Abstract
Hsp110 proteins are relatives of canonical Hsp70 chaperones and are expressed abundantly in the eukaryotic cytosol. Recently, it has become clear that Hsp110 proteins are essential nucleotide exchange factors (NEFs) for Hsp70 chaperones. Here, we report the architecture of the complex between the yeast Hsp110, Sse1, and its cognate Hsp70 partner, Ssa1, as revealed by hydrogen–deuterium exchange analysis and site-specific cross-linking. The two nucleotide-binding domains (NBDs) of Sse1 and Ssa1 are positioned to face each other and form extensive contacts between opposite lobes of their NBDs. A second contact with the periphery of the Ssa1 NBD lobe II is likely mediated via the protruding C-terminal α-helical subdomain of Sse1. To address the mechanism of catalyzed nucleotide exchange, we have compared the hydrogen exchange characteristics of the Ssa1 NBD in complex with either Sse1 or the yeast homologs of the NEFs HspBP1 and Bag-1. We find that Sse1 exploits a Bag-1-like mechanism to catalyze nucleotide release, which involves opening of the Ssa1 NBD by tilting lobe II. Thus, Hsp110 proteins use a unique binding mode to catalyze nucleotide release from Hsp70s by a functionally convergent mechanism.
Keywords: chaperone, nucleotide exchange factor, Sse1, protein folding H/D exchange
Members of the Hsp70 class of chaperones play essential roles in many distinct cellular processes by facilitating protein folding or conformational transitions (1, 2). The Hsp70 chaperone transiently associates with its substrates in a manner controlled by its ATPase cycle. ATP binding to the N-terminal nucleotide-binding domain (NBD) induces conformational changes in the C-terminal substrate-binding domain (SBD) that result in low affinity for substrate. Hydrolysis of ATP converts the Hsp70 to the ADP state, which traps the substrate with higher affinity. Exchange of ADP for ATP and concomitant substrate release completes the chaperone cycle.
The basic chaperone cycle of Hsp70s is modulated by cofactors that regulate either ATP hydrolysis or exchange of ADP for ATP. The hydrolysis of ATP is stimulated by J-domain-harboring co-chaperones. Nucleotide exchange factors (NEFs) accelerate nucleotide release by stabilizing conformational states in the NBD that exhibit low affinity for nucleotides. Rebinding of ATP then triggers dissociation of the NEF. Interestingly, crystallographic studies of the Hsp70 NBD in complex with the 3 structurally unrelated NEFs GrpE, Bag-1, and HspBP1 revealed that each NEF utilizes a unique mode of binding (3–5). Despite distinct binding interfaces, the resulting nucleotide release mechanism triggered by both bacterial GrpE and eukaryotic Bag-1 is similar and involves outward rotation of lobe IIB of the NBD (4). In contrast, HspBP1 embraces NBD lobe II, and a resulting steric conflict displaces lobe I (5).
Recently, several members of the Hsp70 superfamily have been identified as NEFs for canonical Hsp70 chaperones. For example, in the cytoplasm of eukaryotes, the Hsp110 proteins associate with Hsp70s to mediate nucleotide exchange (6, 7). The Hsp110 share their overall structure with canonical Hsp70s but differ by an extended SBD with an acidic loop region inserted between the terminal strands of its β-sheet subdomain and an extended flexible C terminus (8). The yeast Saccharomyces cerevisiae harbors two highly homologous members of the Hsp110 family, the abundant Sse1 and the less expressed Sse2. SSE1 and SSE2 constitute an essential gene pair (9), but viability of null mutant cells can be restored by overexpression of the yeast HspBP1 homolog Fes1 or the Bag-1 domain of yeast Snl1 (7, 10). Given the severity of the sse1 sse2-null phenotype and the high abundance and NEF activity of Sse1/2, Hsp110 appears to constitute the main NEF for cytoplasmic Hsp70s in yeast.
At present, little is understood regarding the association of Hsp110 with Hsp70 and the mechanism of nucleotide release. The Hsp70 NBD has been shown to be sufficient for interaction, whereas on the Hsp110 side both the NBD and SBD could be involved (11, 12). Sse1 is also activated by ATP. Upon ATP binding the protein adopts a stabilized conformation that is required for association with its cognate Hsp70, Ssa1 (12). However, after the complex has formed, nucleotide can be completely removed from Sse1 without any perturbation of the integrity of the complex. The recently published crystal structure of ATP-bound Sse1 thus potentially represents the NEF-active conformation of Sse1 (13). This structure depicts a molecule with extensive contacts between the NBD and both the α-helical and the β-sheet parts of the SBD.
