Abstract
We report here that actin filaments in vitro exist in two populations with significantly different shrinkage rates. Newly polymerized filaments shrink rapidly, primarily from barbed ends, at 1.8/s, but as they age they switch to a stable state that shrinks slowly, primarily from pointed ends, at ≈0.1/s. This dynamic filament stabilization runs opposite to the classical prediction that actin filaments become more unstable with age as they hydrolyze their bound ATP and release phosphate. Upon cofilin treatment, aged filaments revert to a dynamic state that shows accelerated shrinkage from both ends at a combined rate of 5.9/s. In light of recent electron microscopy studies [Orlova A, et al. (2004) Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc Natl Acad Sci USA 101:17664–17668], we propose that dynamic stabilization arises from rearrangement of the filament structure from a relatively disordered state immediately after polymerization to the canonical Holmes helix, a change that is reversed by cofilin binding. Our results suggest that plasticity in the internal structure of the actin filament may play a fundamental role in regulating actin dynamics and may help cells build actin assemblies with vastly different turnover rates.
Keywords: actin dynamics, actin structure, structural plasticity
Actin filaments in diverse cellular assemblies have very different turnover rates, ranging from tens of seconds in lamellipodia and Listeria comet tails to days in cochlear hair cell microvilli (1–3). The actin-binding proteins that cross-link filaments differ in these diverse assemblies, and it has been assumed that they alone determine the differences in turnover rates. Here we argue that plasticity in the internal structure of the actin filament itself may play an important role in controlling filament turnover rates, either by directly controlling filament stability and/or by modulating the binding of proteins that affect filament stability.
The helical structure of the actin filament was determined by fitting high-resolution monomer structures from x-ray crystallography into lower-resolution structures of the filament from x-ray scattering in solution (4, 5). This canonical structure is supported by numerous mutagenesis and EM studies (6). However, EM studies also revealed considerable structural plasticity of filaments (6, 7). Specifically, when pure actin first polymerizes, the helix appears “ragged” or disordered, with many of the subunits rotated and/or tilted a few degrees away from the canonical helix (8, 9). Over time, this disordered initial structure rearranges into the canonical helix. Binding of the actin depolymerization factor cofilin causes the disorder to reappear (10). One might expect a disordered helix to be less stable than an ordered helix. Indeed, cofilin binding has been shown to accelerate depolymerization from filament ends (11, 12), although details of cofilin function remain controversial (13). Here we test the effect of aging of pure actin filaments on endwise depolymerization and reexamine the effect of cofilin binding. A complication in these experiments is that, over time, actin filaments hydrolyze their bound ATP and release phosphate (14). Phosphate release, which occurs over minutes, is thought to destabilize pure actin filaments (15), although a more important consequence in vivo may be to promote cofilin recruitment (11, 16). If phosphate release, causing filament destabilization, is the dominant kinetic effect, we expect older filaments to be less stable than young ones. If, however, structural plasticity dominates, we expect older filaments, with their canonical helical structure, to be more stable. Our data decisively support the latter view.
Results
Actin Filaments Switch to a Slow-Shrinking State with Age in Single-Filament Assays.
We used time-lapse imaging to directly observe dilution-induced depolymerization of individual, fluorescently labeled actin filaments that were immobilized in perfusion chambers using the actin cross-linker filamin (Fig. 1A). Filaments were polymerized in the chamber from rabbit skeletal muscle actin (17). To make a homogenously aged population, monomers were allowed to polymerize for only 1 min before dilution with buffer. Lengths versus time for all filaments in a field were analyzed by using automated segmentation and tracking software (Fig. 1 B and C), preventing possible bias from human selection of filaments for measurement.
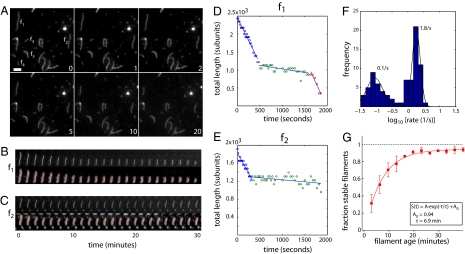
Fig. 1.
