Abstract
Here, we report a previously undescribed approach for controlling metal ion coordination geometry in biomolecules by reorientating amino acid side chains through substitution of L- to D-amino acids. These diastereopeptides allow us to manipulate the spatial orientation of amino acid side chains to alter the sterics of metal binding pockets. We have used this approach to design the de novo metallopeptide, Cd(TRIL12LDL16C)3−, which is an example of Cd(II) bound to 3 L-Cys as exclusively trigonal CdS3, as characterized by a combination of 113Cd NMR and 111mCd PAC spectroscopy. We subsequently show that the physical properties of such a site, such as the high pKa2 for Cd(II) binding of 15.1, is due to the nature of the coordination number and not the ligating group. Further more this approach allowed for the design of a construct, GRANDL12LDL16CL26AL30C, capable of independently binding 2 equivalents of Cd(II) to 2 very similar Cys sites as exclusively 3- and 4-, CdS3 and CdS3O, respectively. Demonstrating that we are capable of controlling the Cd(II) coordination number in these 2 sites solely by varying the nature of a noncoordinating second coordination sphere amino acid, with D-leucine and L-alanine resulting in exclusively 3- and 4-coordinate structures, respectively. Cd(II) was found to selectively bind to the 4-coordinate CdS3O site, demonstrating that a protein can be designed that displays metal-binding selectivity based solely on coordination number control and not on the chemical identity of coordinating ligands.
Keywords: cadmium, coiled-coil peptides, D-amino acids, de novo metallopeptide design
Considering that nearly one-third of all biomolecules contain metal ions (1), it is not surprising that the field of de novo metallopeptides is an area in which significant contributions can be made to an understanding of metallobiochemistry (2–6). Nature exquisitely controls the localization and coordination geometry of metal ions. The de novo design of peptides that fold into well-defined secondary and tertiary structures provides a biological construct in which one can engineer metal-binding sites and assess the factors controlling structure and site specificity. Currently efforts in our research laboratory are directed toward trying to achieve a similar degree of control, in particular for binding Cd(II) in biologically common thiol-rich sites, because Cd(II) is frequently substituted as a probe for Zn(II) (7, 8). A key benefit of the direct chemical synthesis of these constructs is that we are no longer limited to the 20 naturally occurring amino acids and can take full advantage of the extensive library of nonnatural amino acids.
A common biological structural motif is that of the α-helix, and subsequently much de novo design utilizes this scaffold, although β-sheets and mixed α/β-constructs have also been investigated (4, 9–11). Work in our group involves the designed TRI and GRAND peptides based on the heptad repeat, LaKbAcLdEeEfKg, and derivatives thereof (12, 13). These sequences (shown in Table 1) were designed to assemble in aqueous solution into α-helices, which aggregate to form 3-stranded coiled coils at pH values >5.5. Substitution of either an a or d Leu with Cys provides a preorganized homoleptic thiol site in the interior of these coiled coils. The binding of numerous heavy metals such as Hg(II), Bi(III), Pb(II), and As(III) has been explored (12–19), although the most intriguing system has been Cd(II) that binds to TRIL16C as a mixture of 3- and 4-coordinate CdS3 and CdS3O (O from an exogenous water molecule) forms, respectively.
Table 1.
Peptide sequences used in these studies
| Peptide | Sequence* |
|---|---|
| TRI | Ac-G LKALEEK LKALEEK LKALEEK LKALEEK G-NH2 |
| TRIL16C | Ac-G LKALEEK LKALEEK CKALEEK LKALEEK G-NH2 |
| TRIL12AL16C | Ac-G LKALEEK LKAAEEK CKALEEK LKALEEK G-NH2 |
| TRIL16Pen | Ac-G LKALEEK LKALEEK XKALEEK LKALEEK G-NH2 |
| TRIL12LDL16C | Ac-G LKALEEK LKALDEEK CKALEEK LKALEEK G-NH2 |
| GRAND | Ac-G LKALEEK LKALEEK LKALEEK LKALEEK LKALEEK G-NH2 |
| GRANDL16XL26AL30C | Ac-G LKALEEK LKALEEK XKALEEK LKAAEEK CKALEEK G-NH2 |
| GRAND L12LDL16CL26AL30C | Ac-G LKALEEK LKALDEEK CKALEEK LKAAEEK CKALEEK G-NH2 |
Residues in bold and underlined indicate substitutions.
*X = penicillamine; LD = D-leucine.
