Abstract
Homologous to bacteriorhodopsin and even more to proteorhodopsin, xanthorhodopsin is a light-driven proton pump that, in addition to retinal, contains a noncovalently bound carotenoid with a function of a light-harvesting antenna. We determined the structure of this eubacterial membrane protein–carotenoid complex by X-ray diffraction, to 1.9-Å resolution. Although it contains 7 transmembrane helices like bacteriorhodopsin and archaerhodopsin, the structure of xanthorhodopsin is considerably different from the 2 archaeal proteins. The crystallographic model for this rhodopsin introduces structural motifs for proton transfer during the reaction cycle, particularly for proton release, that are dramatically different from those in other retinal-based transmembrane pumps. Further, it contains a histidine–aspartate complex for regulating the pKa of the primary proton acceptor not present in archaeal pumps but apparently conserved in eubacterial pumps. In addition to aiding elucidation of a more general proton transfer mechanism for light-driven energy transducers, the structure defines also the geometry of the carotenoid and the retinal. The close approach of the 2 polyenes at their ring ends explains why the efficiency of the excited-state energy transfer is as high as ≈45%, and the 46° angle between them suggests that the chromophore location is a compromise between optimal capture of light of all polarization angles and excited-state energy transfer.
Keywords: carotenoid antenna, energy transfer, retinal protein, salinixanthin, X-ray structure
Increased efficiency of light harvesting by antennae, such as carotenoids, is common in photosynthetic membranes, large multiprotein systems that contain tens or hundreds of chromophores. The much simpler retinal-based archaeal proton pumps bacteriorhodopsin and archaerhodopsin lack antennae; light collection and proton translocation are performed by a single protein with a single chromophore. Recently, it was shown that a light-harvesting antenna can function also in a retinal protein. Xanthorhodopsin (1), of the eubacterium Salinibacter ruber (2), contains a single energy-donor carotenoid, salinixanthin (3), and a single acceptor, retinal, in a small (25 kDa) membrane protein. Because energy transfer is from the short-lived S2 carotenoid level (4), there must be a short distance and favorable geometry between the 2 chromophores to account for its high (40–50%) efficiency. Close interaction of the 2 chromophores is indicated by dependence of the carotenoid conformation on the presence of the retinal in the protein (1, 5, 6) and spectral changes of the carotenoid during the photochemical transformations of the retinal (1), but, as for the proteorhodopsin family of proteins, no direct structural information has been available (4, 7). Unexpectedly, the crystallographic structure of xanthorhodopsin we report here reveals not only the location of the antenna but also striking differences from the archaeal retinal proteins, bacteriorhodopsin and archaerhodopsin.
The photocycle of xanthorhodopsin (8) and the functional residues in the ion transfer pathway (1) are similar to those of the numerous other eubacterial proton pumps, the proteorhodopsins (9, 10). Proteins homologous to xanthorhodopsin were found recently in the genome of an abundant coastal ocean methylotroph (11) and earlier in the genomes of Gloebacter violaceous, Pyrocystis lunula (12), and others. The proteins in this clade exhibit significantly less homology to the proteorhodopsins (11). For example, 137 residues (50%) are identical in Gloeobacter rhodopsin and xanthorhodopsin, but only 60 residues (22%) in proteorhodopsin and xanthorhodopsin. Although considerable sequence differences separate xanthorhodopsin from the proteorhodopsins (Fig. 1), its structure, the first for a eubacterial proton pump, is likely to be relevant to other eubacterial retinal-based pumps.
Fig. 1.
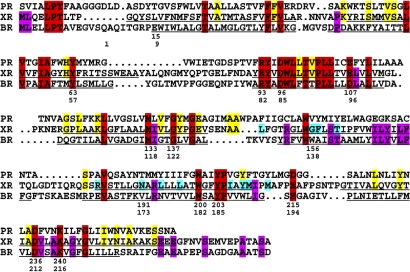
Sequence alignment of green light-absorbing proteorhodopsin (PR), xanthorhodopsin (XR), and bacteriorhodopsin (BR), reevaluated from the one shown in ref. 1 by using information gained from the diffraction structure. Red, conserved residues in all three; purple, conserved residues in xanthorhodopsin and bacteriorhodopsin; yellow, conserved residues in xanthorhodopsin and proteorhodopsins; blue, residues involved with carotenoid binding. Top row of numbers refer to the xanthorhodopsin sequence; bottom row to the bacteriorhodopsin sequence. Underlining indicates residues in transmembrane helices. Proteorhodopsin sequence refers to a species from Monterey Bay, MBP1 (protein accession No. AAG10475).