Actin filaments switch to a stable slow-shrinking state with age. (A) Time-lapse images of actin filaments in buffer. The lower right of each image shows elapsed time in minutes. (Scale bar: 1 μm.) (B and C) Time-lapse images of f1 and f2 (from A) showing raw image (upper) and processed image (lower) overlaid with a segmentation (red). (D and E) Plots of filament lengths of f1 and f2 over time. Straight lines give best linear fits to piecewise segments of the data, used for determining filament shrinkage rates. (F) Histogram of filament shrinkage rates (blue) along with a two-component Gaussian fit (black). Data are shown on a log scale. (G) Plot of the fraction of stable slow-shrinking filaments as a function of filament age, given by time elapsed after start of polymerization (red squares). Data represent mean and SD of 2–4 independent experiments. Smooth curve shows fit of the data to a single exponential of the form S(t) = Aexp(t/τ) + A0, where τ gives a time constant and A0 gives the asymptotic fraction of stable filaments at long times.
At the start of imaging, when filaments were ≈1.5–3 min old, the majority of filaments shortened rapidly (f1–f4 in Fig. 1A). After several minutes, the observed fraction of dynamic fast-shrinking filaments decreased. Some filaments had switched from a fast-shrinking state to a slow-shrinking state (f1 and f2, Fig. 1 A–E), other filaments had already disappeared completely through fast shrinkage (f3 and f4, Fig. 1A), and other filaments had remained in a slow-shrinking state since the onset of imaging (f5, Fig. 1A). Rate histograms showed bimodal shrinkage kinetics for unselected populations (Fig. 1F). The faster population shortened at a mean rate of 1.8/s, whereas the slower population shortened with a mean rate of ≈0.1/s. The barbed-end capping drug cytochalasin D (CytoD) eliminated the faster population without changing the shrinkage rate of the slower population, consistent with fast shrinkage occurring from barbed ends and slow shrinkage occurring from pointed ends [see supporting information (SI) Text Note 1 and Fig. S1]. We measured the fraction of stable slow-shrinking filaments as a function of time after initiating polymerization, identifying stable filaments as those that remained constant in length for 100 s (koff < 1 subunit per second). This analysis revealed that the fraction of stable filaments increased with a timescale of ≈7 min (Fig. 1G). The fraction of stable filaments did not reach unity at long times but converged toward an asymptote of A0 = 0.94 ± 0.01. At the single-filament level, this was due to occasional filaments apparently switching back to fast shrinkage (Fig. 1 B and D). These data are consistent with filaments switching to a stable, slow-shrinking state as they age, with occasional reversion back to a fast-shrinking state.
Actin Filaments in Solution also Stabilize with Age.
Age-dependent stabilization in single-filament assays might represent an artifact of filament attachment (see SI Text Note 2). To test this, we measured depolymerization kinetics in bulk solution by using a standard pyrene actin assay. Polymerization of 3 μM actin was initiated by salt addition. Actin was polymerized for different times and then allowed to depolymerize by addition of a stoichiometric excess of a monomer trap. To control for possible pharmacology artifacts, we tested three different monomer traps, latrunculin B (LatB) (Fig. 2), DNase I, and vitamin D-binding protein (Fig. S2). Immediately after addition of the monomer trap, the pyrene signal decayed rapidly, but a substantial fraction decayed at a much slower rate, persisting for many hours (Fig. 2 Left). The observation that bulk actin depolymerization exhibits a slower phase is not novel; previous studies have also made similar observations and pointed out its inconsistency with a single rate of filament shrinkage (18, 19). However, no attempts were made to experimentally address the cause of the slow phase.
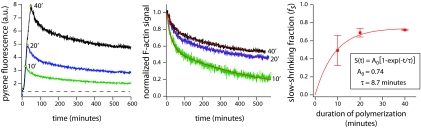
Fig. 2.
Actin filaments in solution stabilize with age. (Left) Pyrene fluorescence traces showing polymerization of 3 μM G-actin (10% pyrene-labeled) initiated by salt addition, followed by depolymerization by addition of 50 μM LatB (yellow circles) at 10 min (green), 20 min (blue), and 40 min (black) after salt addition. The black dashed line represents baseline pyrene signal before salt addition. (Center) F-actin component of the pyrene signal for the different traces in A, aligned such that t = 0 is the onset of depolymerization and normalized such that initial F-actin signal is unity. Red lines denote double exponential fits I(t) = fsexp(−t/τs) + (1 −fs)exp(−t/τf), where fs gives the fraction of the slow-decaying actin population. (Right) Graph showing the fraction of slow-decaying F-actin fs as a function of the duration of polymerization. The smooth line gives the best fit of data to a single rising exponential. Data represent mean and SD of 3 independent experiments.
