Abstract
Functional differences among human cells have been difficult to identify by standard biochemical methods. Loss-of-function shRNA screens provide an unbiased method to compare protein requirements across cell lines. In previous work, we have studied kinase requirements in two settings, either among a panel of cells from numerous tissues or between two cell lines that differ only by the expression of a chosen oncoprotein or tumor suppressor protein. Here we examine the patterns of kinase requirements between two unrelated cells, the cervical carcinoma cell line HeLa and the renal carcinoma cell line 786-O. By using time courses of cell proliferation after shRNA transduction and by introducing different levels of the shRNAs, we were able to carefully compare the kinase requirements. These comparisons identified 10 kinases that were required in HeLa but not 786-O, and 5 kinases that were required in 786-O but not HeLa. The patterns of growth inhibition due to particular sets of shRNAs in a tumor cell line were shown to be similar in some but not all cell lines derived from the same tissue-specific cancer type. Differential kinase requirements promise to be useful in distinguishing important cell-to-cell functional variations and may lead to the identification of fingerprints for different physiological cell states.
Keywords: cervical cancer cells, essential kinases, renal cancer cells, shRNA screens, fingerprints
Traditional biochemical or cell biological approaches are commonly used to study the properties of key regulatory proteins, gathering characteristics of the proteins themselves; identifying the signaling pathways in which they act; and in some cases, measuring the relative activity of their biochemical roles. These methods provide exceptionally strong approaches to identify interaction partners, posttranslational modification events, subcellular location and relocation events, and enzymatic activities. However, these methods are not particularly useful in determining how the functions of these proteins differ between two cells or how they function in the same cell under different conditions. Advances in RNAi methodologies allow functional comparisons of cell lines to be made, and these comparisons promise to provide a deeper understanding of cell-to-cell variations in molecular physiology.
The recent development of high-throughput RNA interference (RNAi) screening has provided the opportunity to study functional roles of proteins in cells in an unbiased and comprehensive fashion. RNAi technology uses synthetic or vector-generated double stranded RNA to degrade mRNAs with homologous sequences (1, for general reviews see refs. 2, 3). Many such screens have been used previously to study various aspects of cell biology (4–15). Our approach involves the systematic down-regulation of each kinase, one at a time, to explore the differences in kinase requirements for proliferation and survival among various cancer cells and then to carefully compare the extent of the responses using RNAi titrations and time courses. In the three earlier papers of this series we have examined how the requirements for kinases vary across commonly used tumor cell lines (16) or how nearly isogenic cell pairs differ in kinase requirements after the expression of a single oncogene or a tumor suppressor gene (17, 18). These studies identified essential kinases in well-known cancer-relevant pathways as well as kinases that previously have not been associated with cancer development. Although most cancer cells in culture show different patterns of kinase requirements, some cell comparisons have more closely related patterns. One comparison in which cells show more closely related patterns is with cell pairs that differ only by the expression of a single gene. When two cells differ in the expression of a cancer protein, the changes in kinase requirements can be linked to the action of the protein. We anticipate that these approaches will provide a powerful method to study the cellular consequences of cancer gene action and that they may identify interesting targets for drug development.
Here we examine a different type of cell-to-cell comparison, one in which two cells that are not obviously related are studied for kinase requirements. These comparisons were made by using titrations of shRNA expression and time courses of shRNA action. In these proof-of-concept experiments, we show that the resulting different patterns of kinase requirements can be used to identify functionally related cells, suggesting that it will be possible to study underlying cell physiologies by assaying for kinase requirements.
Results
To identify essential kinases that might distinguish one cell line from another, we first chose two cell lines that had several similar proliferation properties, but came from different tissues. After preliminary tests looking for cells with similar doubling times, adhesion properties in tissue culture dishes, infectivity by lentiviruses, and media requirement, we selected HeLa and 786-O cells for comparison. HeLa cells are a well characterized cervical carcinoma cell line that expresses human papilloma virus E6 and E7 oncoproteins (19). 786-O cells are a clear cell renal carcinoma line that carries a deletion of the Von Hippel–Lindau (VHL) tumor suppressor gene (20). Both are commonly used in cancer cell biology studies. From our related studies, we know that these two cells have different patterns of kinase requirements characteristic of most lines in culture (16). To compare the kinase requirements of these cells, we used the panel of 100 shRNA “hits” described in Grueneberg et al. (16). These shRNA hits were selected originally from their patterns of requirement for proliferation and viability in either HeLa or 293T cells, and their characterization has been described previously. The majority of kinases were represented by a single shRNA (100 shRNAs for 80 kinases), but the roles of the kinase hits have been carefully confirmed as described below.