Results and Discussion
Xanthorhodopsin was crystallized from bicelles (13), with a type I arrangement of stacked bilayers. The structure was solved to 1.9-Å resolution (Table 1). The P1 unit cell contains 2 molecules of xanthorhodopsin with a head-to-tail arrangement somewhat similar to 2-dimensional crystal forms of bacteriorhodopsin (14) and halorhodopsin (15), as well as 3-dimensional crystals of the D85S bacteriorhodopsin mutant (16), sensory rhodopsin II (17, 18), and Anabaena sensory rhodopsin (19). Considering its function as an ion transporter in the cell membrane, xanthorhodopsin is unlikely to form such dimers in the original Salinibacter cells.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Beamline | 9.1, SSRL, Menlo Park, CA |
| Wavelength, Å | 0.979 |
| Space group | P1 |
| Cell dimensions | a = 52.7 Å, b = 59.5 Å, c = 59.7 Å α = 76.4°, β = 74.9°, γ = 64.1° |
| Resolution range, Å | 45.10–1.90 |
| Total reflections | 166,560 |
| Unique reflections | 46,289 |
| Redundancy | 3.6 (3.5)* |
| Completeness, % | 94.1 (85.5)* |
| Mean I/σ | 8.4 (1.5)* |
| Rsym, % | 5.7 (48.5)* |
| Refinement | |
| Resolution range, Å | 45.10–1.90 |
| No. of reflections used | 46,278 |
| Rwork/Rfree, %† | 24.7/26.5 |
| Rmsd bonds, Å | 0.012 |
| Rmsd angles, ° | 1.35 |
| No. of atoms/avg. B, Å2 | |
| Protein | 3,925/44.2 |
| Waters | 62/53.2 |
| Retinal | 40/33.4 |
| Salinixanthin | 140/73.0 |
| Lipids | 250/67.4 |
| Ramachandran plot (favored/allowed/generously allowed), %‡ | 96.3/3.7/0 |
*Values in parentheses are for the highest-resolution shell.
†Rfree based on a test set size of 4.7%.
‡PROCHECK
Location and Binding Site of Carotenoid Antenna.
The C40 carotenoid lies transverse against the outer surface of helix F at a ≈54° angle to the membrane normal, buried at the protein–lipid boundary (Fig. 2A). Its keto-ring is immobilized by residues at the extracellular ends of helices E and F and by the β-ionone ring of the retinal (Fig. 2B) and rotated 82° from the plane of the methyl side chains of its polyene chain and therefore from the plane of the extended π-system (Fig. 2C). The ring C O is not hydrogen-bonded. The immobilized and acutely out-of-plane orientation of the keto-ring minimizes participation of its 2 double bonds in the conjugated π-system and explains the well-resolved vibronic bands of the carotenoid, the lack of a red-shift of the absorption bands upon binding, and the strong CD bands in the visible region (5). The relatively rigid polyene is wedged in a slot on the outside of helix F, with one side formed by the Leu-194 and Leu-197 side chains and the other by the Ile-205 side chain, and its base by Gly-201. The carotenoid glucoside is hydrogen-bonded to the C
O is not hydrogen-bonded. The immobilized and acutely out-of-plane orientation of the keto-ring minimizes participation of its 2 double bonds in the conjugated π-system and explains the well-resolved vibronic bands of the carotenoid, the lack of a red-shift of the absorption bands upon binding, and the strong CD bands in the visible region (5). The relatively rigid polyene is wedged in a slot on the outside of helix F, with one side formed by the Leu-194 and Leu-197 side chains and the other by the Ile-205 side chain, and its base by Gly-201. The carotenoid glucoside is hydrogen-bonded to the C O and the NH2 of the amide side chain of Asn-191, as well as NH1 of Arg-184. The dependence of the carotenoid spectrum on the retinal is explained by the fact that the retinal β-ionone ring is part of the carotenoid binding site (Fig. 2B).
O and the NH2 of the amide side chain of Asn-191, as well as NH1 of Arg-184. The dependence of the carotenoid spectrum on the retinal is explained by the fact that the retinal β-ionone ring is part of the carotenoid binding site (Fig. 2B).