To test whether the slow-decaying signal was caused by filament annealing, which would decrease the bulk depolymerization rate by lowering the concentration of filament ends, we removed aliquots from bulk solution at different times and imaged them in perfusion chambers. We observed no increase in filament length and found that the great majority of filaments remaining at long times were in the slow-shrinking state, as scored by time-lapse imaging (Fig. S2), suggesting that the slow-decaying signal indeed reflects the existence of a stable filament population that shrinks slowly from ends.
To quantify the relative sizes of the fast- and slow-depolymerizing F-actin populations in this assay, we plotted the F-actin contributions to the pyrene signals, normalized such that their initial values at the onset of depolymerization were unity (Fig. 2 Center). This plot revealed that actin that had been polymerizing for longer times depolymerized to lesser extents after LatB addition. We estimated the contribution of the slow decay to the F-actin signal using double exponential fits of the form I(t) = fsexp(−t/τs) + (1 −fs)exp(−t/τf) (Fig. 2 Center, black lines; see SI Text Note 3). In this two-component exponential fit, the parameters fs and τs give the fraction and decay timescale of the slow-decaying (“stable”) population, and τf gives the decay timescale of the fast-decaying (“dynamic”) population. This analysis showed that fs increases with the duration of polymerization (Fig. 2 Right) and does so with a time constant (τ = 8.7 min) comparable to that observed in single-filament assays (τ = 6.9 min, Fig. 1G). This analysis provides further evidence that filament stabilization, as observed in single-filament imaging, also occurs in bulk solution.
Slow-Shrinking Filaments Have Uncapped Barbed Ends.
Previous authors have attributed unexpected behavior of actin to contamination with trace amounts of CapZ (20), which blocks depolymerization at barbed ends (21). We tested for this possibility in two ways, by directly assaying whether slow-shrinking ends are capped and by further purifying our actin by gel filtration. For these assays, we imaged filaments elongated from Limulus acrosomal fragments, which provide a convenient source of aligned, oriented filaments of similar lengths and ages. Filaments elongated from the barbed ends of immobilized acrosomes were not themselves immobilized, as evident from their blurring out into a fan-shaped fluorescence image (Fig. 3A), so they also avoid possible immobilization artifacts. We allowed these filaments to depolymerize in buffer for different lengths of time. To measure their lengths after depolymerization, we perfused in filamin to bundle them into a parallel array (Fig. 3B). The filament length distribution was rather homogeneous immediately after polymerization (Fig. 3 B Upper and C) but very inhomogeneous after 13 min of depolymerization (Fig. 3 B Lower and D), consistent with switching to the slow-shrinking state also occurring for filaments elongated from the barbed end of acrosome fragments (see SI Text Note 4 and Fig. S3).
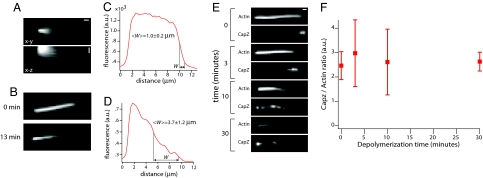
Fig. 3.
Slow-shrinking filaments have uncapped barbed ends. (A) Images of actin filaments elongated from Limulus acrosomal fragments at the onset of depolymerization in the absence of any bundling. Upper shows the bundle in the x-y plane (scale bar: 1 μm), and Lower shows the bundle in the x-z plane (scale bar: 2 μm in z). The diffuse fluorescence arises from the filaments fluctuating in the perfusion chamber. In all images, barbed ends are oriented toward the right. (B) Image of filaments immediately after polymerization (Upper) and 13 min after depolymerization in buffer (Lower). Filaments were bundled by using filamin immediately before imaging. (C) Line scan along the filament bundle shown in B Upper. (D) Line scan along the filament bundle shown in B Lower. 〈W〉 gives the mean half-width of the intensity drop at the barbed end and represents mean and SD of 12 filament bundles from 2 experiments. (E) Images of Alexa Fluor 647 actin and Alexa Fluor 488 CapZ (as labeled) in acrosomal filament bundles allowed to depolymerize for the indicated amounts of time. (F) Graph showing the CapZ/filament ratio as a function of depolymerization time. Each data point represents mean and SD of 10 filament bundles.