Here, we report that Sse1 and Ssa1 use a NEF-binding mode. The NBDs of Sse1 and Ssa1 associate asymmetrically face-to-face, and the C-terminal α-helical domain of Sse1 plays an important role in the interaction. Our data suggest that nucleotide release is triggered by a GrpE/Bag-1-like mechanism.
Results
ATP Induces Extensive Stabilization of the NBD of Sse1.
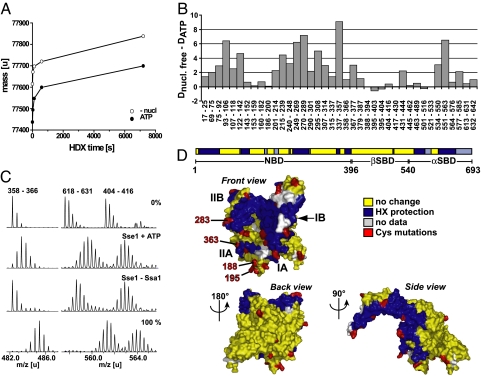
To understand how Hsp110 acts on Hsp70, we first investigated how ATP binding to Sse1 prompts interaction with Ssa1 (12) by applying amide hydrogen exchange (HX) combined with mass spectrometry. With this technique, conformational changes and binding interfaces to partners or ligands can be resolved by monitoring changes in solvent accessibility of amide hydrogen positions in the protein (14). Consistent with previous observations, we found that ATP induces extensive stabilization of Sse1 (Fig. 1A). We mapped the observed changes to precise regions of the molecule by combining our HX setup with online pepsination followed by mass spectrometry [supporting information (SI) Table S1]. We mainly detected two classes of peptides: (i) peptides that exhibited HX protection in the presence of ATP compared with the nucleotide-free condition, and (ii) peptides that did not show any significant difference in exchange properties between the two conditions. Strikingly, 17 of the 24 analyzed segments encompassing the N-terminal NBD exhibited extensive protection upon ATP addition (Fig. 1B, segments 17–25 to 379–387). The changes correspond to 10–77% of the maximal possible deuteron incorporation in the respective peptides. In contrast, ATP induced no pronounced effects on segments derived from the β-sheet subdomain of the SBD (Fig. 1B, segments 395–403 to 521–533). The α-helical subdomain of the SBD also exhibited ATP-induced protection localized to 3 of its 6 segments (Fig. 1B, segments 534–550, 551–563, and 577–585). This protection could reflect a local stabilization consistent with the docking of the α-helical domain against the NBD as observed in the Sse1(ATP) crystal structure (13).
Fig. 1.
Ssa1 association induces HX protection in both the front face of the NBD and the C-terminal α-helical domain of ATP-bound Sse1. (A) Stabilization effect of ATP binding to Sse1, as monitored from the amide proton exchange properties of the protein. Preincubated protein samples in the presence (filled symbols) or absence (open symbols) of ATP were diluted 20-fold into D2O buffer and incubated for different time intervals at 30 °C before quenching the proton–deuteron exchange reaction. The mass increase through deuteron incorporation was determined for Sse1. (B) Difference in deuteron incorporation between nucleotide-free Sse1 and Sse1 + ATP. The data were resolved to individual peptic peptides as indicated by the start and end residue numbers of the corresponding segments (Table S1). (C) HX properties of Sse1 + ATP and Sse1 bound to Ssa1. Mass spectra of representative peptides for protein samples incubated in H2O (0%) or for 2 min in D2O buffer. 100% shows a control spectrum of the same peptide from fully deuterated Sse1. (D) Observed HX protection pattern induced by Ssa1 association, derived from comparison of Sse1 + ATP and Sse1–Ssa1 (see C and Fig. S2) (13). The color code for the peptides is as follows: blue, Ssa1-induced protection; yellow, no change in HX properties upon association with Ssa1; and gray, no data available. The 29 residues that were mutated to cysteine for cross-linking experiments are marked in red (residue numbers 22, 34, 98, 135, 188, 194, 195, 218, 222, 254, 283, 290, 304, 331, 346, 355, 363, 374, 450, 529, 566, 576, 586, 595, 609, 630, 639, 650, and 654). The first projection corresponds to the standard view of Hsp70 NBD, and the domain nomenclature follows Flaherty et al. (23).
