Abstract
Clear cell renal carcinomas are the most common form of kidney cancer and frequently are linked to biallelic inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene. The VHL gene product, pVHL, has multiple functions including directing the polyubiquitylation of the HIF transcription factor. We screened 100 shRNA vectors, directed against 88 kinases, for their ability to inhibit the viability of VHL−/− renal carcinoma cells preferentially compared with isogenic cells in which pVHL function was restored. shRNAs for “hits” identified in the primary screen were interrogated in secondary screens that included shRNA titration studies. Multiple shRNAs against CDK6, MET, and MAP2K1 (also known as MEK1) preferentially inhibited the viability of 786-O and RCC4 VHL−/− cells compared with their wild-type pVHL-reconstituted counterparts. The sensitivity of pVHL-proficient cells to these shRNAs was not restored upon HIF activation, suggesting that loss of an hypoxia-inducible factor (HIF)-independent pVHL function formed the basis for selectivity. A small-molecule Cdk4/6 inhibitor displayed enhanced activity against VHL−/− renal carcinoma cells, suggesting that in some cases hits from shRNA screens such as described here might translate into therapeutic targets.
Keywords: essential kinases, shRNA screens, VHL, kidney cancer, therapeutics
Cancer cells harbor mutations that activate protooncogenes or inactivate tumor suppressor genes. These mutations earmark molecular pathways that are important for cancer genesis and maintenance. Importantly, several approved anticancer drugs target particular kinases, such as Abl, Her2/Neu, and EGFR, that become hyperactive in specific forms of cancer because of gain-of-function mutations. Therefore knowledge of the genetic alterations within a cancer can inform cancer drug discovery.
Mutations that inactivate tumor suppressor genes, especially those causing complete loss of their protein products, present a therapeutic challenge, however, because most drugs also inactivate their protein target. One approach to this problem would be to look for downstream targets that become activated upon tumor suppressor gene inactivation and that play a causal role in tumor maintenance. Another approach, put forth by Hartwell et al. (1), would be to identify genes that are synthetically lethal to the tumor suppressor gene of interest. Two genes (“A” and “B”) are synthetically lethal if mutation of either alone is compatible with viability but mutation of both leads to death (2, 3). If “A” is a cancer-relevant gene, such as a tumor suppressor gene, then in theory inhibitors of the “B” gene product would kill cancer cells harboring the “A” mutation while sparing normal cells. Synthetic lethality therefore provides, in theory, a solution to the loss-of-function problem and to the problem of generating cytotoxic agents that can discriminate between cancer cells and normal cells.
Synthetic lethality has been well studied in the budding yeast Saccharomyces cerevisiae, in which only 20% of the genes are individually essential and synthetic lethal interactions are common among the remaining 80% (4–6). Many of these synthetic lethal interactions would have been difficult to predict a priori and were revealed only by unbiased genetic screens. The tools for performing such genetic screens in human cells did not exist until recently. For these reasons, only a limited number of synthetic lethal interactions with cancer-relevant genes have been described (2, 3).
We chose the von Hippel-Lindau (VHL) tumor suppressor gene, which is mutated in most clear cell renal cell carcinomas (RCC), to explore this paradigm further. The VHL gene product, pVHL, binds hypoxia-inducible factor alpha (HIFα) in an oxygen-dependent manner and targets it for ubiquitin-mediated proteolysis (reviewed in ref. 7). When oxygen levels are low, or pVHL is defective, HIFα accumulates, dimerizes with HIFβ, and activates a suite of genes involved in adaptation to hypoxia including VEGF, PDGF-B, and TGFα. Down-regulation of HIF, particularly HIF2α, is necessary and sufficient for pVHL to suppress clear cell RCC proliferation in vivo (7). HIFα accumulation also occurs in many other solid tumors because of intratumoral hypoxia and usually confers a poor prognosis (8). Importantly, restoration of pVHL function does not affect cell proliferation in vitro under standard cell-culture conditions (9, 10). Differences in proliferation might confound cell-based synthetic lethal screens because many cytotoxic agents kill in a cell cycle-dependent manner.