To test whether the slow-shrinking filaments observed in this assay were capped at barbed ends, we terminated the depolymerization reaction by perfusing in fluorescently labeled CapZ, which binds only to uncapped ends, followed by filamin to bundle and immobilize the now-stable filaments. When CapZ was perfused in immediately after polymerization, the actin intensity along the bundles dropped abruptly at the barbed end, where a bright spot of CapZ localized (Fig. 3E). Thus, all filaments grew to approximately the same starting length, and many could recruit CapZ. When filaments were allowed to depolymerize in buffer before perfusion with CapZ and filamin, the bundle acquired a tapered appearance, indicative of heterogeneity in filament length, and the CapZ distribution become more diffuse (Fig. 3E, 3–10 min). Thirty minutes after initiating depolymerization many actin filaments had depolymerized completely, but those that remained still recruited fluorescent CapZ at their barbed ends (Fig. 3E, 30 min). To quantify the fraction of filaments with free barbed ends at different times after depolymerization, we measured the integrated CapZ signal for individual bundles, excluding the signal at the base of the bundle, which was due to direct binding of CapZ to the barbed end of the acrosome itself. We then divided this quantity by the maximal actin signal in the bundle, which is proportional the filament number. This ratio, which reports on the proportion of actin filaments with uncapped barbed ends, was largely constant during depolymerization (Fig. 3F), indicating that fast- and slow-shrinking filaments are equal in their ability to recruit CapZ.
To further reinforce this point, we repeated these Limulus acrosome dilution assays using actin that was further purified by gel filtration (Fig. S4). These experiments show that gel-filtered actin forms filaments that also stabilize with age and that it does so with kinetics similar to that observed for non-gel-filtered actin (Figs. 1 and 2). Our results here rule out the possibility that the slow-shrinking filament population observed in our assays is due to inactivation of barbed ends by capping protein.
Cofilin Binding Reverts Aged Filaments to a Fast-Shrinking State.
So far, our data showing that aging of pure actin filaments causes them to switch from a fast- to a slow-depolymerizing state are congruent with EM studies showing that aging causes filaments to gradually switch from a disordered state immediately after polymerization to the canonical Holmes helix (8). To further test this congruence, we explored the effect of cofilin on aged filaments, because EM data showed that cofilin binding has the effect of restoring the disordered state (10). We polymerized pure actin in a filamin-coated perfusion chamber for 1 min and let them age in buffer for 30 min. We then perfused these aged filaments with either buffer alone or buffer plus cofilin and observed them using time-lapse imaging. This experimental design removes the complication that cofilin cannot bind strongly to ATP-actin filaments (11, 16); the aged filaments observed here are almost entirely in their ADP-bound form, given known phosphate release kinetics (14). When perfused with buffer, most aged filaments shortened very slowly at the mean rate of 0.1/s (Fig. 4 A and B); occasionally, slow-shrinking filaments switched to a fast-shrinking state during the course of imaging (Fig. 4A Right), as expected (Fig. 1D). The mean fast shrinkage rate for these filaments (2.7/s) was slightly higher than that for filaments imaged immediately after polymerization (1.8/s, Fig. 1F), consistent with a faster shrinkage rate for ADP-actin filaments, considering only those filaments in the fast-shrinking population (ref. 22; see also Table S1). However, fast-shrinking filaments constituted only 14% of the aged population (Fig. 4C), and most filaments remained in a slow-shrinking state during imaging. When aged filaments were perfused with 4 μM cofilin, they severed extensively. In contrast, filaments ≈1–3 min old showed very little severing with cofilin (Fig. S5). Effects of filament aging on severing by cofilin are interesting and may account for some of the discrepancies in the cofilin literature (11, 13).
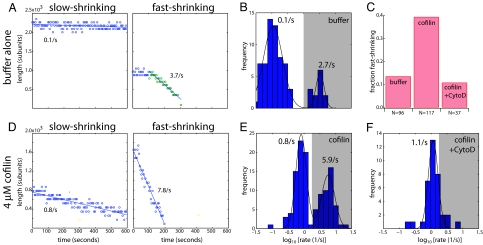
Fig. 4.