Fig. 2.
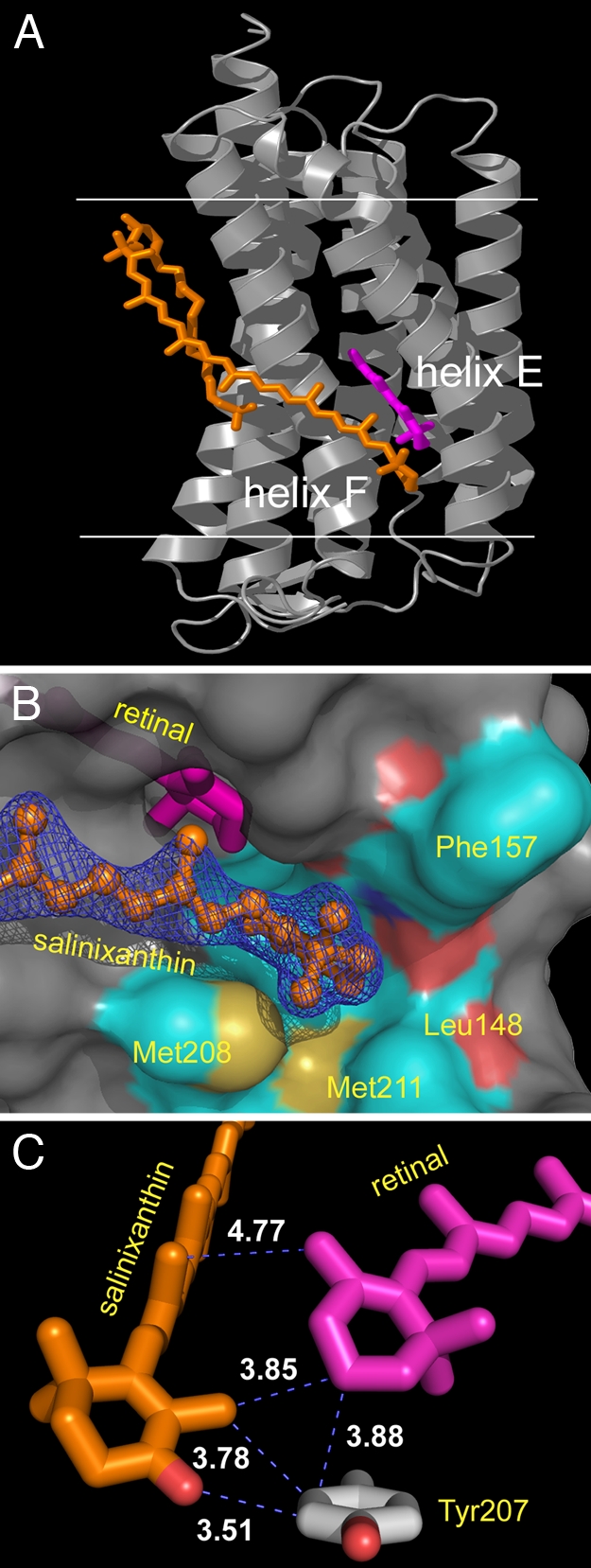
Location of salinixanthin (orange) and retinal (magenta) in xanthorhodopsin. (A) The extended carotenoid is tightly bound on the transmembrane surface of xanthorhodopsin, traversing nearly the entire bilayer, with an inclination of 54° to the membrane normal. Its keto-ring binds in a pocket between helices E and F, very near the β-ionone ring of the retinal. The angle between the chromophore axes is 46°. The angle between the planes of their π-systems is 68°. Horizontal lines indicate the approximate boundaries of the lipid bilayer. Helices E and F are marked. (B) The binding pocket of the keto ring is formed by Leu-148, Gly-156, Phe-157, Thr-160, Met-208, and Met-211, as well as the retinal β-ionone ring. (C) The keto-ring of the carotene is rotated 82° out of plane of the salinixanthin-conjugated system and is in van der Waals distance of the retinal β-ionone and the phenolic side chain of Tyr-207.
The keto-ring of the carotenoid is in the space occupied by Trp-138 in bacteriorhodopsin, one of the bulky side chains that confines the retinal β-ionone ring in that protein. In xanthorhodopsin, it is replaced by a glycine. This residue replacement might be the best diagnostic in a sequence for the possibility of binding a salinixanthin-like carotenoid. Another difference is Glu-141 of xanthorhodopsin, which is alanine in bacteriorhodopsin but a conserved glutamate in proteorhodopsins and involved in spectral tuning (20), whose carboxyl oxygens are ≈4 Å from the retinal β-ionone ring methyls. An intriguing question is whether other retinal proteins might also have an antenna. Of the 12 residues in the xanthorhodopsin carotenoid binding site, 7 are conserved in Gloeobacter rhodopsin (homologs of Gly-156, Thr-160, Asn-191, Leu-197, Ile-205, Tyr-207, and Met-211). Thus, it is probable that this protein can bind the C40 carotenoid of Gloeobacter, echinenone, which contains a keto-ring similar to salinixanthin (21).
The distance between the centers of the 2 linear chromophores is 11.7 Å. The ring moieties are within 5 Å of one another, and part of the retinal β-ionone ring is in van der Waals distance of the carotenoid keto-ring. Both are in contact with the Tyr-207 ring between them (Fig. 2 B and C). This contrasts with the crystal structure of archaerhodopsin (22), a proton pump with a bacterioruberin carotenoid without antenna function (23), where the corresponding center-to-center interchromophore distance is 17 Å, with the closest approach of bacterioruberin to the retinal at 12 Å. In that protein, the carotenoid may have a structural (22) and possibly photoprotective function.
The angle between the axes of the chromophores is 46° (Fig. 2A), somewhat less than the 56 ± 3° estimated from the polarization anisotropy of retinal fluorescence (4). The discrepancy may originate from the off-axis orientation of the transition moment, as in rhodopsin (24). Energy transfer is optimal when the chromophores are parallel. However, where the 2 spectra overlap, the probability of absorbing incident light of all polarization angles is increased when the 2 chromophores are not parallel. This gain would be optimal if the angle between the chromophores were 90°, but that would preclude energy transfer. Thus, the 46° angle in xanthorhodopsin appears to be a compromise between optimal chromophore interaction and optimal collection of light of all polarizations.
In light-harvesting complexes of photosynthetic bacteria, it is the large number of antenna molecules with various orientations (25, 26) that ensures the capture of light of different polarizations.
Structure of Xanthorhodopsin Exhibits Large Differences from Bacteriorhodopsin.