Cell-to-Cell Differences in Response to Kinase Loss.
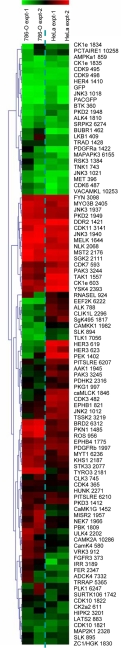
We made lentivirus stocks of each of the 100 hits and tested for proliferation in HeLa and 786-O cells after shRNA expression. Cell number and viability were assayed 5 or 6 days after transduction by the reduction of alamarBlue, a measure of mitochondrial fitness that provides a surrogate endpoint for cell number (21, 22). Even though there were considerable differences between the two cell lines, the assays themselves were robust, showing correlation coefficients of 0.87 among repeats in 786-O cells, and 0.97 among repeats in HeLa cells [supporting information (SI) Fig. S1]. Shown in Fig. 1 are the heat map comparisons using unsupervised hierarchical clustering to examine relationships between shRNAs and cell lines. These comparisons emphasize the difference in kinase requirements between HeLa and 786-O cell lines. Two independent screens were performed for each cell line, and the extent of inhibition was displayed as percent arrest, which was calculated for each individual shRNA response averaged from quadruplicate points and normalized to the value of GFP-expressing lentiviral vector. Percent arrest values were then ranked from the most potent inhibitor of cell growth (in red), to the least effective inhibitor (in green). A complete list showing rank order values in 786-O and HeLa cells are provided in Table S1.
Fig. 1.
Differential kinase requirements in 786-O renal cancer cells and HeLa cervical cancer cells. Two independent screens were performed for each cell line. Reduction in viability induced by each hairpin was calculated relative to value of GFP-expressing lentiviral vector as a nonkilling control and then rank ordered. Color scale ranges represent decreases in viability; red corresponds to the greatest inhibition and green the least. Columns display two different cell lines tested in replicate. Rows display shRNAs. Data were clustered by using an Euclidean distance algorithm.
The heat map showed differences in response at a single dose of lentivirus-expressing shRNAs measured at a single time point. Therefore, the next step was to assay multiple doses of lentivirus-expressing shRNAs at multiple time points after viral transduction. The final criteria for assigning a kinase as better killers in HeLa than 786-O (group 1 shRNAs) or as better killers in 786-O than HeLa (group 2 shRNAs) was based on their ability to sustain differential growth activities under several assay conditions. We required an shRNA response with multiple shRNAs, over a range of viral shRNA concentrations, at different time points following shRNA expression, and with corresponding changes in mRNA levels.
For these tests, we started with a smaller subset of shRNAs. We originally established this subset in other studies designed to look for kinases that showed pronounced differences among cell lines and that we hoped might become interesting candidates for drug development strategies. Specifically, the set was chosen based on the following criteria: (i) a representation of shRNAs that showed strong differential activities in our previous test panel of 21 cell lines, and (ii) kinases that have been poorly studied to date (16). Because we were not attempting to examine all differential killers, we chose 36 kinases as a smaller shRNA collection that enabled more in-depth studies with multiple shRNAs for each kinase (Table S2). As shown in Fig. S2, they were representative of the full range of responses for the 100 hits in HeLa and 786-O cells. For our studies, we examined 5 shRNAs for each of these kinases to follow the responses from different levels of shRNA knockdown. This test set therefore contains 173 shRNAs (≈5 shRNAs for each kinase).