Aged filaments revert to a fast-shrinking state upon cofilin treatment. (A) Length versus time plots for aged filaments in buffer, showing a slow-shrinking filament (Left) and a fast-shrinking filament (Right). Straight lines give best linear fits. (B) Shrinkage rate histogram for filaments in buffer. (C) Bar chart showing fraction of fast-shrinking filaments in buffer (left bar), 4 μM cofilin (center bar), and 4 μM cofilin and 1 μM CytoD (right bar). Fast-shrinking filaments were identified as those whose shrinkage rates fell into the gray shaded regions of the shrinkage rate histograms in B, E, and F. (D) Length versus time plots for aged filaments treated with cofilin, showing a slow-shrinking filament (Left) and fast-shrinking filament (Right). (E) Shrinkage rate histogram for filaments treated with 4 μM cofilin. (F) Shrinkage rate histogram for filaments treated with 4 μM cofilin and 1 μM CytoD.
Here we focus only on the effects of cofilin on filament shrinkage from ends. To eliminate indirect effects on shrinkage rate due to severing, we did not analyze any new filament ends that were generated as a result of severing, measuring shrinkage rates only from preexisting ends. When we added cofilin to aged filaments, we again found slow- and fast-shrinking filament populations, with mean rates of 0.8/s and 5.9/s, respectively (Fig. 4 D and E). As in case for buffer alone, CytoD addition removed the fast-shrinking population without affecting the slow-shrinking population (Fig. 4 C and F), implying that fast-shrinking filaments in cofilin depolymerize primarily from barbed ends, and slow ones from pointed ends. Moreover, the fraction of filaments in the fast-shrinking population was 39%, significantly higher than that in buffer alone (Fig. 4 C and E; χ2 = 17.5, d.f. = 1, P < 0.01). Therefore, in addition to promoting severing, cofilin had three effects on aged filaments: it increased the pointed-end shrinkage rate (from ≈0.1/s to 0.8/s, Fig. 4 B and E), consistent with previous reports (11), increased the barbed-end shrinkage rate (from 2.6/s to 5.1/s, Fig. 4 B and E, estimated from the rate difference between fast and slow populations) (see SI Text Note 5 and Fig. S6), and increased the fraction of filaments shrinking rapidly from barbed ends from ≈14% to ≈39% (Fig. 4C). These three effects are all indicative of decreased filament stability in cofilin and are congruent with EM data showing increased disorder in aged filament after cofilin binding (10).
Nonmuscle Actin also Stabilizes with Age.
To test whether age-dependent stabilization is a conserved aspect of actin dynamics, we purified actin from bovine thymus, which consists of a physiological mixture of β- and γ-actin isoforms (23). We found strong evidence for age-dependent stabilization of nonmuscle actin in single-filament assays (Fig. S7), suggesting that this process is indeed conserved across evolution (24). Kinetic differences were observed: a higher fraction of the nonmuscle filaments appeared to polymerize directly into the stable form, and the depolymerization rates of both the slow- and fast-depolymerizing forms of young filaments (0.3/s and 6.4/s, respectively) were faster than their muscle equivalents.
Discussion
In this article we show that young actin filaments depolymerize more rapidly than older ones, contradicting the classic prediction that filaments become more unstable as they age because of hydrolysis of bound ATP and subsequent phosphate release. We have ruled out obvious sources of potential artifactual stabilization, including surface interaction and contamination with capping protein. In light of EM data (8–10), we interpret age-dependent stabilization as resulting from structural relaxation of young filaments, where many subunits are in a tilted conformation, into the canonical Holmes helix.
Our observations seem to contradict years of kinetic studies, which concluded that actin dynamics could be explained by a single-filament structure, with unique on and off rates at each end, that depend only on bound nucleotide state. However, it is important to realize that most previous kinetic experiments did not measure shrinkage rates directly, but instead inferred them indirectly by extrapolation of polymerization data (15, 25); recent single-filament experiments that did measure shrinkage rates directly chose only fast-shrinking filaments for analysis, under the assumption that slow-shrinking filaments were an immobilization artifact (26).
Although dynamic filament stabilization contradicts the textbook view of actin dynamics, we find considerable precedent in the kinetics literature. Previous bulk studies of dilution-induced actin depolymerization had already noted incomplete decay of the F-actin signal (18, 19), inconsistent with classical theory but in agreement with our experiments (Fig. 2 Left). A study that used sonication to continuously break filaments, thus promoting very rapid actin polymerization, measured a very high critical concentration for fast-polymerizing actin (27). Those data were interpreted as showing that ATP-actin dissociates rapidly from an ATP-actin lattice, which contradicts the classical model, but our data suggest that this article may have measured the critical concentration of a pure population of young filaments, which is higher than that of an aged population. More recent studies observed that shrinkage rates for filaments that were first polymerized from ATP-actin monomer and then aged (1.4/s from ref. 26) were slower than those for filaments polymerized directly from ADP-actin monomer (5.4/s from ref. 22) but did not account for their observations. Taken together, these observations suggest that the phenomenon we report has been seen often in the past but ignored because it does not fit into the established view of how actin behaves.