We have shown that subtle sequence modification allowed for the complete control of Cd(II) coordination number and site preference in these peptides. By removal of steric bulk above the plane of the Cys (replacing a Leu with an Ala, TRIL12AL16C), a cavity for a water molecule was achieved yielding a fully 4-coordinate CdS3O species (20). However, efforts to obtain 3-coordinate Cd(II) bound to Cys were unsuccessful (20, 21). Ultimately, a pure CdS3 environment was realized by resorting to the nonnatural amino acid analogue of Cys, penicillamine (Pen), in which bulky methyl groups replace the β-hydrogen atoms and are thought to exclude exogenous water from binding to the Cd(II). The application of 113Cd NMR and 111mCd PAC spectroscopies confirmed the structural assignment of these derivatives. Subsequently, we demonstrated that heterochromic peptides, which simultaneously bound Cd(II) in one site exclusively as a CdS3O structure and in a second site in a CdS3 environment, could be prepared using a similar strategy. Despite these successes, we were disappointed that a simple cysteine derivative could not be found that could enforce 3 coordination upon the metal center.
The use of D-amino acids in peptide design has much potential, because these are the common “nonnatural” amino acids, and chemically the side chains remain the same as their “natural” L-amino acid analogues. Modifications can vary from single mutations that may be used to introduce hairpin turns or terminate α-helixes (22, 23) or inversion of all amino acids to yield a mirror image structure (24, 25). Single, double, and even triple D-amino acid substituted α-helixes have been investigated and found to be tolerated (26–28), and heteromeric coiled coils containing both left- and right-handed helixes made up of D- and L-amino acid building blocks, respectively, have been prepared (29). A major benefit of D-amino acids containing peptides is that they possess enhanced proteolytic stability, which has significant implications for therapeutic applications (30).
Initial modeling studies encouraged us to believe that we could use an approach to increase the effective steric bulk above the Cys site by reorienting the side chain in the residue directly above the Cys plane by altering the chirality of 1 backbone carbon. If successful, we should be able to prepare peptides capable of binding Cd(II) to a Cys site in an exclusively trigonal, 3-coordinate geometry. We reasoned that substitution of a single L-Leu by D-Leu, TRIL12LDL16C, would reposition the β-isopropyl moiety toward the C terminus, as illustrated in Fig. 1.
Fig. 1.
Schematic representation of the L- to D-Leu mutation at position 12. Reorientation of the amino acid side chain serves to increase the steric bulk in the plane above the Cys ligands.
One can envision that, in antiparallel coiled coils, commonly found in nature, the residues belonging to the antiparallel strand may have the side chains orientated in such a way that their topology may resemble that of D-amino acids, and as such, enforce similar steric constraints on metal ion sites. Such observations are likely to provide a clearer understanding of how properties vary as a function of coordination geometries adopted by metals with almost identical first-coordination spheres in biomolecules.
In this article, we demonstrate an approach for controlling metal ion coordination geometries within a biological construct. Furthermore, using diastereopeptides we address 3 important issues regarding the use of D-amino acids in predominantly L-amino acid designed metallopeptides. First, we assess whether TRIL12LDL16C can enforce a 3-coordinate structure on Cd(II) using the natural L-Cys as a ligand. Second, we demonstrate that using a single L- to D-mutation, a short peptidic sequence containing 2 very similar Cys sites can be designed that is capable of independently binding 2 equivalents of Cd(II) as exclusively 3- and 4-coordinate structures. Finally, we demonstrate that this strategy provides an alternative vehicle for achieving Cd(II) binding-site specificity.
Results
CD Spectroscopy.
To determine whether introduction of a single D-amino acid in the interior of an otherwise all L-amino acid sequence would still allow for the well-structured designed coiled coil to form in solution, it was necessary to monitor the peptide region of the CD that yielded the characteristic coiled-coil spectra with minima at 208 and 222 nm. The molar ellipticity values obtained at 222 nm and pH 8.5 of −34 053 and −36 858 deg·dmol−1·cm2 for TRIL12LDL16C and GRANDL12LDL16CL26AL30C, respectively, are consistent with folded coiled-coil structures.
Cd(II)-Binding Stoichiometry.