The xanthorhodopsin structure extends the architecture of the growing family of rhodopsins (27). Remarkably, there are greater differences from the main chain of bacteriorhodopsin than in any of the crystallized microbial rhodopsins, including halorhodopsin, archaerhodopsin, sensory rhodopsin II, and Anabena sensory rhodopsin. Helices A and G are longer by 4 and 9 residues, respectively, and their tilt and rotation, particularly of helix A, are considerably different (Fig. 3A). The 28 residues that comprise helix B are 4 residues shifted in the sequence toward the C terminus (i.e., toward the extracellular side). In bacteriorhodopsin, the interhelical B–C antiparallel β-sheet interacts with the D–E loop, whereas in xanthorhodopsin it reorients dramatically to interact with the Arg-8 peptide C O near the N terminus, where it forms a mini 3-stranded β-sheet. As a result, the tip of the B–C loop is displaced, by 30 Å, toward the periphery of the protein (Fig. 3). Unexpected in a heptahelical membrane protein, this produces a large cleft that extends far into the interior and brings functional residues, buried in other rhodopsins, near the aqueous interface (Fig. 3B).
O near the N terminus, where it forms a mini 3-stranded β-sheet. As a result, the tip of the B–C loop is displaced, by 30 Å, toward the periphery of the protein (Fig. 3). Unexpected in a heptahelical membrane protein, this produces a large cleft that extends far into the interior and brings functional residues, buried in other rhodopsins, near the aqueous interface (Fig. 3B).
Fig. 3.
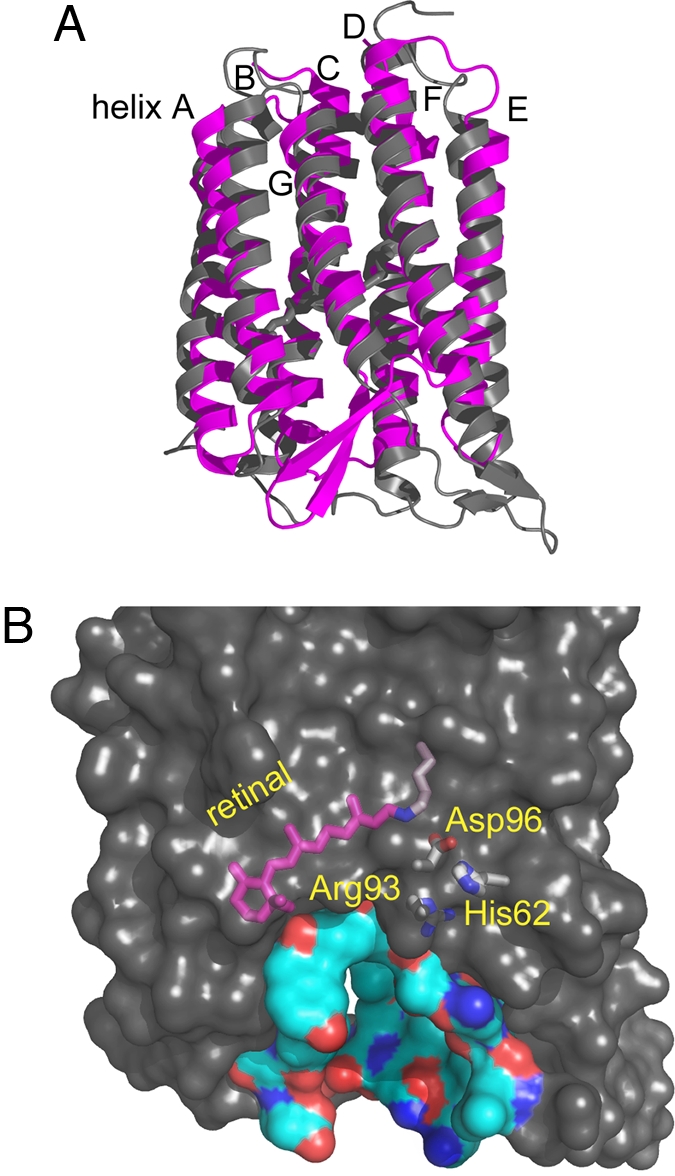
Displacements of the B–C and F–G interhelical segments expose a deep cavity that extends from the extracellular side halfway toward Schiff base. (A) Comparison of xanthorhodopsin and bacteriorhodopsin. The antiparallel β-sheet of the B–C segment of xanthorhodopsin (gray) packs against the N terminus (Bottom Right), whereas in bacteriorhodopsin (magenta) this segment packs against the F–G loop (Bottom Left). Helices A–E are viewed from the front; F and G are in the back, as marked. (B) The resulting hydrophilic cavity in xanthorhodopsin extends from the extracellular surface to Arg-93 and other buried functional groups. In bacteriorhodopsin, this region is occupied by the protein and includes the proton release group composed of Glu-194, Glu-204, and 3 ordered water molecules, absent in xanthorhodopsin. Buried residues with functional roles in transport are shown to illustrate their proximity to the aqueous interface.
In bacteriorhodopsin, Wat-402 receives a hydrogen bond from the protonated retinal Schiff base and donates hydrogen bonds to the 2 anionic residues Asp-85 and Asp-212 (28, 29), and this arrangement is conserved in xanthorhodopsin (Fig. 4). However, the carboxylate of the homolog of Asp-85, Asp-96 in this protein, is severely rotated, and the hydrogen-bonded aqueous network of water molecules in the extracellular region that facilitate proton release in the photocycle (30) is replaced by hydrogen-bonded residues that are likely to be more resistant to rearrangement than an aqueous network. In bacteriorhodopsin, a pair of glutamate residues, Glu-194 and Glu-204, stabilizes a hydrogen-bonded aqueous network (31), which is the source of the proton released to the extracellular surface after the retinal Schiff base is deprotonated. Sequence alignment (Fig. 1) shows that the eubacterial pumps, proteorhodopsin and xanthorhodopsin, contain only 1 of these acidic groups. Further, in xanthorhodopsin, at least, the single glutamate is far removed from Arg-93 (>18 Å vs. 7.3 Å in bacteriorhodopsin). In bacteriorhodopsin, release of the proton is triggered by the movement of the positively charged arginine (Arg-82 in this protein) side chain toward the glutamate pair, upon protonation of Asp-85 (30). This is less likely to occur in the xanthorhodopsin photocycle, because NH1 and NH2 of Arg-93 are both hydrogen-bonded to the peptide carbonyl of Gln-229 instead of water molecules.