Kinases play important roles in biology, frequently are deregulated in cancer, and can, as described earlier, be inhibited with drug-like small molecules. VHL loss leads, indirectly, to activation of kinases that are important for renal carcinoma tumorigenesis (reviewed in ref. 7), including kinases present within the tumor cells themselves, such as EGFR (11, 12), c-Met (13–16), and cyclin D1-associated kinases (17, 18), and those associated with blood vessels, such as kinase insert domain receptor and platelet-derived growth factor receptor. Kidney cancer is refractory to most conventional chemotherapeutics and radiotherapy but often responds, at least temporarily, to drugs that inhibit kinase insert domain receptor.
Here we report a proof-of-concept synthetic lethal genetic screen that revealed differential kinase requirements of isogenic renal carcinoma cells that differ only in their VHL status.
Results
786-O and RCC4 are clear cell renal carcinoma lines in which both VHL alleles have been inactivated mutationally. We previously infected these lines with a retroviral vector encoding hemagglutinin (HA)-tagged wild-type pVHL or the backbone vector (10, 19), which also contains a puromycin resistance cassette. These reconstituted cells were maintained as polyclonal pools in the presence of puromycin and were used for the experiments described below. We confirmed that the cells producing wild-type pVHL accumulated lower levels of HIFα, as expected (20), and HIF-responsive gene products such as Glut1 [supporting information (SI) Fig. S1A]. In keeping with previous reports (9, 10), reintroducing wild-type pVHL into 786-O and RCC4 cells did not affect their ability to proliferate in vitro over a period of 5 days under standard cell-culture conditions, nor was their cell-cycle distribution significantly altered (Fig. S1 B–D and data not shown).
Lentiviral short hairpin libraries (shRNA) have been used recently to conduct loss-of-function screens in mammalian cells (21–24). We decided to interrogate these two isogenic renal cell line pairs with a focused small hairpin library in hopes of identifying kinases that become especially important for the viability of cells lacking pVHL. The library consisted of 100 lentiviral shRNA vectors targeting 88 distinct kinases (some kinases were represented with two shRNAs) (Table S1). Eighty of these kinase shRNAs were chosen because they affected the viability of HeLa and 293T cells in preliminary screens (25). Note that because HeLa and 293T are both VHL+/+, kinase genes that are truly synthetically lethal to VHL would not be present among the 80 chosen for this pilot study. Nonetheless, we reasoned that VHL inactivation might alter quantitatively the requirements for particular kinases, leading to differential sensitivity to kinase inhibition.
Each cell line was grown in duplicate 96-well plates and was infected with a lentiviral vector (one vector per well). Viral stocks were produced in 96-well plate format and used at an arbitrary titer that was sufficient, in pilot experiments, to infect all the cells in 96-well plates containing puromycin-sensitive HeLa and 293T cells as test lines (see Methods). The renal carcinoma cells were assayed for viability 5 days after viral transduction using alamarBlue, which measures the number of metabolically active cells (26). In duplicate independent screens we observed that the raw alamarBlue values were highly reproducible (correlation coefficients > 0.90; Fig. S2 and data not shown). As expected, viability was not inhibited by a scrambled shRNA or GFP cDNA, whereas shRNAs against some essential kinases, such as DDR2, killed both VHL−/− cells and their pVHL-restored counterparts (Fig. 1A and Fig. S3). Notably, inhibition by some kinase shRNAs seemed to be influenced by VHL status (Fig. 1A and Fig. S3).
Fig. 1.
VHL status affects sensitivity to shRNA-mediated kinase inhibition. (A) Raw alamarBlue fluorescence values for 786-O cells infected with lentiviruses encoding kinase shRNAs. Red and black bars depict values for 786-O cells infected with a retrovirus encoding HA-pVHL or with the empty vector, respectively. The x-axis indicates rank order of shRNAs based on corresponding alamarBlue values after infection of the empty vector cells. See Table S2 for gene names. (B and C). Difference in percentage loss of viability (Δ %LOV), after normalization (see text) for 786-O cells (B) and RCC4 cells (C) infected with lentiviruses encoding kinase shRNAs. shRNAs that preferentially inhibit VHL−/− cells appear on the right, and the pVHL-restored cells to the left. See Tables S4 and S5 for gene names. Lentiviruses that scored positively in 786-O cells (Δ %LOV > 30% in B) are indicated with yellow bars in C.