We asked whether the HX properties of Sse1 correlated with those of the characterized Hsp70 DnaK (15). We found that overall, the ATP-induced stabilization of the NBD of Sse1 was more extensive than for DnaK. Moreover, upon comparing ADP and ATP conditions for Sse1, we found an identical pattern of nucleotide-induced HX protection (Fig. S1). We conclude that Sse1 does not display conformational changes characteristic of Hsp70 chaperones in response to nucleotide binding. Therefore, Sse1 lacks ATP-dependent interdomain communication.
Taken together, these data indicate that the Sse1 NBD requires ATP to fold and compact, inducing a conformation active in nucleotide exchange. From these data, it seems likely that the NBD of Sse1 represents an important interaction site for Hsp70 and that the α-helical subdomain might also be involved.
Ssa1 Inhibits HX in Sse1 at the Front Face of the NBD and at the α-Helical SBD Subdomain.
ATP-bound Sse1 undergoes further HX protection when it associates with Ssa1 (12) as a consequence of direct shielding of amide hydrogens at the interaction interface or of locally decreased dynamics of the molecule. To identify the interaction interface, we compared HX properties of monomeric Sse1 + ATP or Sse2 + ATP with those of stable Sse1/2–Ssa1 complexes at the level of peptides (Tables S2 and S3 and Fig. S2). Fig. 1C shows representative spectra of Sse1 peptides harboring protection (peptides 358–366 and 618–631) or no change upon Ssa1 interaction (peptide 404–416). Because the Sse1 and the Sse2 datasets featured a very similar exchange behavior (88% of overlapping segments showing identical results), we pooled the two datasets and mapped the peptides exchange characteristics on the recently published crystal structure of ATP-bound Sse1 (Fig. 1D) (13). From here on, we will use the Hsp70 nomenclature and perspective presented in Fig. 1D as a reference point when referring to different regions of Sse1 and the NBD of Ssa1.
The protected peptides in Sse1/Sse2 showed an average protection ranging from 0.4 to 4.9 amide protons and localized to both the NBD (regions 17–68, 122–142, 250–289, 358–366, and 379–394) and the α-helical subdomain of the SBD (regions 551–585 and 606–653). Within the NBD, the protected peptides mapped to the front face of the NBD, whereas we did not observe any Ssa1-induced protection at the backside of the NBD (Fig. 1D). Thus, our HX data indicate that both the front face of the NBD and the α-helical subdomain play important roles in the interaction of Sse1/2 with the NBD of Ssa1 and might potentially represent direct contact points. In contrast, direct contacts on the back of the NBD, including the β-sheet subdomain, seem improbable.
Positions at Sse1 NBD Subdomain IIA and IIB Cross-Link to Ssa1.
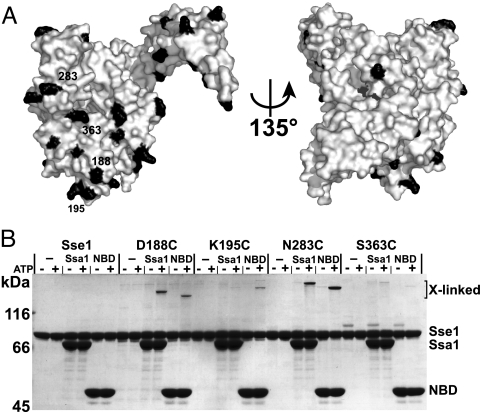
We next tested the putative Ssa1 interaction sites identified with HX by using site-specific cross-linking of Sse1. In total, we introduced 29 single cysteine mutations in the functional cysteine-free allele sse1Δcys (Fig. S3), mainly at positions located within or adjacent to regions of Ssa1-induced HX protection (in red in Fig. 1D). Each protein was purified and labeled at its unique cysteine with the heterobifunctional UV-activated cross-linker benzophenone-4-iodoacetamide (BPIA) (length 10 Å). Four of the 29 proteins formed detectable cross-links with Ssa1 and with the isolated NBD of Ssa1 (Fig. 2). The use of the isolated Ssa1 NBD excluded the possibility that the observed interactions are those of Hsp70 with substrate. Furthermore, appearance of the cross-links was strictly dependent on the presence of ATP in the reaction, a requirement for Sse1–Ssa1 association (11, 12). The four cross-linked positions D188C, K195C, N283C, and S363C are all located in the front outer rim of NBD lobe II (Fig. 1D). We conclude that this surface of the Sse1 NBD is a bona fide Ssa1 interaction site.
Fig. 2.
Positions at Sse1 NBD subdomain IIA and IIB outer rim cross-link to Ssa1. Each mutant protein (see Fig. 1D) was BPIA-labeled on its introduced cysteine and tested for the ability to cross-link to full-length or the NBD of Ssa1 as described in Materials and Methods. Four BPIA-labeled cysteines formed ATP-dependent cross-links to Ssa1.