Fig. 4.
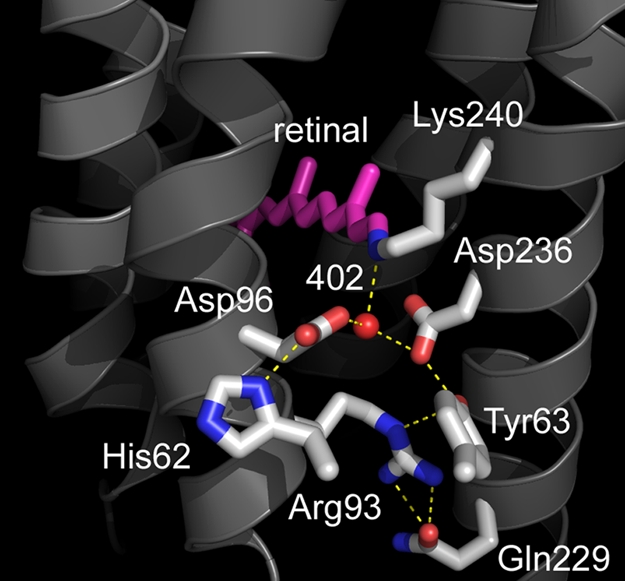
Structure of the retinal, the Schiff base counterion, and the extracellular region. The counterion to the Schiff base is an aspartate–histidine complex. The network of water molecules that leads to the extracellular surface in bacteriorhodopsin is missing, and Arg-93 interacts primarily with protein side chains.
An Asp–His Hydrogen-Bonded Pair Is Part of Counterion to Retinal Schiff Base.
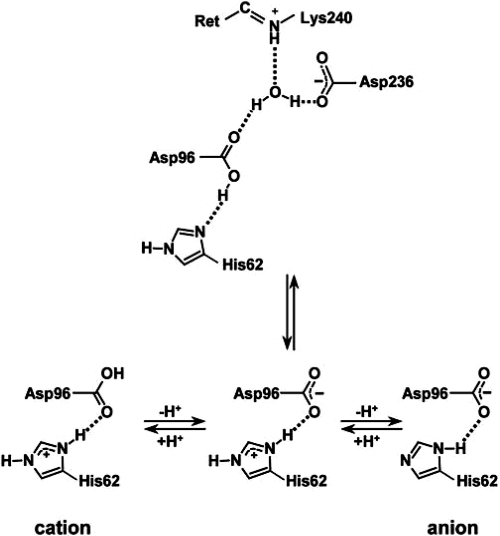
One of the distinguishing features of eubacterial proton pumps is that the pKa of the primary proton acceptor is not as low as 2.5 in bacteriorhodopsin, but near 7 (32, 33). The origin of the high pKa, which makes these proteins functional as pumps only at alkaline pH (34), has been an unsolved problem. In xanthorhodopsin, ND1 of His-62 is hydrogen-bonded to OD1 of Asp-96. At 2.42/2.55 Å (in the 2 molecules of the asymmetric unit), this is a very short hydrogen bond, such as found at active sites of numerous proteins (35). Thus, its proton may be shared by the imidazole ring and the carboxylate in a single-well, strong hydrogen bond, and the complex, with an expected pKa higher than the aspartate alone (36), must be regarded as the Schiff base counterion. If analogy with bacteriorhodopsin holds, the anionic, rather than the neutral complex (Fig. 5), is the proton acceptor of the Schiff base in the photocycle. However, we cannot exclude the possibility that the neutral form is the proton acceptor, because the carboxylate may accommodate another proton to yield a cationic complex.
Fig. 5.
Protonation states of the aspartate–histidine counterion complex. The pH of the crystallization (5.6), is below the observed (8) spectral transition between the protonated and deprotonated forms of the Schiff base counterion (presumably neutral/zwitterionic and anionic). Thus, it seems likely that the crystallographic structure contains the neutral/zwitterionic counterion.
A histidine at this position is highly conserved in the proteorhodopsins (see Fig. 1 and ref. 37), making it likely that the aspartate–histidine complex is a general characteristic of eubacterial pumps. It is likely to account not only for the observed high pKa of the counterion in these retinal proteins, but also for the smaller red shift of the chromophore maximum upon the protonation of the counterion in xanthorhodopsin than in bacteriorhodopsin (3–5 nm vs. 40 nm) (8) because sharing of the proton with the histidine would leave a partial charge on the aspartate. However, the much smaller red shift in xanthorhodopsin than in proteorhodopsin (≈30 nm) (33) suggests that there must be structural differences that influence the delocalization of the proton between the carboxylate and the indole ring.