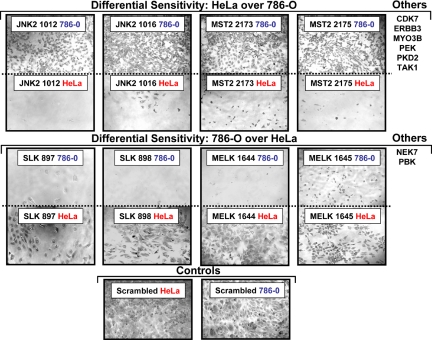
Two sets of comparative experiments were designed. First, a time course of shRNA knockdown was used to study the specific kinases required for cell proliferation. HeLa and 786-O cells were transduced with the subset of shRNA lentiviruses, and viability was assessed by alamarBlue at Days 1, 3, 6, and 9 after transduction. We looked for cases among the 36 kinases in which there were differential sensitivities between the two cell lines. Fig. 2 shows photomicrographs of cultures infected with two different shRNAs for several representative kinases that showed differential sensitivities between HeLa and 786-O cells. The predicted loss of 8 kinases inhibited the growth of HeLa cells dramatically, but did not affect the proliferation of 786-O cells. Conversely, the predicted loss of 4 kinases affected 786-O more profoundly than HeLa cells. Other shRNAs either showed little inhibition, or the patterns between HeLa and 786-O cells were similar. For the kinases in which differences were seen, there were two or more shRNAs per kinase that gave similar results.
Fig. 2.
Differential kinase requirements in HeLa and 786-O cells assayed by time course. Photomicrographs of HeLa and 786-O cell lines transduced with viruses show differential sensitivities to kinase down-regulation. Images of crystal violet stained cells were taken 6 days posttransduction. Additional shRNAs that demonstrate similar differential growth inhibition in this assay are listed at the right.
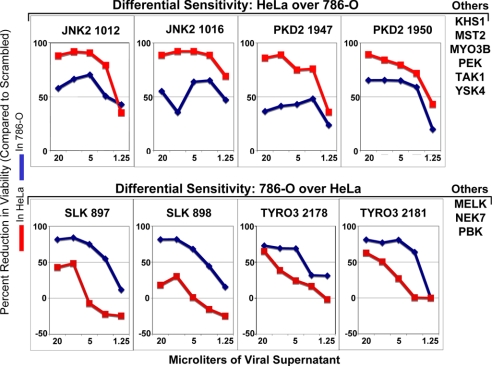
Extremely high levels of perturbants, such as small molecule inhibitors or retroviral shRNA inhibitors, may give nonspecific inhibitions. To be in the linear range of the assay to extract the most useful information, we used a wide range of viral MOIs, from a very high dose of 20 μl in a 96-well format to a very low dose of 1.25 μl, to deliver shRNAs to cells. AlamarBlue readings were made at 6 days posttransduction, and values were plotted by using percent growth inhibition, which was calculated for each viral concentration by normalizing to a scrambled nonkilling control at the same concentration (Fig. 3). At higher concentrations there is nonspecific growth inhibition in control wells, which translates into an artificial decrease in percent growth inhibition at the highest viral concentrations, as seen for JNK2, PKD2, and SLK. At lower concentrations, the curves shown in Fig. 3 depict kinases in which shRNA expression inhibited the proliferation of one cell relative to the other. Eight kinases were identified in which HeLa cells were more sensitive to kinase loss than 786-O cells, and 5 kinases in which 786-O cells were more sensitive than HeLa cells.
Fig. 3.
Differential kinase requirements in HeLa and 786-O cells assayed by viral titration. HeLa and 786-O cells were transduced with viruses at concentrations ranging from 20 μl to 1.25 μl. Cells were grown in the absence of puromycin. AlamarBlue measurements were taken 6 days posttransduction, and the reduction in viability was compared to wells infected with an shRNA containing a scrambled target sequence as a nonkilling control. shRNAs that induce differential killing effects between the two cell lines are shown.
Based on time course and titration experiments, 15 kinases showed cell-type specific effects on down-regulation. Ten kinases were identified as being more important for HeLa viability than for 786-O, and their shRNAs are referred to as group 1 shRNAs. These were CDK7, ERBB3, JNK2, KHS1, MST2, MYO3B, PEK, PKD2, TAK1, and YSK4. Five kinases, MELK, NEK7, PBK, SLK, and TYRO3, were found to be more important for 786-O viability than for HeLa, and their shRNAs are referred to as group 2 shRNAs. There was extensive overlap of the kinases identified by the time course and titration approaches. Both approaches were able to identify shRNAs whose actions suggested differential kinase requirements in these cells, and therefore we kept any kinase that scored in either assay for future comparisons.