The ability to bind Cd(II) and the number of Cd(II) bound per trimer was determined by titrating aliquots of Cd(II) into a solution of known concentration of peptide at a pH that allows the peptide to fully bind the Cd(II). The steady appearance of the characteristic ligand-to-metal charge-transfer (LMCT) transition at 235 nm due to the formation of Cd-S bonds in the complex Cd(TRIL12LDL16C)3−, in Fig. 2A for the titration of CdCl2 into a 30 μM solution of TRIL12LDL16C at pH 8.5, abruptly plateaus at 1 equivalent Cd(II) (see Fig. 2B). The total metal concentration was used to calculate an extinction coefficients of λ235 23 600 M−1·cm−1 for the complex Cd(TRIL12LDL16C)3−. In contrast, λmax occurs at 230 nm, and 2 equivalents of Cd(II) are required in the analogous experiment with 30 μM GRANDL12LDL16CL26AL30C at pH 8.5 (see Fig. 2B).
Fig. 2.
UV-visible spectra for (A) the titration of CdCl2 into a solution of TRIL12DLL16C at pH 8.5 with the characteristic LMCT at λmax 235 nm and (B) the titration curve of absorbance at λmax vs. equivalents of Cd(II) added per trimer for both 10 μM (TRIL12DLL16C)3 and (GRANDL12DLL16CL26AL30C)3 (λmax 230 nm) at pH 8.5.
pH Dependence.
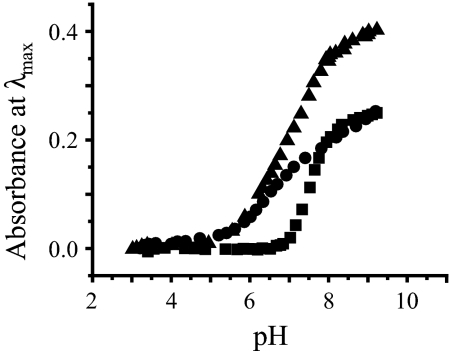
The pH of a solution containing 10 μM CdCl2 and 40 μM TRIL12LDL16C was varied from approximately pH 4 to 10 and resulted in the growth of the characteristic LMCT band at 235 nm. The intensity of the band at 235 nm increased with increasing pH, reaching a plateau above approximately pH 8.5. A plot of the absorbance at 235 nm as a function of pH is shown in Fig. 3. A fit of the experimental data to the model published for the simultaneous release of 2 protons on binding Cd(II) to 3 thiolates (16) yields a pKa2 of 15.1. Analogous experiments performed with 30 μM GRANDL12LDL16CL26AL30C in the presence of 1 equivalent of Cd(II) per trimer resulted in the growth of the LMCT band at 230 nm at a lower pH (see Fig. 3). The pH profile of GRANDL12LDL16CL26AL30C in the presence of 2 equivalents of Cd(II) per trimer (Fig. 3) led to a profile that would match the summation of the former 2 experiments.
Fig. 3.
UV-visible pH titrations of Cd(II) binding. pH dependence of 1 equivalents of Cd(II) binding (■) to 40 μM TRIL12DLL16C; and 1 (●) and 2 (▴) equivalents of Cd(II) binding to 30 μM GRANDL12DLL16CL26AL30C as a function of pH.
111mCd Perturbed Angular Correlation (PAC) Spectroscopy.
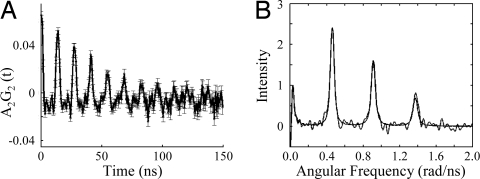
111mCd PAC spectroscopy was used to determine the Cd(II) coordination geometry in the complex 111mCd(TRIL12LDL16C)3−. The Fourier transform of the experimental data for 111mCd and TRIL12LDL16C in a 1:12 ratio 300 μM peptide monomer concentration, at pH 9.1 and 1 °C shows the presence of 1 species as indicated by 3 peaks in Fig. 4B. The parameters for this species are reported in Table 2. Notably the ω0 value of 0.4563 rad/ns is consistent with a trigonal Cd(II) species bound to 3 thiolates.
Fig. 4.
PAC data for 111mCd(TRIL12DLL16C)3−. (A) The perturbation function and (B) Fourier transform (experimental data and fits are shown overlaid) at pH 9.1, 1 °C, and a TRIL12DLL16C:111mCd ratio of 12:1, and a peptide monomer concentration of 300 μM.
Table 2.
Parameters fitted to PAC data*
| Peptide | pH | ω0 (rad/ns) | η | Δω0/ω0 ×100 | 1/τc us−1 | A ×100 | χr2 |
|---|---|---|---|---|---|---|---|
| TRIL16Pen† | 8.7 (1°C) | 0.4540 (9) | 0.02 (10) | 1.6 (4) | 19 (2) | 6.9 (3) | 0.91 |
| TRIL12LDL16C‡ | 9.1 (1°C) | 0.4563 (4) | 0.10 (1) | 0.7 (2) | 14 (1) | 8.2 (2) | 1.18 |
*The numbers in parentheses are the standard deviations of the fitted parameters.