α-Helical SBD Subdomain Is Important for the Interaction with Ssa1.
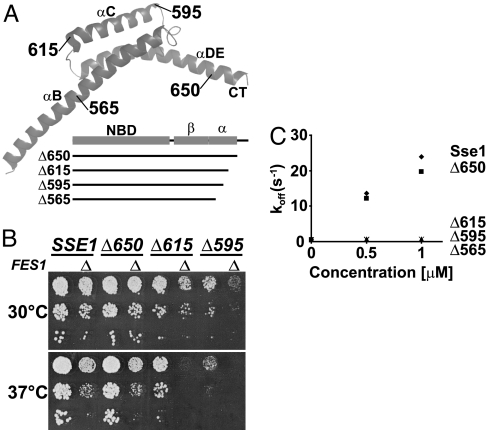
We further tested the importance of the C-terminal α-helical subdomain by using genetic analysis. The Sse1 α-helical subdomain consists of 3 α-helices (αB, αC, and αDE; see Fig. 3A). Based on the crystal structure of Sse1, we designed 4 C-terminal truncation alleles of SSE1 (Fig. 3A) that removed sequences comprising the flexible C-terminal tail (sse1Δ650), and in addition the last helix αDE (sse1Δ615), the helix αC (sse1Δ595), and the protruding part of αB (sse1Δ565). Three of the 4 alleles supported growth of sse1Δ sse2Δ cells and allowed further genetic characterization (Fig. 3B), whereas sse1Δ565 was not functional (data not shown). The cells harboring sse1Δ650 grew indistinguishably from SSE1 control cells at 30 °C and 37 °C, whereas sse1Δ615 cells exhibited a mild, and sse1Δ595 a severe, slow growth phenotype, respectively. Consistent with a defect in the chaperone machinery, the phenotypes of sse1Δ595 and sse1Δ615 became more severe when the cells were grown under heat stress at 37 °C. Moreover, these two sse1 alleles exhibited strong synthetic interactions with a deletion in FES1, the gene coding for the other known cytoplasmic NEF of yeast cells, indicating that the growth defect is specifically linked to a deficiency in NEF activity (Fig. 3B).
Fig. 3.
The α-helical SBD subdomain is important for the interaction with Ssa1. (A) Schematic representation of the truncation mutants of Sse1. The α-helical subdomain (α) is drawn as a ribbon structure (13) with names of helices and the last residue of each truncation mutant indicated. (B) Ten-fold serial dilutions of sse1Δ sse2Δ or sse1Δ sse2Δ fes1Δ (Δ) yeast strains carrying SSE1, or derived truncated alleles, in the context of its endogenous locus on a single-copy plasmid were spotted onto YPD medium and incubated at 30 °C or 37 °C for 2 days. (C) MABA-ADP dissociation rate [koff (s−1)] from Ssa1 (0.5 μM) by the indicated concentrations of Sse1 or derived truncation mutants. The Sse1 proteins were preincubated with 3 mM ATP before measuring the release rates with a stopped-flow instrument.
Cells adapt to protein folding stress by inducing the expression of chaperones, for example, by up-regulating the heat shock response (16). We found that the yeast cells compensated for the activity defects of the sse1 truncation alleles by up-regulating the inducible Hsp70 chaperones Ssa3/4 (Fig. S4). Thus, it is likely that the growth phenotypes actually do not fully reflect the impact the truncation mutations have on the NEF activity of Sse1. To address this directly, we purified the truncated proteins and analyzed their NEF activity in vitro in N8-(4-N′-methylanthraniloylaminobutyl)-8-aminoadenosine 5′-diphosphate (MABA-ADP) release assays (Fig. 3C). Correlating with the full complementation exhibited by sse1Δ650, Sse1650 had 90% of Sse1 activity at equimolar concentration with Ssa1. In contrast, Sse1615, Sse1595, and Sse1565 exhibited only very low NEF activity (<1% of Sse1). In summary, our genetic and biochemical analyses support the conclusion that the α-helical subdomain is important for the NEF activity of Sse1, in agreement with previous reports (6, 17).
Sse1 Interaction Induces Stabilization of Ssa1 Lobe I and Destabilization of Lobe II.