Once protonated in the photocycle, the His-62–Asp-96 complex would be a good candidate for the origin of the proton released to the medium upon deprotonation of the retinal Schiff base, but at neutral pH, at least, such early proton release does not occur. Our unpublished measurements of transient proton release and uptake with the pH indicator dye pyranine (performed as described in refs. 32 and 38) indicate that unlike in bacteriorhodopsin, but like its mutants that lack the specialized proton release complex (38, 39) and as in proteorhodopsin (32), there is no release of a proton in the xanthorhodopsin photocycle at the time the Schiff base becomes deprotonated. Instead, the sequence of proton release to the extracellular side and uptake from the cytoplasmic side is reversed: proton uptake in the cycle occurs first, evidently by Glu-107 after it has reprotonated the retinal Schiff base, and the release to the extracellular surface is delayed until the final photocycle step. The rationale for keeping counterion protonated until the end of the photocycle might be the same as in bacteriorhodopsin for which it was demonstrated (40) that a neutralized counterion facilitates thermal retinal isomerization, an obligatory step in the reaction cycle.
Hydrogen-Bonded Aqueous Network in the Cytoplasmic Domain Needs Less Rearrangement for Proton Transfer.
In the cytoplasmic region of bacteriorhodopsin, the proton donor is in an anhydrous environment that constitutes the hydrophobic barrier in the cytoplasmic half of the protein. This, and the fact that OD1 of Thr-46 is an acceptor of its proton (28), raises its pKa. The aspartic acid becomes a proton donor to the Schiff base during the photocycle only after the Glu–Thr hydrogen bond is broken and this region is hydrated so as to create a hydrogen-bonded chain of 4 water molecules to connect the proton donor to its acceptor (41). In xanthorhodopsin, as in the proteorhodopsins, these residues are replaced by a glutamic acid and a serine (Fig. 1), and they are not hydrogen-bonded to one another. The carboxyl is hydrogen-bonded to Wat-502 that connects to the peptide carbonyl of Lys-240 (Fig. 6). It appears, therefore, that in xanthorhodopsin part of the cytoplasmic hydrogen-bonded chain of water molecules between the retinal and the proton donor is in position already before photoisomerization of the retinal.
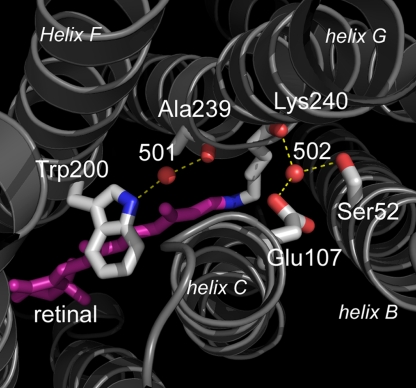
Fig. 6.
Cytoplasmic region, with the proton donor Glu-107 and its link via Wat-502 to the retinal region. As in the other microbial rhodopsins, Wat-501 is hydrogen-bonded tightly between the tryptophan just above the retinal (Trp-200) and the main-chain carbonyl of the residue in the π-bulge of helix G (Ala-239). Helices B, C, F, and G are marked.
As in the other structurally characterized microbial rhodopsins, the other cytoplasmic water, Wat-501, is hydrogen-bonded tightly between the tryptophan located immediately on the cytoplasmic side of the retinal (Trp-200 in xanthorhodopsin) and the main-chain carbonyl of the residue at the apex of the π-bulge of helix G (Ala-239), a connection that is broken during the photocycle as the movement of the retinal C20 displaces the side chain of Trp-200 by >1 Å (30). Wat-501 possesses one of the lowest B factors of any atom in the model, and it is noteworthy that its presence appears to induce a deviation from planarity in the nearby tyrosine side chain.
Materials and Methods
A membrane fraction enriched in xanthorhodopsin was prepared by washing S. ruber cell membranes 4 times with distilled water, followed by washing 3 times with 0.01% dodecyl maltoside in 100 mM NaCl and 5 mM N,N-bis(2-hydroxyethyl)glycine (pH 8). The resulting membrane fraction was resuspended in 30 mM phosphate (pH 5.6) and 1 mM sodium azide and concentrated to contain 5 mg/ml xanthorhodopsin (as estimated from absorption at 560 nm). The protein was solubilized in a bicelle-type medium (13) by adding 1 volume of a 16.7% (wt/wt) dimyristoyl phosphatidylcholine in 20% nonyl maltoside to 3 volumes of the xanthorhodopsin preparation, vortexing, and incubating overnight at 4 °C.
Crystals (≈30 × 30 × 150 μm) were grown at 22 °C over 4–5 months in sitting drops (on Cryschem plates; Hampton Research), containing 10 μL of solubilized xanthorhodopsin, 3 μL of 3 M sodium phosphate (pH 5.6), and 2 μL of 2.5 mM sodium azide. The reservoirs contained 1 mL of 2.5 M or 3 M sodium phosphate (pH 5.6). After 5-min equilibrations with 5%, 10%, and then 15% ethylene glycol, the crystals were frozen rapidly in liquid nitrogen. Data were collected at 100 K on beamline 9.1 at the Stanford Synchrotron Research Laboratory (SSRL) as 360 frames with 1° rotations. Data reduction statistics are listed in Table 1.