We next normalized the raw alamarBlue values to the average of scrambled shRNA value and the value for a lentiviral vector encoding GFP for that plate and converted to percentage loss of viability (%LOV) where
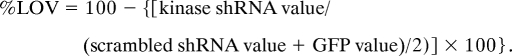 |
Differences in cell viability between the VHL−/− and pVHL-restored cells in response to kinase knockdown was calculated using the formula:
 |
Notably, there were more shRNAs that preferentially inhibited the viability of 786-O VHL−/− cells relative to their pVHL-restored counterparts than shRNAs in which VHL loss was protective (Figs. 1B and 2A and Table S2). This finding suggests that VHL loss, in general, has decreased the fitness of 786-O cells with respect to kinase inhibition. Secondly, the effects observed in RCC4 cells were less dramatic than in 786-O cells (Figs. 1C and 2A and Table S3), probably in part because these cells are less infectable with lentiviruses than are 786-O cells, as determined by fluorescence of cells infected with the GFP lentivirus (data not shown). We therefore defined “hits” arbitrarily as those shRNAs associated with a Δ %LOV ≥ 35% and 20% for the 786-O cells and RCC4 cells, respectively.
Fig. 2.
Kinase shRNAs that preferentially inhibit VHL−/− renal carcinoma cells. (A) Heat map depicting the effects of the 100 shRNA vectors on the two cell-line pairs after normalization to the GFP cDNA and scrambled shRNA controls (see text). Hierarchical clustering analysis using the Manhattan clustering method was done using the TM4 program (35). Experiments or shRNAs with relatively similar patterns are clustered together. shRNAs causing > 50% loss of viability are indicated in red, and those with < 50% loss of viability are in green. (B) List of shRNA vectors (kinase name followed by TRC collection shRNA number) that scored positively (see text for criteria) when tested in 786-O and RCC4 isogenic cell-line pairs that did (VHL) or did not (vector) produce wild-type pVHL. (C) Photomicrographs (100 × magnification) of crystal violet-stained 786-O cells 5 days after infection with lentiviruses encoding the indicated shRNAs. scr = scrambled shRNA vector.
Knockdown of 18 kinases (represented by a total of 20 hairpins) scored as hits in 786-O cells (Fig. 2B, Left column), whereas 15 of the 88 kinases satisfied the hit criteria for RCC4 cells (Fig. 2B, Right column). Only five of the total hits (CDK6, MAP2K1, MET, IRR, and HER4) overlapped between the two cell lines and were tested further. To minimize the possibility that differences in cell survival between the isogenic cell-line pairs were caused by off-target shRNA effects, we tested four or five independent shRNAs for each of these five kinases from the original Broad Institute TRC shRNA library (21). For all five kinases we observed preferential inhibition of VHL−/− cells (Δ %LOV > 20%) with at least two independent shRNAs in both cell-line pairs, suggesting that the observed phenotypes were caused by modulation of the intended targets.
For practical reasons the screen was conducted at an arbitrary viral titer. Next we performed viral titration experiments over a 8-fold range of viral MOIs. At high viral titers the shRNAs vectors inhibited the viability of both renal carcinoma lines irrespective of VHL status (Fig. 3, Fig. S4, and data not shown). At lower titers the CDK6, MET, and MAP2K1 shRNAs preferentially inhibited the VHL−/− cells over several 2-fold dilutions (Fig. 3A, Fig. S4, and data not shown). Similar results were obtained with a second independent shRNA for all three kinases (data not shown). Immunoblot analysis confirmed that the degree of target knockdown in the wild-type cells equaled or exceeded that observed in the VHL−/− cells (Fig. 3B and data not shown). Therefore the increased sensitivity of the VHL−/− cells was unlikely to reflect more efficient target knockdown. In some experiments we noted increased basal levels of c-MET in VHL−/− cells compared with pVHL-restored cells (Fig. 3B), in keeping with a recent report (13).
Fig. 3.
Kinase shRNAs that preferentially inhibit 786-O VHL−/− cells over a range of viral titers. (A) Viability of 786-O cells infected with indicated amounts of lentiviruses encoding shRNAs directed against CDK6, MET, or MAP2K1. Cell number was assayed in triplicate using alamarBlue 5 days after infection and normalized to scrambled hairpin control. Circles and squares depict values for 786-O cells infected with a retrovirus encoding HA-pVHL or with the empty vector, respectively. Error bars = 1 SEM. (B) Immunoblot analysis of cells treated with 2.5 μl, 5 μl, and 10 μl of virus as in A. scr = scrambled shRNA vector.