We next sought to understand how association with Sse1 affects the NBD of Ssa1 and causes the release of nucleotide. Upon direct comparison of the HX properties of nucleotide-free Ssa1 NBD alone or in complex with Sse1, we found that Sse1 induced protection in lobe I of Ssa1 (Fig. 4A, average protection of 1.0–2.7 amide protons; see also Table S5 and Fig. S5). In contrast, the other half of the NBD, lobe II, appears to become largely destabilized by the interaction with Sse1 (average deprotection of 0.9–2.3 amide protons). However, one segment that localizes to the top part of lobe II (subdomain IIB) exhibited protection. These effects on the NBD were also observed in Sse1-associated full-length Ssa1 (Fig. S5), indicating that the presence or absence of the Ssa1-SBD does not affect the interaction with Sse1.
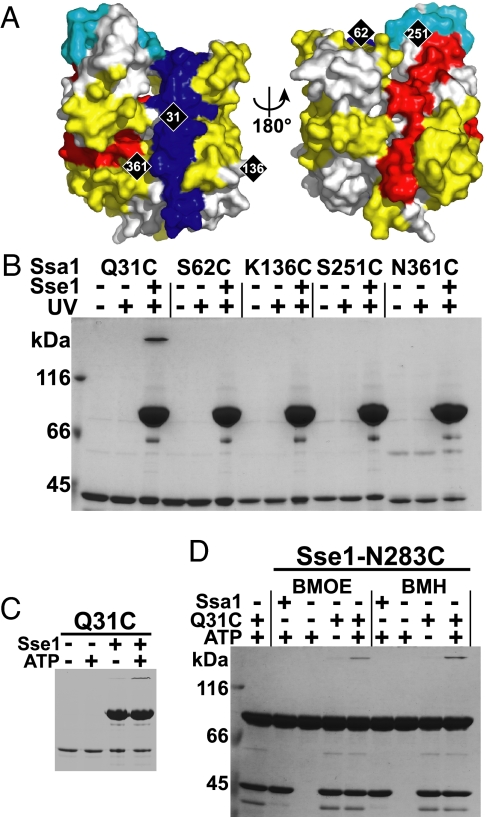
Fig. 4.
Residue 31 on the front face of Ssa1 cross-links with residue 283 on the front face of Sse1. (A) Ssa1 NBD modeled onto the structure of the Hsc70 NBD (23) as viewed from the front and back. The molecule is colored according to the observed pattern of Sse1-induced HX protection (blue), weak protection (cyan), and deprotection (red) (Fig. S5). Yellow indicates no change in HX properties, and gray, no data available. Positions of the 5 introduced cysteines are marked in black. (B) Each mutant protein was BPIA-labeled on its introduced cysteine and tested for the ability to cross-link to Sse1 (Upper). (C) The cross-link at Q31C was tested for ATP dependence (Lower). (D) Sse1-N283C was mixed with the control protein Ssa1 NBD (Ssa1) or the Ssa1 NBD carrying the Q31C mutation (Q31C) in the presence or absence of 1 mM ATP and tested for its ability to form a cysteine–cysteine cross-link with BMOE and BMH.
The protection pattern induced by Sse1 suggests that Sse1 binds to lobe I of Ssa1. In contrast, lobe II is destabilized, suggesting an opening of the NBD structure that might trigger nucleotide release. In support of this notion, ATP binding induces the opposite effect in Ssa1, namely, strong stabilization of lobe II (Fig. S6 and Table S4). The Sse1-induced stabilization of the upper tip of lobe II suggests an additional contact point that might be important to detach the two lobes from each other.
NBD of Ssa1 Interacts Face-to-Face with the NBD of Sse1.
We tested whether the protected region in Ssa1 NBD subdomain I directly contacts Sse1 by attempting to cross-link BPIA-labeled cysteines positioned in the NBD of Ssa1 to Sse1 (Fig. 4A). We found that Q31C-BPIA, situated in a conserved central loop of subdomain IB, formed ATP-dependent cross-links to Sse1 (Fig. 4 B and C). Notably, Q31 of Ssa1 is positioned in a surface loop that is unique to Hsp70s and is essential for the interaction between bacterial DnaK and its NEF GrpE (18). We tested the possibility that also the conserved loop in Ssa1 interacts directly with the NBD of Sse1. We attempted to cross-link Ssa1-NBD-Q31C to our 29 purified Sse1 cysteine mutants by using cysteine–cysteine reactive bismaleimide cross-linkers bis(maleimido)ethane (BMOE), 8 Å; and bis(maleimido)hexane (BMH), 13 Å. Only Sse1-N283C formed ATP-dependent cross-links to Ssa1-NBD-Q31C (Fig. 4D). The constraints given by the localization of the 2 cysteines on each respective NBD and the length of the shortest cross-linker (8 Å) indicate that the 2 NBDs interact face-to-face. Accordingly, the front outer rim of lobe II of Sse1 forms extensive contacts with the inner side of lobe I in Ssa1.