A More Complex Actin Dynamics.
Our results, taken together with EM structural data of the actin filament and previously ignored kinetic observations, argue that the structure and stability of an actin filament is not determined uniquely by the state of its bound nucleotide; rather, it is determined by a complex set of chemical and physical changes that have strong history dependence and antagonistic consequences. This additional complexity may account for previous observations that the turnover of filaments at steady state is not well described by a single rate of subunit association or dissociation from ends (28). We note that the time in which filaments stabilize (≈10 min, Figs. 1–3) is similar to that in which they release their bound phosphate (14), suggesting that these two processes may even be coupled. More work is required to determine how these two effects influence each other and what factors determine the steady-state outcome.
Physiological Implications.
One interpretation of our data is that dynamic stabilization itself enables the actin molecule to build filaments with vastly different stabilities in vivo. The unstable conformation of newly polymerized filaments may account for the turnover of dynamic actin assemblies in Listeria comet tails or lamellipodia (2, 3). Conversely, the stabilizing structural rearrangements that occur with aging may confer enhanced stability on long-lived actin assemblies, such as microvilli in cochlear hair cells (1). Aβ amyloid fibers, which are long-lived in vivo, also undergo stabilizing conformational changes after polymerization (29), indicating that dynamic stabilization by structural relaxation may be a general property of protein polymers and not so surprising to find in the case of actin.
A slightly different interpretation of our data is that dynamic stabilization, as observed with pure actin, is an epiphenomenon. It reflects the ability of the filament to change conformation but does not directly regulate physiological dynamics. In this view, rearrangements in filament structure are more important for controlling the recruitment, and/or mediating the effects, of cofilin and other filament-binding proteins. In the cell, phosphate release from actin leads to cofilin recruitment, at least for filaments in fast-turnover assemblies, which in turn rejuvenates the filament in terms of both structure (8) and kinetics (Fig. 4). The more important question for physiological regulation may be: why do some actin filaments recruit cofilin and turn over rapidly, whereas others, which still hydrolyze ATP, turn over slowly? Recruitment of tropomyosin antagonizes cofilin (30), and we hypothesize that this antagonism may in part be due to preferred binding of tropomyosin to the canonical Holmes helix and of cofilin to the tilted state. The highly cooperative nature of the transitions between alternate structural states (31) may enable entire filaments and filament assemblies to interact with the same binding partners in cells.
Both of our interpretations point to a central role of structural plasticity in regulation of actin turnover; they differ only in whether filament structure controls subunit dissociation rate directly or acts indirectly to mediate the binding and function of accessory proteins that regulate dissociation rate. In either case, further investigation of the role of structural plasticity in cells will be important to understand actin biology.
Materials and Methods
Protein Purification.
Skeletal muscle actin was purified following the method of ref. 17. Purified F-actin was then subject to two depolymerization/polymerization cycles and labeled on lysines by using Alexa Fluor 647 NHS-ester (Molecular Probes/Invitrogen) following the procedure of ref. 32. Before the imaging experiments, 60 μM G-actin was treated with 1 mM EGTA and 0.2 mM MgCl2 on ice for 5 min to convert Ca-ATP-actin to Mg-ATP-actin. Actin was then diluted to 12 μM in G buffer (2 mM Tris·Cl/0.2 mM CaCl2/0.2 mM ATP/0.1% 2-mercaptoethanol, final pH 8.0), incubated overnight at 4 °C, and centrifuged at 100,000 rpm in a TLA100 rotor for 20 min to remove aggregates. Cofilin was purified as described in ref. 33. Filamin was purified as described in ref. 34. Murine CapZ was purified as described in ref. 35 and labeled by incubating with a 10-fold molar excess of Alexa Fluor 488 maleimide (A-10254; Invitrogen) at 4 °C for 3 h. Unconjugated Alexa Fluor 488 was then separated by gel filtration and dialysis.
Imaging and Analysis of Single Actin Filaments.