In contrast to CDK6, MET, and MAP2K1, a differential for the IRR and HER4 shRNA vectors between VHL−/− and pVHL-restored cells was observed only at a single MOI (data not shown). Therefore these kinases were not studied further.
To determine whether the alamarBlue readings were a true representation of the number of viable cells, we stained cells with crystal violet 5 days after infection with the different kinase shRNA vectors (Fig. 2C). shRNAs against DDR2 and a scrambled shRNA served as positive and negative controls, respectively. This assay confirmed that inhibition of CDK6, MET, and MAP2K1 preferentially inhibited VHL−/− renal carcinoma cells relative to the pVHL-restored cells.
Moreover, we recapitulated our shRNA findings with synthetic siRNAs that were designed using the sequences of the most effective shRNAs targeting CDK6, MET, and MAP2K1 (Fig. S5). Collectively, these results indicate that the shRNA phenotypes we observed were caused by the specific inhibition of their intended kinase targets and were not an artifact of viral infection.
Suppression of HIFα is the best-documented pVHL activity. We next asked whether increased HIFα was required for the decreased viability of VHL−/− renal carcinoma cells following inhibition of CDK6, MET, or MAP2K1. 786-O VHL−/− cells produce HIF2α but not HIF1α (27) (Fig. S1A). Our laboratory created 786-O cells that produce both wild-type pVHL and a HIF2α variant that escapes pVHL-dependent degradation because two proline residues that are critical for pVHL binding have been replaced with alanine (HIF-2α dP→A) (19). As expected, HIF-2α protein levels were comparable in the 786-O VHL−/− cells (vector) and the wild-type pVHL cells producing the HIF2α dP→A variant (Fig. 4A). Furthermore, HIFα signaling was activated in these cells, as shown by induction of downstream targets such as the glucose transporter, Glut1, and the prolyl hydroxylase EglN3 (Fig. 4A).
Fig. 4.
Deregulation of HIF2α does not fully explain the increased requirement of 786-O VHL−/− cells for CDK6, MAP2K1, and MET. (A) Immunoblot analysis of 786-O cells infected to produced HA-pVHL alone (VHL) or HA-pVHL and a stabilized version of HIF2α (VHL + dP →A). Parental cells infected with an empty virus (vector) served as controls. (B) Percentage loss of viability (% LOV) of cells (normalized to scrambled shRNA) infected as in A after subsequent infection with 5 μl (gray bars) or 10 μl (black bars) of lentiviruses encoding shRNAs targeting CDK6, MET, or MAP2K1. Cells were grown in 96-well plates and were assayed for survival in triplicate using alamarBlue 5 days post-infection. Error bars = 1 SEM.
Interestingly, the HIF2α dP→A variant did not sensitize the wild-type pVHL cells to the CDK6 or MET shRNAs and had only a modest effect on sensitivity to the MAP2K1 shRNA (Fig. 4B). Therefore, the increased requirement of VHL−/− cells for these kinases is caused, at least in part, by the loss of a HIF-independent pVHL function.