Sse1 Utilizes a Bag-1-Type NEF Mechanism.
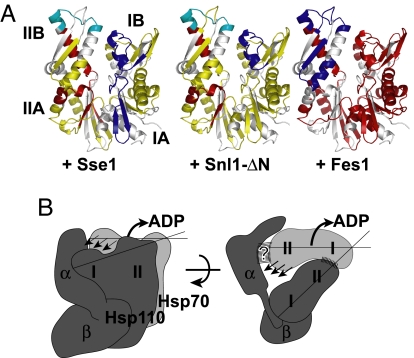
The Sse1-induced deprotection observed in lobe II of Ssa1 suggests an opening of the NBD and a possible mechanism of nucleotide release. To characterize this mechanism further, we investigated whether Sse1 shares any mechanistic features with Bag-1-type or HspBP1-type NEFs. Using HX analysis, we compared the nucleotide-free NBD of Ssa1 in complex with either the HspBP1 homolog Fes1 or the Bag-1 domain of Snl1 (Snl1-ΔN) with the results of our HX measurements of Sse1-associated NBD (Figs. S7 and S8 and Tables S5 and S6).
Fes1 induced massive destabilization of the Ssa1 NBD (Fig. 5A and Fig. S8); most peptides exhibited exchange properties characteristic of unstructured or highly dynamic segments (>80% of available amide hydrogens exchanged within 10 s). In contrast to the overall behavior of the NBD, subdomain IIB became protected in the presence of Fes1. Our HX data on Fes1 are consistent with the published crystal structure of the homolog HspBP1 interacting with a fragment of NBD subdomain IIB of Hsp70 (5). Moreover, we find that Fes1 association with subdomain IIB results in a near loss of tertiary structure of the rest of the interacting NBD.
Fig. 5.
Sse1 utilizes a Bag-1-type NEF mechanism. (A) Pattern of NEF-induced HX protection and deprotection mapped onto a model of Ssa1-NBD. The effects of Sse1 (Left), Snl1-ΔN (Center), and Fes1 (Right) were compared in 10-s HX reactions (Fig. S8). For the color code, see Fig. 4A. (B) Schematic model for Hsp110 (dark gray) and Hsp70 (light gray) interaction and the mechanism of catalyzed nucleotide release. The lobes (I/II) of the 2 respective NBDs and the subdomains of the Hsp110 SBD (α/β) are indicated. Striped areas mark interaction surfaces. Arrows denote outward rotation of Hsp70 NBD lobe II and the triggered release of ADP. See Discussion for details.
Snl1-ΔN induced protection in subdomain IB and at the top of subdomain IIB and deprotection of two segments in Ssa1 NBD lobe II. These exchange patterns are in good agreement with the binding site of Bag-1 derived from the crystal structure and with the described outward rotation of subdomain IIB (4). The HX-pattern induced by Sse1 is remarkably similar to the pattern evoked by Snl1-ΔN. Specifically, we observe that binding of Sse1 and Snl1-ΔN to Ssa1 similarly results in protection in subdomain I and deprotection of identical peptides in subdomain II, at the interface of subdomains A and B. Thus, we predict a similar outward rotation of subdomain IIB as described for Bag-1. However, the Sse1-induced protection in subdomain I covers a larger protein segment than what we observe for Snl1-ΔN, consistent with an extended interaction area that includes lobe IA. In summary, our HX data exclude the possibility that Sse1 exploits an HspBP1-like NEF mechanism and instead suggest that Hsp110 proteins employ a Bag-1-like mechanism.
Discussion
This work outlines how the Hsp110 Sse1 associates with the Hsp70 Ssa1 and how this interaction triggers nucleotide release. Our understanding of the Sse1–Ssa1 interaction is illustrated schematically in a model presented in Fig. 5B. The main contact between the 2 molecules is mediated via their NBDs that orient to face each other, so that lobe I of Ssa1 contacts lobe II of Sse1. The protruding α-helical subdomain in the C terminus of Sse1 plays an important role for a productive interaction, perhaps by contacting Ssa1 at the upper periphery of subdomain IIB. As a result, the NBD of Ssa1 undergoes a conformational switch, resulting in a state that is incompatible with nucleotide binding.