Time-lapse imaging of single actin filaments was performed by using the assay described in detail in ref. 36. Briefly, perfusion chambers were incubated with 10 μg/mL filamin for 10 min and then with blocking solution (5 mg/mL casein/0.2% Tween 20/0.1% Pluronic F-127) for 5 min. Alexa Fluor 647 Mg-ATP-actin was then polymerized in perfusion chambers in assay buffer (50 mM KCl/2 mM MgCl2/2 mM ATP/100 mM K+-Hepes, pH 7.8). Unpolymerized monomer was then washed out by using assay buffer supplemented with oxygen scavengers (4.5 mg/mL glucose/0.2 mg/mL glucose oxidase/35 μg/mL catalase) and 10% blocking solution. Attached filaments were either imaged directly or perfused with cofilin immediately before imaging. Single actin filaments were identified and segmented in time-lapse images by using an algorithm described in ref. 36. Segmented filaments were tracked over successive time-lapse images by using a semiautomated tracking routine written in MATLAB. Length versus time traces for single filaments were then analyzed by linear regression to obtain shrinkage rates.
Pyrene Actin Assay.
Skeletal muscle actin, prepared as described above, was mixed with pyrene actin (AP05; Cytoskeleton) and diluted in G buffer (60 μM 10% pyrene labeling). This pyrene actin was then converted to its Mg-ATP form by incubation with 1 mM EGTA and 0.2 mM MgCl2 for 5 min, diluted to 3 μM in G buffer with 0.25 mM EGTA and 0.05 mM MgCl2 to prevent reversion of actin to its Ca-ATP form, incubated for 2 h at 4 °C, and centrifuged at 100,000 rpm in a TLA120.2 rotor for 20 min to remove aggregates. Actin polymerization was induced by addition of 10× assay buffer (0.5 M KCl/20 mM MgCl2/20 mM ATP/1 M K+-Hepes, pH 7.8) and monitored over time by measuring emission at 365 nm by using a fluorimeter (Cary Eclipse; Varian). Depolymerization was induced by addition of 50 μM Latrunculin B.
Imaging of Filaments Elongated from Limulus Acrosomal Bundles.
Imaging of actin filaments grown off Limulus acrosomal actin bundles was performed as described in ref. 36. For experiments in Fig. 3 A–C, Limulus acrosomal bundles in the chamber were incubated with 3 μM Alexa Fluor 647 actin monomer for 5 min to promote filament elongation. The chamber was perfused with assay buffer to wash out unpolymerized monomer and then perfused with assay buffer with oxygen scavengers, 10% blocking solution, and 70 μg/mL filamin to bundle filaments together for imaging. Filament bundle depolymerization was then imaged by using time-lapse fluorescence microscopy. For experiments in Fig. 3 D and E, Limulus acrosomal bundles were incubated with 3 μM Alexa Fluor 647 actin monomer for 8 min. Filaments were depolymerized in buffer alone for varying times; depolymerized filaments were perfused with 1 μM Alexa Fluor 488 CapZ for 1 min before perfusion with filamin. To estimate the Alexa Fluor 488 CapZ/actin ratio, the integrated CapZ fluorescence in a single-filament bundle was measured in a region of interest surrounding the filament bundle. Background was measured in a neighboring region of interest where no filament bundles were present. The relative number of actin filaments in the bundle was estimated by taking maximal fluorescence intensity along the filament bundle length. The integrated CapZ fluorescence was divided by estimated actin filament number to obtain the Alexa Fluor 488 CapZ/actin ratio.
Supplementary Material
Acknowledgments.
We thank Guichy Waller and Paul Matsudaira (Massachusetts Institute of Technology, Cambridge, MA) for Limulus acrosomal sperm processes. We also thank Marc Kirschner, John Hartwig, Michael Brenner, and Tom Pollard for useful advice and the T.J.M. laboratory for discussions. We also thank Sophie Dumont for critical reading of the manuscript and our anonymous reviewers for useful comments. This work was supported by National Institutes of Health Grant GM 23928. H.Y.K. is a Howard Hughes Medical Institute predoctoral fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807394105/DCSupplemental.