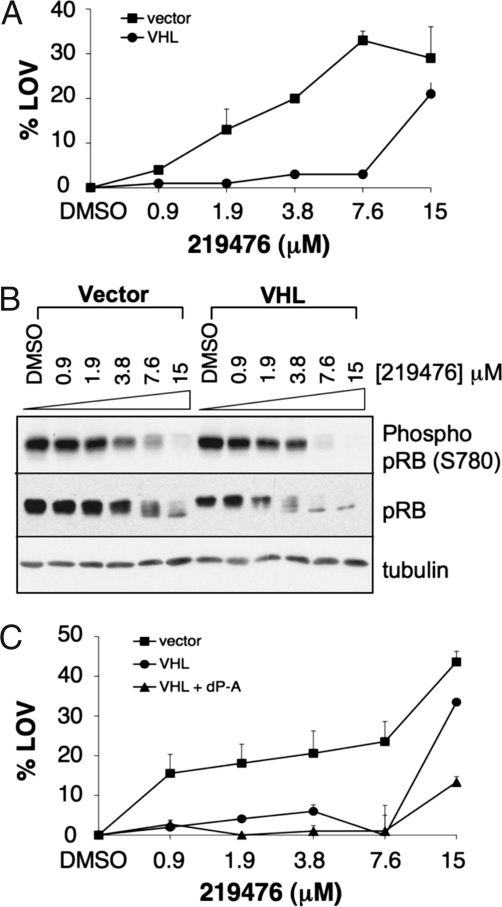
The biological output of a catalytically inactive kinase bound to a small organic molecule need not be the same as seen after elimination of that kinase. In a final set of experiments we treated the isogenic cell-line pairs with varying concentrations of a commercially available Cdk4/6 inhibitor (Calbiochem catalog no. 219476). As shown in Fig. 5A, 786-O VHL−/− cells were more sensitive than their pVHL-restored counterparts to this agent. The drug concentration required to inhibit proliferation approximated the concentration required to inhibit pRB phosphorylation, as determined by increased pRB electrophoretic mobility and immunoblot analysis with an antibody specific for phosphorylated Ser-780, which is a Cdk4/6 phosphorylation site (28) (Fig. 5B). Notably, pRB phosphorylation was affected in both the VHL−/− cells and wild-type cells, consistent with comparable levels of target inhibition in both contexts. In keeping with the findings in Fig. 4, the HIF2α dP→A variant did not sensitize and, at higher concentrations, protected wild-type cells treated with the Cdk4/6 inhibitor (Fig. 5C). RCC4 VHL−/− cells also were more sensitive than wild-type cells to 219476 (Fig. S6). Like most small-molecule kinase inhibitors, 219476 may have other targets in addition to Cdk4 and Cdk6. Nonetheless, these results suggest that hits obtained in screens such as the one we used might, in some cases, translate into potential drug targets.
Fig. 5.
A CDK4/6 inhibitor preferentially inhibits VHL−/− cells. (A) Percentage loss of viability (%LOV) of 786-O cells infected with a retrovirus encoding HA-pVHL (circles) or empty vector (squares) and then treated with Cdk4/6 inhibitor (Calbiochem catalog no. 219476) at indicated concentrations for 48 h in 96-well plates. Viable cell number was determined in triplicate by XTT assay and normalized to DMSO-treated controls. Error bar = 1 SEM. (B). Immunoblot analysis of cells treated as in A. (C) Percentage loss of viability (%LOV) of 786-O cells infected to produced HA-pVHL alone (VHL; circles) or HA-pVHL and a stabilized version of HIF2α (VHL + dP →A; triangles) and then treated with Cdk4/6 inhibitor (Calbiochem catalog no. 219476). Parental cells infected with an empty virus (vector; squares) served as controls, and viability was determined as in A.
Discussion
A conceptually attractive approach to treating a cancer would be to exploit vulnerabilities created by the genetic and epigenetic alterations that took place in that cancer over time (1, 2). These alterations might include the changes responsible for the transformed phenotype as well as passenger mutations. In this pilot study we found that inactivation of the VHL tumor suppressor gene, which is common in clear cell renal carcinomas, led to decreased fitness following down-regulation of specific kinases. In particular, we found that VHL−/− renal carcinoma cells were more sensitive to loss of MET, CDK6, and MAP2K1 than were their pVHL-restored counterparts.
The kinase shRNA collection we used in this pilot study primarily included kinases that affect the viability of HeLa and/or 293T cells, both of which are VHL+/+. For this reason, our screen was biased against the identification of kinases that are truly dispensable for wild-type cells but are required for VHL−/− cells (synthetically lethal to VHL). Nonetheless, we were able to show that loss of pVHL quantitatively alters the requirements for specific kinases. Whether these quantitative differences are large enough to exploit therapeutically is uncertain, however. For this reason, and others, it will be important to expand this screen to the remainder of the kinome and, in time, to other “druggable” proteins.
As is true for many screens, the screening approach described here can yield false negatives and false positives. With respect to false negatives, we found that differences between VHL−/− cells and their wild-type pVHL counterparts could be masked at very high MOIs. It therefore is possible that screening at different MOIs might have revealed additional hits. Complicating matters, the degree to which each vector in the TRC shRNA collection down-regulates its intended target is not known, nor is it practical to normalize rigorously the number of infectious particles generated in the high-throughput lentiviral production schema we used. A trivial explanation for false negatives would be failure to achieve target knockdown. This failure might be minimized by the incorporation of multiple shRNA vectors against the same target in the primary screen, even at the expense of covering fewer targets.