Our results from both HX and site-specific cross-linking experiments are consistent with a face-to-face interaction of the NBDs. Specifically, the constraints given by the length of the cross-linkers (10 Å for BPIA and 8 Å for BMOE) and the architecture of the NBDs indicate that the interaction occurs between the outer periphery of lobe II of Sse1 and lobe I of Ssa1. Our data do not clearly differentiate between either a parallel or an oblique positioning of the NBDs toward each other, but the lack of positive BPIA cross-links from positions at lobe I of Sse1 suggests an angled interaction. An angled interaction would also resolve a potential steric conflict with the α-helical domain of Sse1 that is protruding from the front face of lobe IB of the Sse1 NBD (13). Our HX data as well as genetic and biochemical analyses indicate that the α-helical subdomain is important for NEF activity. Assuming that the recently published crystal structure of Sse1–ATP (13) reflects the active conformation of the molecule, we predict that the α-helical subdomain directly contacts the upper periphery of subdomain IIB in Ssa1.
We have directly compared the HX properties of the NBD of Ssa1 in complex with all known NEFs of the yeast cytosol, namely Sse1, the HspBP1 homolog Fes1, and the Bag-1 homolog Snl1-ΔN. Our data indicate that Fes1 binds to lobe II and triggers a global unfolding event that involves almost the entire NBD. This nucleotide release mechanism is distinct from a proposed model for HspBP1 NEF function, which involves an opening of the NBD by displacing lobe I (5). However, the authors based their model on a crystal structure of truncated HspBP1 in complex with the isolated lobe II of the Hsp70 NBD and could therefore not directly extract structural information regarding lobe I. We find that Sse1 employs a nucleotide release mechanism very similar to the one of Snl1-ΔN. This functional convergence is reminiscent of bacterial GrpE and eukaryotic Bag-1, which, despite distinct interaction modes, both trigger a analogous conformational shift in Hsp70 that leads to nucleotide release (4). Thus, we propose to distinguish two types of nucleotide release mechanisms: (i) catalysis of nucleotide release through global loss of tertiary structure of the NBD (HspBP1/Fes1), and (ii) catalysis of nucleotide release through opening of the nucleotide-binding cleft by tilting lobe II (Bag-1/GrpE/Hsp110). Apparently, the limited conformational repertoire of the Hsp70 NBD forces evolution to use similar nucleotide release mechanisms despite different NEF-binding modes.
Sse1 harbors a multidomain architecture characteristic of the Hsp70 superfamily, raising the possibility that Sse1 might play a role as a chaperone. The classical Hsp70 chaperones require the tight allosteric coupling of the NBD and SBD that controls the folding cycle (2). Our HX data show that this allostery is not present in Sse1; both ATP and ADP induce similar effects in the molecule. Specifically, we do not observe any opening of the substrate-binding pocket upon ATP binding nor protection in the linker region, in contrast to what has been reported for DnaK (15). Thus, a classical Hsp70 chaperone mechanism can be ruled out, and any potential substrate-binding and chaperone activity of Hsp110 will therefore function differently.
Materials and Methods
Protein Expression and Purification.
Ssa1 (including Ssa11–378, called Ssa1 NBD), Sse1, and Sse1 carrying cysteine mutations or C-terminal deletions were expressed with an Ulp1 cleavable N-terminal His6-Smt3 tag and purified as described in ref. 12. The same strategy was used for Fes1 (pCA707). Purification of Snl1-ΔN and stoichiometric complexes of Ssa–His10 and Sse1/2–Strep–Tag II were purified as described (10, 12). Briefly, Sse1–Ssa1 complexes were formed in the presence of nucleotide, purified via Ni-IDA chromatography, followed by StrepTactin chromatography, and finally depleted from any residual nucleotide by phosphatase treatment.
Hydrogen–Deuterium Exchange Experiments and Mass Spectrometry.
HX experiments were performed in a manner similar to that described earlier (15, 19). Briefly, 100–200 pmol of purified proteins or protein complexes was preincubated for 3–10 min at 30 °C in the presence or absence of nucleotide and then diluted 10- (Table S6 and Fig. S7 and S8) or 20-fold (all other experiments) into D2O-based buffer [25 mM Hepes-KOH (pH 7.4), 50 mM KCl, 5 mM MgCl2] to initiate amide proton–deuteron exchange. The exchange reaction was stopped by the addition of 1 volume of ice-cold quench buffer [0.4 M potassium phosphate buffer (pH 2.2)]. Quenched samples were immediately injected into the HPLC setup, subjected to online peptic digest, and analyzed on an electrospray ionization quadrupole time-of-flight mass spectrometer (QSTAR Pulsar; Applied Biosystems) as described (15, 20). The data processing was performed according to ref. 15. Tables S1–S6 contain the mass of all identified peptides and average number of deuterons incorporated under all experimental conditions. D2O buffer for the HX experiment was prepared by using 99.85% D2O (Euriso-top), lyophilized, and redissolved five times in fresh D2O volumes.