References
- 1.Schneider ME, Belyantseva IA, Azevedo RB, Kachar B. Rapid renewal of auditory hair bundles. Nature. 2002;418:837–838. doi: 10.1038/418837a. [DOI] [PubMed] [Google Scholar]
- 2.Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- 3.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 4.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 5.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 6.Reisler E, Egelman EH. Actin's structure and function: What we still do not understand. J Biol Chem. 2007;282:36133–36137. doi: 10.1074/jbc.R700030200. [DOI] [PubMed] [Google Scholar]
- 7.Schoenenberger CA, Bischler N, Fahrenkrog B, Aebi U. Actin's propensity for dynamic filament patterning. FEBS Lett. 2002;529:27–33. doi: 10.1016/s0014-5793(02)03267-2. [DOI] [PubMed] [Google Scholar]
- 8.Orlova A, et al. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc Natl Acad Sci USA. 2004;101:17664–17668. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinmetz MO, Goldie KN, Aebi U. A correlative analysis of actin filament assembly, structure, and dynamics. J Cell Biol. 1997;138:559–574. doi: 10.1083/jcb.138.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlier MF, et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J Cell Biol. 1997:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriyama K, Yahara I. Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 1999;18:6752–6761. doi: 10.1093/emboj/18.23.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Carlier MF, Pantaloni D. Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry. 1986;25:7789–7792. doi: 10.1021/bi00372a001. [DOI] [PubMed] [Google Scholar]
- 15.Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchoin L, Pollard TD. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J Biol Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- 17.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 18.Bryan J, Coluccio LM. Kinetic analysis of F-actin depolymerization in the presence of platelet gelsolin and gelsolin-actin complexes. J Cell Biol. 1985;101:1236–1244. doi: 10.1083/jcb.101.4.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh TP, Weber A, Higgins J, Bonder EM, Mooseker MS. Effect of villin on the kinetics of actin polymerization. Biochemistry. 1984;23:2613–2621. doi: 10.1021/bi00307a012. [DOI] [PubMed] [Google Scholar]
- 20.Casella JF, Barron-Casella EA, Torres MA. Quantitation of Cap Z in conventional actin preparations and methods for further purification of actin. Cell Motil Cytoskeleton. 1995;30:164–170. doi: 10.1002/cm.970300208. [DOI] [PubMed] [Google Scholar]
- 21.Caldwell JE, Heiss SG, Mermall V, Cooper JA. Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry. 1989:8506–8514. doi: 10.1021/bi00447a036. [DOI] [PubMed] [Google Scholar]
- 22.Fujiwara I, Vavylonis D, Pollard TD. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc Natl Acad Sci USA. 2007;104:8827–8832. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galkin VE, VanLoock MS, Orlova A, Egelman EH. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin's sequence and structure. Curr Biol. 2002;12:570–575. doi: 10.1016/s0960-9822(02)00742-x. [DOI] [PubMed] [Google Scholar]
- 25.Lal AA, Korn ED, Brenner SL. Rate constants for actin polymerization in ATP determined using cross-linked actin trimers as nuclei. J Biol Chem. 1984;259:8794–8800. [PubMed] [Google Scholar]
- 26.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlier MF, Pantaloni D, Korn ED. Polymerization of ADP-actin and ATP-actin under sonication and characteristics of the ATP-actin equilibrium polymer. J Biol Chem. 1985;260:6565–6571. [PubMed] [Google Scholar]
- 28.Fujiwara I, Takahashi S, Tadakuma H, Funatsu T, Ishiwata S. Microscopic analysis of polymerization dynamics with individual actin filaments. Nat Cell Biol. 2002;4:666–673. doi: 10.1038/ncb841. [DOI] [PubMed] [Google Scholar]
- 29.Esler WP, et al. Alzheimer's disease amyloid propagation by a template-dependent dock-lock mechanism. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- 30.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlova A, Prochniewicz E, Egelman EH. Structural dynamics of F-actin: II. Cooperativity in structural transitions. J Mol Biol. 1995;245:598–607. doi: 10.1006/jmbi.1994.0049. [DOI] [PubMed] [Google Scholar]
- 32.Kellogg DR, Mitchison TJ, Alberts BM. Behaviour of microtubules and actin filaments in living Drosophila embryos. Development. 1988;103:675–686. doi: 10.1242/dev.103.4.675. [DOI] [PubMed] [Google Scholar]
- 33.Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K. Filamin, a new high-molecular-weight protein found in smooth muscle and nonmuscle cells. Purification and properties of chicken gizzard filamin. Biochemistry. 1977;16:1857–1865. doi: 10.1021/bi00628a015. [DOI] [PubMed] [Google Scholar]
- 35.Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155:251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kueh HY, Mitchison TJ, Brieher WM. Actin filament disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.