With respect to false positives, we found no evidence that VHL status affected the degree of target knockdown achieved with our lentiviral vectors. We also required that the same phenotype be observed with multiple independent shRNA vectors in secondary screens to minimize the contribution of off-target effects. Because VHL−/− renal carcinomas harbor mutations at other loci, we also required that selectivity be manifest in two different cell-line pairs so as to avoid scoring genetic interactions that, rather than being robust, might be peculiar to the unique molecular context provided by a single line. In this regard, we initially were surprised that few of the 20 shRNA hits identified in the 786-O cells also scored as hits in the RCC4 cells. However, this finding probably results, at least in part, from technical factors related to the infectivity of RCC4 cells and the arbitrary thresholds used to define hits. We note that 14 of the 20 hits identified in the 786-O cells preferentially inhibited the growth of VHL−/− RCC4 cells compared with their pVHL-restored counterparts and that 9 the 20 were among the 25 shRNAs that showed the greatest selectivity for VHL−/− cells in the RCC4 context (Fig. 1C). This apparent correlation between the two cell lines was significant (P < 0.01) using the Spearman's rank correlation, suggesting that the overlap between these two lines is driven by VHL biology rather than by chance.
One goal of screens such as this one is to identify potential therapeutic targets in cancer. Several caveats apply, however. First, the biological consequences of eliminating a kinase genetically might differ from those induced by specifically inactivating the catalytic function of a kinase with a drug. Moreover, drugs, unlike genetic tools, usually do not discriminate between closely related paralogs. In this regard, although we achieved preferential inhibition of VHL−/− renal carcinoma cells with a small-molecule CDK6 inhibitor (Fig. 5), we did not attain selectivity with a small-molecule MEK inhibitor (Fig. S7). Finally, we do not know yet whether the genetic interactions we identify in the context of renal carcinoma cells will hold true in other cell types and tissues. In short, a kinase that was especially important for the viability of VHL−/− renal carcinoma cells also might be critical for some VHL+/+ normal cells.
Synthetic lethal interactions often are difficult to predict, and genome-wide screens in model organisms indicate that they can be difficult to rationalize even a posteriori, as is true also in the work described here (4, 5). It nonetheless is intriguing that MET, CDK6, and MAP2K1 have been linked previously to kidney cancer biology. Germline MET mutations cause papillary renal carcinoma, and there is recent evidence that VHL loss, which is linked tightly to clear cell histology, also leads to c-MET activation (13–16). VHL loss in renal epithelial cells, but not in other cell types tested, leads to up-regulation of Cyclin D1 (17, 18), which is a partner protein for Cdk6 and Cdk4. Finally, loss of MAP2K1, via an unclear mechanism, impairs HIFα transactivation function (29–32). In short, our studies suggest that renal carcinoma cells remodel signaling networks upon VHL inactivation so that they become more dependent on MET, CDK6, and MAP2K1. The fact that we identified three hits with fairly direct links to kidney cancer suggests that expansion of this screen to additional enzymes might provide additional insights into kidney cancer biology and perhaps reveal new drug targets for this disease.
Materials and Methods
Cell Lines.
786-O and RCC4 human renal carcinoma cell line derivatives (10, 19) were maintained in DMEM containing 10% FBS in the presence of 1 μg/ml puromycin.
Lentivirus Production.
High-throughput lentiviral production was as described in detail elsewhere (32). Briefly, DNA was prepared using a large-scale plasmid purification kit (Qiagen). 293T packaging cells were co-transfected with shRNA-encoding replication-deficient lentiviral vectors and the necessary helper plasmids for virus production. The virus was pseudotyped with the envelope glycoprotein from vesicular stomatitis virus as described previously (21, 33, 34). Successful production of virus was measured by two methods: direct titering of a subset of the viruses in National Institutes of Health (NIH) 3T3 cells and measurement of viral genomic RNA sequences for the puromycin acetyl transferase gene from viral supernatants harvested during a production run. In an NIH 3T3 colony-forming assay, formal viral titers typically were 2 × 106 cfu/ml, and comparisons of RNA levels of the puromycin acetyl transferase gene showed no more than a 2-fold variation from the mean values, suggesting that production of most shRNA expression vectors was similar.