Nucleotide Release Analysis.
Nucleotide release was measured by using MABA-ADP (TriLink Technologies, Inc.) that is a fluorescently labeled analog of ADP, and the stopped flow instrumentation SX-18MV from Applied Photophysics as described (12, 21).
Cross-Linking Experiments.
Cysteine mutants of Sse1 and Ssa1 NBD were labeled with the heterobifunctional cross-linker BPIA (Invitrogen) by incubating the proteins on ice at 30 μM with 1 mM BPIA and 5 mM tris(2-carboxyethyl)phosphine (TCEP). After 30 min, the reactions were stopped by the addition of 10 mM β-mercaptoethanol, and efficiency of labeling was verified by using mass spectrometry. BPIA cross-linking was initiated by incubating 6 μM BPIA-labeled Sse1 or Ssa1NBD with the unlabeled interaction partner (12 μM) in the presence of 1 mM ATP on ice overnight. Subsequently, the mixture was irradiated with UV light (365 nm, 100 W) at a distance of 3 cm for 5 min and analyzed by SDS/PAGE.
Complexes for cross-linking with BMOE and BMH (Thermo Scientific) were obtained by incubating 9 μM Sse1 with 9 μM Ssa1NBD overnight on ice in buffer supplemented with 1 mM ATP and 6.5 μM TCEP. Cross-linking was carried out on ice by adding 74 μM BMOE or BMH and stopped after 2 h by the addition of SDS/PAGE sample buffer containing 50 mM DTT.
Yeast Strains.
All yeast strains are isogenic descendants of the S288C-derived strain, BY4743 (22). Details on construction of yeast strains are given in the Supplementary Text.
Supplementary Material
Acknowledgments.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB638 and the Fonds der Chemischen Industrie (B.B). J.F. was supported by a Human Frontier Science Program long-term fellowship, and H.R. was supported by the Boehringer Ingelheim Fonds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804187105/DCSupplemental.
References
- 1.Craig EA, Huang P. In: Protein Folding Handbook. Buchner J, Kiefhaber T, editors. Wiley; 2005. pp. 490–515. [Google Scholar]
- 2.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 4.Sondermann H, et al. Structure of a Bag/Hsc70 complex: Convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 5.Shomura Y, et al. Regulation of Hsp70 function by HspBP1: Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easton DP, Kaneko Y, Subjeck JR. The Hsp110 and Grp170 stress proteins: Newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trott A, Shaner L, Morano KA. The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics. 2005;170:1009–1021. doi: 10.1534/genetics.105.043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadlish H, et al. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS ONE. 2008;3:e1763. doi: 10.1371/journal.pone.0001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075–15084. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andréasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J Biol Chem. 2008;283:8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 15.Rist W, Graf C, Bukau B, Mayer MP. Amide hydrogen exchange reveals conformational changes in Hsp70 chaperones important for allosteric regulation. J Biol Chem. 2006;281:16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- 16.Morano KA, Liu PC, Thiele DJ. Protein chaperones and the heat shock response in Saccharomyces cerevisiae. Curr Opin Microbiol. 1998;1:197–203. doi: 10.1016/s1369-5274(98)80011-8. [DOI] [PubMed] [Google Scholar]
- 17.Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J Biol Chem. 2004;279:21992–22001. doi: 10.1074/jbc.M313739200. [DOI] [PubMed] [Google Scholar]
- 18.Buchberger A, Schröder H, Buttner M, Valencia A, Bukau B. A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Nat Struct Biol. 1994;1:95–101. doi: 10.1038/nsb0294-95. [DOI] [PubMed] [Google Scholar]
- 19.Rist W, Jørgensen TJ, Roepstorff P, Bukau B, Mayer MP. Mapping temperature-induced conformational changes in the Escherichia coli heat shock transcription factor σ32 by amide hydrogen exchange. J Biol Chem. 2003;278:51415–51421. doi: 10.1074/jbc.M307160200. [DOI] [PubMed] [Google Scholar]
- 20.Rist W, Rodriguez F, Jørgensen TJ, Mayer MP. Analysis of subsecond protein dynamics by amide hydrogen exchange and mass spectrometry using a quenched-flow setup. Protein Sci. 2005;14:626–632. doi: 10.1110/ps.041098305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theyssen H, Schuster HP, Packschies L, Bukau B, Reinstein J. The second step of ATP binding to DnaK induces peptide release. J Mol Biol. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- 22.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70-K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.