The structure was solved by iterative molecular replacement, by using a composite model that consisted of helices A and B of Anabaena sensory rhodopsin [Protein Data Bank (PDB) ID code 1XIO, residues 4–51] and helices C–G of bacteriorhodopsin (PDB ID code 1C3W, residues 81–231) with the program PHASER (42). The first rotation function exhibited low signal-to-noise ratio with a top Z score of 4.77. The correct solution was peak 5 with a Z score of 4.73. After 10 cycles of restrained refinement with the program REFMAC (43), 1 molecule of the resulting model was used for a second round of molecular replacement, yielding much-improved signal-to-noise ratio with Z scores of 7.32 and 7.10 for the two rotation functions, respectively. Maps were improved by 2-fold averaging with the program DM (44). Electron density for the salinixanthin carotenoid was readily visible at this stage. The model was improved by iterative cycles of model building and refinement with the program COOT (45) and refinement with the program CNS (46), initially with and later without NCS restraints. The rmsd values between the final model (molecule A/B) and the search models, bacteriorhodopsin and Anabaena sensory rhodopsin, are 1.91/1.88 Å and 2.00/1.98 Å, respectively. Sequence identities with bacteriorhodopsin and Anabaena sensory rhodopsin are 24.6% and 21.5%, respectively. In addition to the carotenoid, numerous lipids and lipid fragments have been included in the model. All residues fall within the allowed regions of the Ramachandran plot. Refinement statistics are listed in Table 1. The coordinate file, 3DDL, has been deposited at the Protein Data Bank.
Acknowledgments.
We thank T. Poulos and H. Li (University of California, Irvine) for sharing transportation of crystals and beam time and for help with remote data collection. This work was supported by National Institutes of Health Grant GM29498 and Department of Energy Grant DEFG03-86ER13525 (to J.K.L.), U.S. Army Research Office Grant W911NF-06-1-0020 (to S.P.B.), and National Institutes of Health Grant GM56445 (to H.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3DDL).
References
- 1.Balashov SP, et al. Xanthorhodopsin: A proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antón J, et al. Salinibacter ruber gen nov, sp nov, a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol. 2002;52:485–491. doi: 10.1099/00207713-52-2-485. [DOI] [PubMed] [Google Scholar]
- 3.Lutnaes BF, Oren A, Liaaen-Jensen S. New C40-carotenoid acylglycoside as principal carotenoid in Salinibacter ruber, an extremely halophilic eubacterium. J Nat Prod. 2002;65:1340–1343. doi: 10.1021/np020125c. [DOI] [PubMed] [Google Scholar]
- 4.Balashov SP, Imasheva ES, Wang JM, Lanyi JK. Excitation energy-transfer and the relative orientation of retinal and carotenoid in xanthorhodopsin. Biophys J. 2008;95:2402–2414. doi: 10.1529/biophysj.108.132175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balashov SP, Imasheva ES, Lanyi JK. Induced chirality of light-harvesting carotenoid salinixanthin and its interaction with the retinal of xanthorhodopsin. Biochemistry. 2006;45:10998–11004. doi: 10.1021/bi061098i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imasheva ES, et al. Chromophore interaction in xanthorhodopsin: Retinal dependence of salinixanthin binding. Photochem Photobiol. 2008;84:977–984. doi: 10.1111/j.1751-1097.2008.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanyi JK, Balashov SP. Xanthorhodopsin: A bacteriorhodopsin-like proton pump with a carotenoid antenna. Biochim Biophys Acta. 2008;1777:684–688. doi: 10.1016/j.bbabio.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imasheva ES, Balashov SP, Wang JM, Lanyi JK. pH-dependent transitions in xanthorhodopsin. Photochem Photobiol. 2006;82:1406–1413. doi: 10.1562/2006-01-15-RA-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Béjà O, et al. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: An array of physiological roles? Nat Rev Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- 11.Giovannoni SJ, et al. The small genome of an abundant coastal ocean methylotroph. Environ Microbiol. 2008;10:1771–1782. doi: 10.1111/j.1462-2920.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- 12.Mongodin EF, et al. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci USA. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faham S, Bowie JU. Bicelle crystallization: A new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 14.Michel H, Oesterhelt D, Henderson R. Orthorhombic 2-dimensional crystal form of purple membrane. Proc Natl Acad Sci USA. 1980;77:338–342. doi: 10.1073/pnas.77.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunji ERS, von Gronau S, Oesterhelt D, Henderson R. The 3-dimensional structure of halorhodopsin to 5 Å by electron crystallography: A new unbending procedure for 2-dimensional crystals by using a global reference structure. Proc Natl Acad Sci USA. 2000;97:4637–4642. doi: 10.1073/pnas.080064697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouhani S, et al. Crystal structure of the D85S mutant of bacteriorhodopsin: Model of an O-like photocycle intermediate. J Mol Biol. 2001;313:615–628. doi: 10.1006/jmbi.2001.5066. [DOI] [PubMed] [Google Scholar]