Viral Infections in 96-Well Format.
786-O and RCC4 cells were seeded at 2,000 and 3,000 cells per well respectively, in a final volume of 100 μl per well in 96-well plates. Twenty-four hours later, 50 μl of media was removed, and different amounts of viral supernatant were added (1.25–20 μl, depending on the experiment), all in the presence of 8 μg/ml polybrene. Plates were spun at 1,178 × g for 30 min at room temperature in an SX4750 μPlate Carrier (Beckman Coulter). Infected cells were washed 12–16 h after infection. Cells were analyzed 5 days later.
Immunoblot Analysis.
Cell extracts were made using EBC buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40) supplemented with complete protease inhibitor mixture (Roche Molecular Biochemicals), resolved by SDS/PAGE, and transferred to nitrocellulose membranes (Whatman). After blocking in Tris-buffered saline with 5% nonfat dry milk, the membranes were probed with the following primary antibodies: anti-HA mouse monoclonal antibody (HA-11, Covance Research Products), anti-HIF2α mouse monoclonal antibody (Yoji Minamishima and W.G.K, unpublished data), anti-HIF1α mouse monoclonal antibody (BD Transduction Laboratories), anti-Glut1 rabbit polyclonal antibody (GT111-A, Alpha Diagnostic), anti-phospho Rb (S780) (Cell Signaling Technology), anti-Rb (BD Biosciences), and anti-tubulin and anti-vinculin mouse monoclonal antibody (Sigma). Bound antibody was detected with HRP-conjugated goat anti-rabbit or goat anti-mouse (Thermo Scientific) and SuperSignal West Pico or Dura chemiluminescent substrate (Thermo Scientific).
In Vitro Cell Proliferation Assays.
In vitro cell proliferation assays were performed using a Cell Proliferation Kit II (XTT) (Roche Diagnostics) according to the manufacturer's instructions. Briefly, 1,000 cells were seeded per well in 96-well plates. At the indicated time points, XTT-labeling reagent/electron-coupling reagent was added to the cells. Four hours later the spectrophotometrical absorbance at 450 nm was measured using a microtiter plate reader (PerkinElmer Life and Analytical Sciences).
alamarBlue Assay.
Five days after infection, medium was removed from 96-well plates and alamarBlue™ reagent (Biosource, Invitrogen), diluted 1:10 in supplemented DMEM, was added to each well. Plates then were incubated for 4–8 h at 37°C before reading on a Spectrafluor Plus microtiter well plate reader (Tecan) at 595 nm.
Crystal Violet Staining.
After the medium was removed, the cells were washed with PBS and fixed with 10% acetic acid and 10% methanol. After 30 min, cells again were washed with PBS and then were incubated for 2 h with 0.4% crystal violet in 20% ethanol, followed by two final PBS wash steps. Phase-contrast images were acquired with an inverted microscope (Nikon).
siRNA Transfections.
siRNA sequences used were: CDK6: gaagaagactggcctagagat; MET: cagaatgtcattctacatgag; MAP2K1: gagggagaagcacaagatcat; and GL3: cttacgctgagtacttcga.
786-O and RCC4 cells were plated at 1.5 × 103 cells per well in 96-well plates, and 10 pmol of siRNA oligos were transfected using Dharmafect 1 (Dharmacon) according to the manufacturer's protocol.
Supplementary Material
Acknowledgments.
We thank Donna Neuberg for help with statistical analysis. A.B.-R. was supported by the American Foundation for Urologic Disease and the Tisch Family Fund for research in solid tumors. W.G.K. is a Howard Hughes Medical Institute investigator and was supported by the Murray Foundation. This work is supported by National Institutes of Health Grant R21CA104940 (to W.G.K and A.B-R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806574105/DCSupplemental.