There was also good overlap of the kinases identified by the heat map versus time course and titration approaches, however, there were some differences. We understand the reasons for these differences and explain the limitations of the initial screening results in the discussion section.
Validation of shRNA Effects.
Three sets of experiments were used to demonstrate that the differential effects of shRNA expression in HeLa and 786-O cells were due to the specific down-regulation of kinase mRNA and that down-regulation occurred irrespective of the functional outcome. The first test was to show that similar phenotypes were induced by multiple shRNAs that target different regions of the kinase mRNA. Table S3 lists the various shRNA constructs for each of the 15 kinases. In each case, there are multiple shRNAs that show similar phenotypic changes, making it unlikely that the resulting phenotypes were due to off-target effects. Photomicrographs of five shRNAs each for three representative kinases showing the extent of inhibition in HeLa are presented in Fig. S3.
A second, more direct test was to measure the knockdown levels of mRNA after the introduction of shRNAs into HeLa and 786-O cells. First we show starting mRNA levels in both cell lines for JNK2, MST2, SLK, and TAK1 (Fig. S4). The starting RNA levels for these kinases were somewhat higher in 786-O cells, but this did not affect the sensitivities, as SLK was more essential in 786-O cells. Similar patterns were seen for other kinases; the initial mRNA level did not indicate which cell would be more sensitive to its loss (data not shown). To expand on this notion, we have included microarray-based RNA expression data for 13 out of 15 kinases in each of the cell lines (Fig. S5). For the majority of kinases, the RNA expression levels are higher in 786-O cells. Even so, levels do not significantly differ, further supporting our notion that differences in expression levels are not the major reason for determining sensitivity.
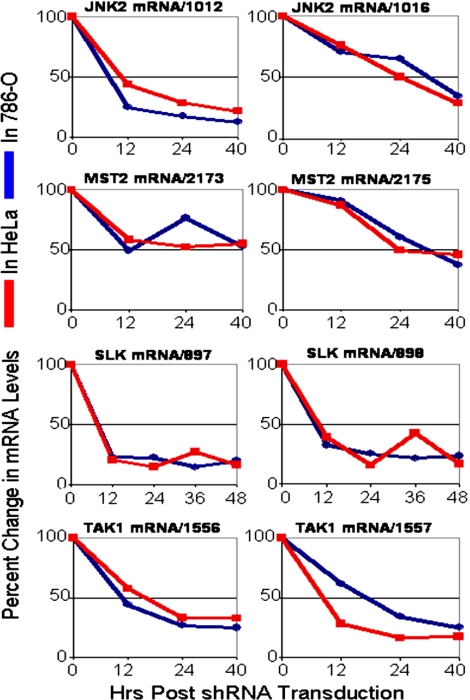
Next, we measured the knockdown mRNA levels for JNK2, MST2, SLK, and TAK1 kinases after transduction by two independent shRNAs into HeLa and 786-O cells (Fig. 4). After shRNA viral transduction, the decay profiles in HeLa and 786-O cells are similar between the two cell lines over time, even though the inhibition of proliferation or survival is seen only in one cell. These data suggest that the timing and extent of decay is due to the specific shRNA sequence and that, at least for these four pairs of two shRNAs per kinase, the RNAi machinery works similarly in the two cell lines tested. Fig. S6 shows the profile of GAPDH mRNA during lentivirus transductions. GAPDH mRNA levels do not change significantly during transduction, arguing that the specific decay seen in kinase levels is due to shRNA action and is not a consequence of the impending cell death.
Fig. 4.
Target mRNA levels in HeLa and 786-O cells. Panomics QuantiGene Assays were used to quantify mRNA levels in HeLa and 786-O cells. Target transcripts were measured before transduction (0 h) and at indicated time points after transduction. Gene-specific transcripts were normalized to GAPDH, and percent decay was calculated relative to scrambled shRNA control. The results indicate similar dose-response of target knockdowns between the two cell lines.
