Lett. 2007;90 081905. [Google Scholar]
- 21.Morozov V, Morozova T. Electrospray deposition as a method for mass fabrication of mono- and multicomponent microarrays of biological and biologically active substances. Anal Chem. 1999;71:3110–3117. doi: 10.1021/ac981412h. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs H, Welle AM. A printing of organic and inorganic nanomaterials using electrospray ionization and coulomb-force-directed assembly. Appl Phys Lett. 2005;87:263119. [Google Scholar]
- 23.Kim KH, Moldovan N, Espinosa HD. A nanofountain probe with sub-100 nm molecular writing resolution. Small. 2005;1:632–635. doi: 10.1002/smll.200500027. [DOI] [PubMed] [Google Scholar]
- 24.Moldovan N, Kim KH, Espinosa HD. Design and fabrication of a novel microfluidic nanoprobe. J MEMS. 2006;15:204–213. [Google Scholar]
- 25.Moldovan N, Kim K-H, Espinosa HD. A multi-ink linear array of nanofountain probes. J Micromech Microeng. 2006;16:1935–1942. [Google Scholar]
- 26.Wu B, Ho A, Moldovan N, Espinosa HD. Direct deposition and assembly of gold colloidal particles using a nanofountain probe. Langmuir. 2007;23:9120–9123. doi: 10.1021/la7011952. [DOI] [PubMed] [Google Scholar]
- 27.Kim K-H, et al. Direct delivery and submicrometer patterning of DNA by a nanofountain probe. Adv Mater. 2008;20:330–334. [Google Scholar]
- 28.Browning-Kelley M, Wadu-Mesthrige K, Hari V, Liu G. Atomic force microscopic study of specific antigen/antibody binding. Langmuir. 1997;13:343–350. [Google Scholar]
- 29.Brewer S, et al. Probing BSA binding to citrate-coated gold nanoparticles and surfaces. Langmuir. 2005;21:9303–9307. doi: 10.1021/la050588t. [DOI] [PubMed] [Google Scholar]
- 30.Weeks BL, DeYoreo JJ. Dynamic meniscus growth at a scanning probe tip in contact with a gold substrate. J Phys Chem B. 2006;110:10231–10233. doi: 10.1021/jp0615914. [DOI] [PubMed] [Google Scholar]
- 31.Jang JY, Schatz GC, Ratner MA. Liquid meniscus condensation in dip-pen nanolithography. J Chem Phys. 2002;116:3875–3886. [Google Scholar]
- 32.Peters TJ. All About Albumin: Biochemistry, Genetics, and Medical Applications. San Diego: Academic; 1995. [Google Scholar]
- 33.Hendrickson W, et al. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc Natl Acad Sci USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silverton E, Navia M, Davies D. Three-dimensional structure of an intact human immunoglobulin. Proc Natl Acad Sci USA. 1977;74:5140–5144. doi: 10.1073/pnas.74.11.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brakke KA. The surface evolver. Exp Math. 1996;1:141–165. [Google Scholar]
- 36.Lin SC, Tseng F, Chieng CC. Numerical simulation of protein stamping process driven by capillary force. IEEE Trans Nanobiosci. 2002;1:121–128. doi: 10.1109/tnb.2003.809463. [DOI] [PubMed] [Google Scholar]
- 37.Weeks B, Vaughn M, DeYoreo J. Direct imaging of meniscus formation in atomic force microscopy using environmental scanning electron microscopy. Langmuir. 2005;21:8096–8098. doi: 10.1021/la0512087. [DOI] [PubMed] [Google Scholar]
- 38.Fisher LR, Israelachvili JN. Experimental studies on the applicability of the Kelvin equation to highly curved concave menisci. J Colloid Interface Sci. 1981;80:528–541. [Google Scholar]
- 39.Stifter T, Marti O, Bhushan B. Theoretical investigation of the distance dependence of capillary van der Waals forces in scanning force microscopy. Phys Rev B. 2000;62:13667–13673. [Google Scholar]
- 40.Cho N, et al. Phase of molecular ink in nanoscale direct deposition processes. J Chem Phys. 2006;124 doi: 10.1063/1.2147139. 024714. [DOI] [PubMed] [Google Scholar]
- 41.Melcher JR. Continuum Electromechanics. Cambridge, MA: MIT Press; 1981. [Google Scholar]
- 42.Chen J, Hsieh W. Electrowetting-induced capillary flow in a parallel-plate channel. J Colloid Interface Sci. 2006;296:276–283. doi: 10.1016/j.jcis.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 43.Mann M, Wilm M. Electrospray mass spectrometry for protein characterization. Trends Biol Sci. 1995;20:219–224. doi: 10.1016/s0968-0004(00)89019-2. [DOI] [PubMed] [Google Scholar]
- 44.Hanson E. Recent Progress in Ink Jet Technologies II. Springfield, VA: Society for Imaging Science and Technology; 1999. [Google Scholar]
- 45.Lin X, Creuzet F, Arribart H. Atomic-force microscopy for local characterization of surface acid–base properties. J Phys Chem. 1993;97:7272–7276. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.