References
- 1.Hartwell L, Szankasi P, Roberts C, Murray A, Friend S. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 3.Kaelin WG., Jr Cell biology: Divining cancer cell weaknesses. Nature. 2006;441:32–34. doi: 10.1038/441032a. [DOI] [PubMed] [Google Scholar]
- 4.Sharom JR, Bellows DS, Tyers M. From large networks to small molecules. Curr Opin Chem Biol. 2004;8:81–90. doi: 10.1016/j.cbpa.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 6.Wong SL, et al. Combining biological networks to predict genetic interactions. Proc Natl Acad Sci USA. 2004;101:15682–15687. doi: 10.1073/pnas.0406614101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin WG., Jr von Hippel-Lindau Disease. Annual Review of Pathology: Mechanisms of Disease. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 8.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 9.Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Tumor suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 10.Li L, et al. Hypoxia-inducible factor linked to differential kidney cancer risk seen with type 2A and type 2B VHL mutations. Mol Cell Biol. 2007;27:5381–5392. doi: 10.1128/MCB.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Paulsen N, et al. Role of transforming growth factor-alpha in VHL−/− clear cell renal carcinoma cell proliferation: A possible mechanism coupling von Hippel-Lindau tumor suppressor inactivation and tumorigenesis. Proc Natl Acad Sci USA. 2001;98:1387–1392. doi: 10.1073/pnas.031587498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith K, et al. Silencing of epidermal growth factor receptor suppresses hypoxia-inducible factor-2-driven VHL−/− renal cancer. Cancer Res. 2005;65:5221–5230. doi: 10.1158/0008-5472.CAN-05-0169. [DOI] [PubMed] [Google Scholar]
- 13.Pennacchietti S, et al. Hypoxia promotes invasive growth by transcriptional activation of the Met protooncogene. Cancer Cell. 2003;3:347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 14.Koochekpour S, et al. The von Hippel-Lindau tumor suppressor gene inhibits hepatocyte growth factor/scatter factor-induced invasion and branching morphogenesis in renal carcinoma cells. Mol Cell Biol. 1999;19:5902–5912. doi: 10.1128/mcb.19.9.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakaigawa N, et al. Inactivation of von Hippel-Lindau gene induces constitutive phosphorylation of MET protein in clear cell renal carcinoma. Cancer Res. 2006;66:3699–3705. doi: 10.1158/0008-5472.CAN-05-0617. [DOI] [PubMed] [Google Scholar]
- 16.Peruzzi B, Athauda G, Bottaro DP. The von Hippel-Lindau tumor suppressor gene product represses oncogenic beta-catenin signaling in renal carcinoma cells. Proc Natl Acad Sci USA. 2006;103:14531–14536. doi: 10.1073/pnas.0606850103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bindra RS, Vasselli JR, Stearman R, Linehan WM, Klausner RD. VHL-mediated hypoxia regulation of cyclin D1 in renal carcinoma cells. Cancer Res. 2002;62:3014–3019. [PubMed] [Google Scholar]
- 18.Zatyka M, et al. Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease. Cancer Res. 2002;62:3803–3811. [PubMed] [Google Scholar]
- 19.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxwell P, et al. The von Hippel-Lindau gene product is necessary for oxygen-dependent proteolysis of hypoxia-inducible factor a subunits. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 21.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 22.Berns K. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 23.Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 24.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 25.Grueneberg DA, et al. Kinase requirements in human cells: I. Comparing kinases requirements across various cell types. Proc Natl Acad Sci USA. 2008;105:16472–16477. doi: 10.1073/pnas.0808019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama GR, Caton MC, Nova MP, Parandoosh Z. Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J Immunol Methods. 1997;204:205–208. doi: 10.1016/s0022-1759(97)00043-4. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell P, et al. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci USA. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa M, et al. The consensus motif for phosphorylation by cyclin D1-cdk4 is different from that for phosphorylation by cyclin A/E-cdk2. EMBO. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 29.Sang N, et al. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J Biol Chem. 2003;278:14013–14019. doi: 10.1074/jbc.M209702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 31.Lee E, Yim S, Lee SK, Park H. Two transactivation domains of hypoxia-inducible factor-1alpha regulated by the MEK-1/p42/p44 MAPK pathway. Mol Cells. 2002;14:9–15. [PubMed] [Google Scholar]
- 32.Hur E, Chang KY, Lee E, Lee SK, Park H. Mitogen-activated protein kinase kinase inhibitor PD98059 blocks the trans-activation but not the stabilization or DNA binding ability of hypoxia-inducible factor-1alpha. Mol Pharmacol. 2001;59:1216–1224. doi: 10.1124/mol.59.5.1216. [DOI] [PubMed] [Google Scholar]
- 33.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 34.Stewart S, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.