A third test compared the extent of kinase mRNA knockdown with the severity of the phenotype. Fig. S7 shows mRNA levels of MST2 and TAK1 kinases in HeLa cells 24 h after transduction and photomicrographs of the infected cells 6 days after transduction. The degree of growth inhibition correlated with the level of kinase mRNA knockdown. Interestingly, for TAK1 the severity of the phenotype is not directly proportional to the level of knockdown of the mRNA. Rather there appears to be a threshold of mRNA knockdown, and when mRNA loss exceeds this threshold, cell viability is compromised.
Patterns of Kinase Dependence in One Cell Can Predict Patterns of Response in Other Closely Related Lines.
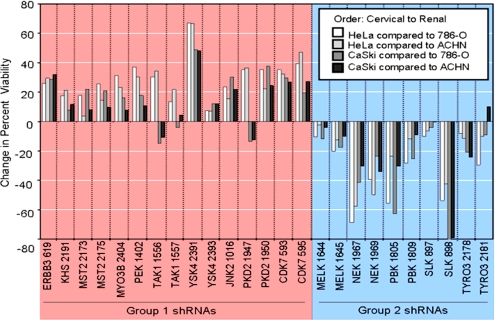
To test whether the sensitivities to the loss of 15 kinases identified above would have any predictive value, we tested other cells lines from the same tissue of origin as HeLa and 786-O for the effects of kinase down-regulation. We knew from previous experiments that tissue and cell type of origin can lead to similar kinase requirements (16); therefore, we tested how three additional cervical carcinoma cell lines and three additional clear cell renal carcinoma lines responded to kinase loss (Fig. S8A and Fig. S8B). In both cervical and renal cell carcinomas, we were able to identify one additional line that responded to kinase loss in manner very similar to that of the reference cell line. For cervical cell lines, the patterns with CaSki cells were very similar to HeLa's, whereas the other two cervical cell lines tested, SiHa and C33, were quite different. For renal carcinomas, ACHN cells were very closely related to 786-O, whereas A498 were less similar and RCC4 were quite different from 786-O cells. Fig. 5 shows all of the possible comparisons of the two most closely related cervical lines and the two most closely related renal lines. With minor variations, the shRNAs originally identified as better killers in HeLa cells inhibited the CaSki cell line better than either 786-O or ACHN cells, whereas the shRNAs identified as better killers in 786-O inhibited the ACHN line better than HeLa or CaSki cells. Based on a limited number of shRNAs and a limited number of cells line, our initial findings suggest that the response to inhibition of kinases in one cell line may be predictive of the functional state of other related cell lines.
Fig. 5.
Differential kinase requirements in cervical and renal cell lines. HeLa (cervical), CaSki (cervical), 786-O (renal), and ACHN (renal) cells were transduced with 25 shRNA-expressing viruses targeting 15 genes previously identified as preferential killers in HeLa (group 1 shRNAs) or 786-O (group 2 shRNAs) cells. AlamarBlue measurements were taken six days after transduction, and difference in percent reduction in viability was calculated between pairs of cervical and renal cell lines.
Discussion
In the four papers of this series (16, 17, 18, and the present article), we have examined methods to compare the kinases requirements in various human cancer cell lines. The work has demonstrated that cells come from different tissues or experience different mutations during tumor development often have different rate limiting steps for proliferation and survival after the reduction of kinase levels through shRNA treatments (16). When cells are closely related and differ only by expression of a single oncogene or tumor suppressor gene, shRNA methods can be used to detect differences that stem from the action of these proteins (17, 18). In the proof-of-concept experiments presented here, we use similar strategies to examine how cancer cells that arise from different tissue sources and that suffer different tumorigenic mutations respond to perturbation by shRNA treatment. These focused shRNA-based screens identified 15 kinases that showed differential requirements in two commonly used human cell lines, the cervical HeLa and renal 786-O carcinoma cell lines. These cells proliferate at similar rates, grow in the same media, and are similarly susceptible to lentivirus shRNA transduction. The set of shRNAs that were used for these screens was a small group of 100 shRNA hits identified in earlier screens. This set was chosen not to be exhaustive in finding differences in kinase requirements but rather to be a useful set to develop strategies for careful comparisons of shRNA treatments. Our goals were twofold: to learn how differences in kinase requirements could be teased out and confirmed, and to determine whether the identified kinases might have interesting patterns of requirements in other cell lines. Central to the approaches developed here was measuring the cellular responses to titrations of shRNA levels and following the shRNA responses over time. We have found that simple comparisons in our primary screens can be confounded by off-target effects or limited resolution due to single viral doses or single time point measurements, but that carefully identified differences using methods such as titrations and time courses can identify patterns of kinase requirements that may help categorize the a cell's functional state.