- 17.Luecke H, Schobert B, Lanyi JK, Spudich EN, Spudich JL. Crystal structure of sensory rhodopsin II at 2.4 Ångströms: Insights into color tuning and transducer interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Royant A, et al. X-ray structure of sensory rhodopsin II at 2.1-Å resolution. Proc Natl Acad Sci USA. 2001;98:10131–10136. doi: 10.1073/pnas.181203898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogeley L, et al. Anabaena sensory rhodopsin: A photochromic color sensor at 2.0 Å. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kralj JM, et al. Protonation state of Glu-142 differs in the green- and blue-absorbing variants of proteorhodopsin. Biochemistry. 2008;47:3447–3453. doi: 10.1021/bi7018964. [DOI] [PubMed] [Google Scholar]
- 21.Takaichi S, Mochimaru M. Carotenoids and carotenogenesis in cyanobacteria: Unique ketocarotenoids and carotenoid glycosides. Cell Mol Life Sci. 2007;64:2607–2619. doi: 10.1007/s00018-007-7190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimura K, Kouyama T. Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2. J Mol Biol. 2008;375:1267–1281. doi: 10.1016/j.jmb.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 23.Boichenko VA, Wang JM, Antón J, Lanyi JK, Balashov SP. Functions of carotenoids in xanthorhodopsin and archaerhodopsin, from action spectra of photoinhibition of cell respiration. Biochim Biophys Acta. 2006;1757:1649–1656. doi: 10.1016/j.bbabio.2006.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgakopoulou S, Cogdell RJ, van Grondelle R, van Amerongen H. Linear-dichroism measurements on the LH2 antenna complex of Rhodopseudomonas acidophila strain 10050 show that the transition dipole moment of the carotenoid rhodopsin glucoside is not collinear with the long molecular axis. J Phys Chem B. 2003;107:655–658. [Google Scholar]
- 25.Green BR, Parson WW, editors. Light-Harvesting Antennas in Photosynthesis. Dordrecht: Kluwer; 2003. [Google Scholar]
- 26.Polivka T, Sundström V. Ultrafast dynamics of carotenoid excited states: From solution to natural and artificial systems. Chem Rev. 2004;104:2021–2071. doi: 10.1021/cr020674n. [DOI] [PubMed] [Google Scholar]
- 27.Spudich JL, Jung K-H. In: Handbook of Photosensory Receptors. Briggs WR, Spudich JL, editors. Darmstadt: Wiley; 2005. pp. 1–23. [Google Scholar]
- 28.Luecke H, Schobert B, Richter H-T, Cartailler J-P, Lanyi JK. Structure of bacteriorhodopsin at 1.55-Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 29.Belrhali H, et al. Protein, lipid, and water organization in bacteriorhodopsin crystals: A molecular view of the purple membrane at 1.9-Å resolution. Structure. 1999;7:909–917. doi: 10.1016/s0969-2126(99)80118-x. [DOI] [PubMed] [Google Scholar]
- 30.Luecke H, Schobert B, Richter H-T, Cartailler J-P, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2-Å resolution. Science. 1999;286:255–260. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 31.Garczarek F, Gerwert K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 32.Dioumaev AK, et al. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry. 2002;41:5348–5358. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- 33.Imasheva ES, Balashov SP, Wang JM, Dioumaev AK, Lanyi JK. Selectivity of retinal photoisomerization in proteorhodopsin is controlled by aspartic acid-227. Biochemistry. 2004;43:1648–1655. doi: 10.1021/bi0355894. [DOI] [PubMed] [Google Scholar]
- 34.Dioumaev AK, Wang JM, Balint Z, Váró G, Lanyi JK. Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic. Biochemistry. 2003;42:6582–6587. doi: 10.1021/bi034253r. [DOI] [PubMed] [Google Scholar]
- 35.Cleland WW, Frey PA, Gerlt JA. The low-barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 36.Rangarajan R, Galan JF, Whited G, Birge RR. Mechanism of spectral tuning in green-absorbing proteorhodopsin. Biochemistry. 2007;46:12679–12686. doi: 10.1021/bi700955x. [DOI] [PubMed] [Google Scholar]
- 37.Man-Aharonovich D, et al. Characterization of RS29, a blue-green proteorhodopsin variant from the Red Sea. Photochem Photobiol Sci. 2004;3:459–462. doi: 10.1039/b316071h. [DOI] [PubMed] [Google Scholar]
- 38.Balashov SP, et al. Glutamate-194 to cysteine mutation inhibits fast light-induced proton release in bacteriorhodopsin. Biochemistry. 1997;36:8671–8676. doi: 10.1021/bi970744y. [DOI] [PubMed] [Google Scholar]
- 39.Brown LS, et al. Glutamic acid-204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. J Biol Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]
- 40.Balashov SP, Imasheva ES, Govindjee R, Ebrey TG. Titration of aspartate-85 in bacteriorhodopsin: What it says about chromophore isomerization and proton release. Biophys J. 1996;70:473–481. doi: 10.1016/S0006-3495(96)79591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schobert B, Brown LS, Lanyi JK. Crystallographic structures of the M and N intermediates of bacteriorhodopsin: Assembly of a hydrogen-bonded chain of water molecules between Asp-96 and the retinal Schiff base. J Mol Biol. 2003;330:553–570. doi: 10.1016/s0022-2836(03)00576-x. [DOI] [PubMed] [Google Scholar]
- 42.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Cowtan KD, Zhang KYJ. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Brünger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]