After analysis of the responses to shRNA treatment from the 100 hits, we chose 36 likely kinase candidates for more extensive studies. We used five shRNAs for each kinase, examining continued cell proliferation in the presence of shRNA-induced kinase knockdown and measuring proliferation in the presence of increasing levels of shRNA. Fifteen kinases whose roles are required in one cell but appear to be more dispensable in another were identified. There was extensive overlap among the kinases identified by the time course and titration approaches, and these assays suggest that HeLa cells are more sensitive to the continued expression of CDK7, ERBB3, KHS1, JNK2, MST2, MYO3B, PEK, PKD2, TAK1, and YSK4 kinases for proliferation, as compared to 786-O. Conversely, 786-O cells are more dependent on the presence of MELK, NEK7, PBK, SLK, and TYRO3 kinases, as compared to HeLa cells. The roles of these kinases were confirmed by showing similar phenotypic responses to two or more shRNAs to each kinase mRNA, by following mRNA decay over time induced by multiple shRNAs, and by correlating the extent of phenotypic response to mRNA knockdown.
Cells Respond Differently to Kinase Down-Regulation.
Our data demonstrate that RNAi screens can be used to identify kinases that are more important in one cell line than in another. These differences identify rate-limiting points for proliferation and/or survival in these cells. Based on earlier results, we expected that the HeLa and 786-O cells would show differences because of their origin from cervical keratinocytes and renal epithelial cells, respectively. When we used the 15 kinase differences to examine other cervical and renal carcinoma cell lines, we found one additional cervical tumor cell line and one additional renal carcinoma cell line that had similar patterns of response to loss of test kinases, whereas other cells, even from the same tissue, were more distantly related. Some of the lines derived from the same tissue appeared no more closely related than cells from other tissues. This suggests that some cancer cells from the same tissue origin may have related patterns of kinase requirements, whereas others may have developed under different pressures and may be functionally unrelated as judged by shRNA sensitivities, at least from this small panel of shRNAs.
Origins of Differential Kinase Requirements.
We do not know at present what the molecular origins of the differential kinases requirements for HeLa and 786-O are. As mentioned above, there are no obvious connections to known signaling pathways among the 15 kinases identified here, nor are there biochemical activities that would suggest common reasons for their selection. Also, none of these kinases has been identified as being frequently mutated in human cancers.
One can imagine several events that would contribute to differential requirements. As mentioned above, it is likely that tissue and cell lineage of origin is an important variable. We base this on two observations. First, primary cells from the same tissue and lineage have very similar patterns of kinase requirements, but the kinase requirement from different cell lineages are quite distinct (16). Second, we have not yet seen similar patterns of kinase requirements between any two tumor cells that arise from different sites (16 and data not shown). A second source of change in kinase requirements is the various oncogene or tumor suppressor gene mutations or epigenetic changes that occur during tumorigenesis. We know that single oncogene or tumor suppressor mutations induce a small set of discernable changes (17, 18). We anticipate that these changes will be context and order specific, and the compendium of tumorigenic changes that drive cancer development will lead to complicated differences in kinase requirements.
A third source of differences will be changes that are selected based on adaptation to tissue culture. In addition, other random genetic or epigenetic changes might occur as genomic instability develops during tumorigenesis. Together these changes will contribute to the array of differential responses we have seen.
Kinase Differences and Results in Primary Screens.
After identifying the 15 kinase differences described above, we reexamined how these kinases behaved in our initial screen. For the data depicted in the heat map of Fig. 1, only 8 of the final list of 15 kinases were correctly identified as members of final groups 1 and 2. Most discordant were the single shRNAs for KHS1 (shRNA 2187) and SLK (shRNA 894), which showed differential activity in the opposite direction than we eventually determined based on the criteria above. This arose from several confounding events. In both cases the single shRNA from Fig. 1 continued to score against the cell line first identified in the heat map data, but no additional confirmatory shRNAs could be identified. This suggested that the result was reproducible but called into question whether it was a reliable on-target effect. This result was confounded as more detailed studies using different shRNAs at various concentrations and measured over different time points were examined. Multiple shRNAs that met our criteria for determining a positive were found that now affected the comparative cell line. These results were uncovered only as the data set was expanded. We feel confident that the criteria of multiple shRNAs, time courses of shRNA knockdown, and correlation of phenotypic response with level of mRNA knockdown is sufficient to identify these kinases as differentially required, and we are left to propose that the original single shRNAs scored only through hitting unintended targets. This is clear support for the growing appreciation in the RNAi field that off-target effects are common and that follow-up experiments are essential to gain confidence in the selection of hits. It also supports the advantages of using assays such as titrations and time courses to expand the range at which responses are measured and highlights the problem of relying on a single time point or shRNA concentration for screening results. These observations also argue that primary screening data need to be approached in one of two general strategies: either initial hits must be selected as having two or more shRNAs to identify a positive or, if single strong hits will be followed, the secondary screens must be able to distinguish between off-target and on-target effects. In this case, if the original heat map was our only screening tool, the initial comparison would have misclassified these two kinases as required in the two cell lines, while more detailed studies sorted out the actual preference in these cells.
Other differences from the comparison of the final classification and original heat map arose from examples that showed no differential effects in the heat map, but kinases were ultimately assigned to group 1 or 2. This included CDK7, MELK, MST2, MYO3B, and PKD2. These were missed in the original data because the differential effects were seen only on titration of virus and through following the time course of shRNA effects.
In conclusion, we were not able to discern 7 of identified 15 kinases as differentially required based on our original data sets, but on closer examination the patterns were teased out in a careful stepwise fashion using titration and time course experiments. In simple terms, primary screens done with single viral concentrations and at one time point may be too crude to find the correct time and titration at which to identify a comprehensive list of kinases important for cell proliferation or survival.
Identifying Promising Candidates for Drug Targets.
Although it is not known how well shRNA knockdowns will predict the effectiveness of an eventual small molecule inhibitor, the identification of kinases whose loss leads to selective inhibition of cell proliferation or viability of tumor cells provides a reasonable method to identify interesting candidates for consideration in drug development. Among the kinases that were studied in detail in our studies, several have characteristics that make them promising candidates for cancer chemotherapeutic studies. These characteristics include strong lethal phenotypes and preferential killing of one cell type over the other. The essential nature of these kinases coupled with their infrequent mutation in cancer cells raises an interesting feature of RNAi screening. Unlike straight mutant hunts for proteins whose alterations drive cancer development, RNAi screens provide an unbiased approach to identify all essential proteins, including both the mutated cancer genes themselves and those that become essential only after the action of other cancer mutations. One can imagine the identification of proteins, in our case kinases, that become essential only after cells suffer cancer mutations. Although one can't know how powerful a target class such proteins would be, the possibility of finding a large number of such proteins and finding them in a rapid and reliable manner should be a powerful approach.
Materials and Methods
Tissue Culture.
Human cervical cancer cell lines HeLa, CaSki, SiHa, and C33A were a gift from the lab of Dr. Karl Munger. Human renal cancer cell lines 786-O, ACHN, A498, and RCC4 were a gift from the lab of Dr. William Kaelin. All cell lines were cultured in DMEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 4 mM L-glutamine, and 10% FBS at 37°C, with humidified atmosphere and 5% CO2.
One Hundred Hits DNA and Virus Production.
High quality DNA preparations for 100 hits were obtained using a large-scale plasmid purification kit (Qiagen). For high-throughput lentiviral production in a 96-well format, 293T packaging cells were cotransfected with shRNA-encoding replication deficient viral vectors and the necessary helper plasmids for virus production. The virus was pseudotyped with the envelope glycoprotein from vesicular stomatitis virus (VSV-g) as previously described (13, 23, 24).
Transductions in a 96-Well Format.
Cells were seeded between 2,000 and 5,000 cells/well by using a 96-well format in a final volume of 100 μl/well. Twenty-four hours later, 50 μl of media was removed and different amounts of viral supernatant were added depending on the experiment (ranging from 1.25 μl to 20 μl) in the presence of 8 μg/ml polybrene final concentration. Plates were spun at 1,178 × g for 30 min. Infected cells were washed between 12–16 h posttransduction, and 24 h later, 2 μg/ml puromycin was added to select wells. Cells were harvested 6 days posttransduction for alamarBlue measurements and for crystal violet image acquisition.
AlamarBlue Assay.
Five or 6 days posttransduction, media were removed from 96-well plates and alamarBlue reagent (Biosource/Invitrogen) diluted 10-fold in supplemented DMEM was added to each well. Plates were then incubated for 2–4 h at 37°C before reading on a microtiter well plate reader at 595 nm.
Crystal Violet Staining.
Six days posttransduction, media were removed. Cells were washed with PBS and fixed with 10% acetic acid and 10% methanol. After 30 min, cells were washed with PBS and incubated for 2 h with 0.4% crystal violet in 20% ethanol, followed by two final PBS wash steps. Phase contrast images were acquired at a magnification of 100×.
mRNA and Genomic RNA Analysis.
For mRNA quantitation of transduced HeLa and 786-O cells, the transduction protocol was performed in 12-well plates with 120 μl of viral supernatant. After transduction, plates were spun at 1,178 × g for 30 min. Infected cells were washed 12 h posttransduction and harvested at different time points by lysing cells in buffer supplied with Panomics QuantiGene assay according to the manufacturer's recommended procedure. Target-specific probes were incubated with 80 μl of cell lysate. GAPDH control-specific probes were incubated with 5 μl of cell lysate that diluted 1:10. To calculate the percent knockdown, we normalized the target values to GAPDH values and then to scrambled shRNA control. To assess the relative titers of viruses produced in a 96-well format, we measured the puromycin-N-acetyl transferase (PAC) genomic RNA levels from viral supernatants according to the manufacturer's protocol. In brief, 5 μl of virus supernatant was diluted in 75 μl of complete medium and lysed with 40 μl of lysate buffer. The samples were processed as above.
Supplementary Material
Acknowledgments.
We would like to thank Drs. Karl Munger (Brigham and Women's Hospital, Boston, MA) and William Kaelin (Dana Farber Cancer Institute, Boston, MA) for the cervical and renal carcinoma cell lines, respectively. We are very grateful to Drs. John Doench and Andrew Conery for critical reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806578105/DCSupplemental.
References
- 1.McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huppi K, Martin SE, Caplen NJ. Defining and assaying RNAi in mammalian cells. Mol Cell. 2005;17:1–10. doi: 10.1016/j.molcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Tomari Y, Zamore PD. Perspective: Machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 4.Kittler R, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 5.Mackeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 6.Neumann B, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 7.Pelkmans L, et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- 8.Zheng L, et al. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc Natl Acad Sci USA. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 10.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 11.Kolfschoten IG, et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Westbrook TF, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 13.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 14.Schlabach MR, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva JM, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grueneberg DA, et al. Kinase requirements in human cells: I. Comparing kinases requirements across various cell types. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0808019105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin A, et al. Kinase requirements in human cells: II. Genetic interaction screens identify alterations in kinase requirements following HPV16 E7 expression in cancer cells. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0806195105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bommi-Reddy A, et al. Kinase requirements in human cells: III. Altered kinase requirements in VHL-/- renal carcinoma cells detected in a pilot synthetic lethal screen. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0806574105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seedorf K, Oltersdorf T, Krammer G, Rowekamp W. Identification of early proteins of the human papilloma viruses type 16 (HPV 16) and type 18 (HPV 18) in cervical carcinoma cells. EMBO J. 1987;6:139–144. doi: 10.1002/j.1460-2075.1987.tb04731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Tumour suppression by the human von Hippel-Lindau gene product. Nature Medicine. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 21.O'Brien J, Wilson I, Orton T, Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama GR, Caton MC, Nova MP, Parandoosh Z. Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J Immunol Methods. 1997;204:205–208. doi: 10.1016/s0022-1759(97)00043-4. [DOI] [PubMed] [Google Scholar]
- 23.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 24.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.