†Data from ref. 21.
‡LD = D-leucine.
113Cd NMR Spectroscopy.
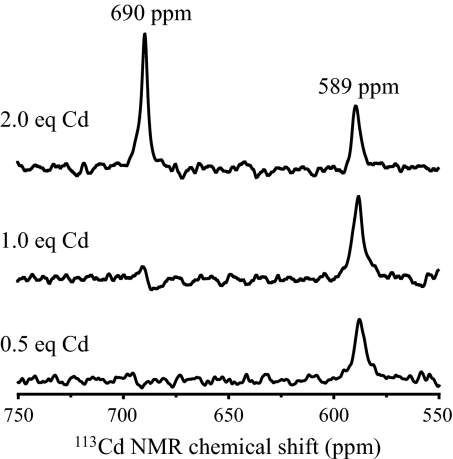
A single resonance was observed in the 113Cd NMR spectrum of 4.0 mM (TRIL12LDL16C)3 recorded at pH 9.2 in the presence of 1.1 equivalents of 113Cd(NO3)2, with a chemical shift of 697 ppm. The spectrum recorded of a 3.7 mM solution of (GRANDL12LDL16CL26AL30C)3 at pH 9.4 in the presence of half an equivalent of 113Cd(NO3)2 resulted in a single peak with chemical shift of 589 ppm. Another spectrum was recorded in the presence of 1 equivalent of 113Cd(NO3)2 at pH 9.3, which again resulted in a single peak with a chemical shift of 589 ppm. The spectra of fully loaded Cd2(GRANDL12LDL16CL26AL30C)32−, in the presence of 2 equivalents of 113Cd(NO3)2 at pH 9.4, contains 2 peaks with chemical shifts of 690 and 589 ppm, respectively, see Fig. 5.
Fig. 5.
113Cd NMR spectra of solutions of (GRANDL12DLL16CL26AL30C)3 in the presence of different amounts of 113Cd(NO3)2 at pH ≈9.4.
Discussion
Our objective in this study is to assess whether one can exploit the alternate side-chain conformations of D-amino acids to alter the coordination number preference of a metal bound in the interior of a 3-stranded coiled coil by a single residue substitution. If successful, we should be able to use this strategy to prepare a peptide capable of binding Cd(II) to 3 Cys ligands exclusively as trigonal CdS3. Furthermore, we should be able to prepare a single short sequence capable of binding 2 equivalents of Cd(II) to 2 similar Cys sites, separated by ≈20 Å, exclusively as 3- and 4-coordinate complexes, respectively. We have achieved this latter objective using Pen (21, 31); however, the applicability of the prior studies is limited by the fact that penicillamine alters both steric and inductive effects of the metal ligand. Clearly, achieving such a construct solely with L-Cys will have significant implications for interpreting real metal ion sites in biomolecules. In this article, peptides containing a single D-amino acid in an otherwise all L-amino acid sequence are termed diastereopeptides, and those capable of binding 2 equivalents of Cd(II) in 2 different coordination geometries but with almost identical metal-binding sites, will be termed heterochromic diastereopeptides.
Apo Diastereopeptides.
Our design hypothesis was that changing the chirality at a single carbon backbone (L- to D-Leu mutation) would serve to reorient the Leu side chain toward the C terminus (Fig. 1) increasing the steric bulk directly above the Cys plane. It is proposed that this bulk would prevent exogenous water from binding to the Cd(II) in Cd(TRIL12LDL16C)3−. Before we examined the metal-binding capability of the peptides, we first needed to assure ourselves that the introduction of a single D-amino acid into an otherwise all L-amino acid sequence did not alter the designed secondary and tertiary structure of our peptide aggregates. The CD spectrum of the apopeptides confirmed that these constructs were well folded coiled coils. Although up to three D-amino acid substitutions in a 19-aa α-helix has been reported to be tolerated (27), this is a previously unreported example of D-amino acids substitutions in the interior of an otherwise all L-amino acid coiled coil construct.
Characterizing Trigonal Cd(II)Cys3.
Having established that insertion of 1 D-leu into the sequence had negligible effect on the 3-stranded coiled-coil structure, we next examined the binding of Cd(II) to TRIL12LDL16C. Titration of Cd(II) into a solution of TRIL12LDL16C led to a UV chromophore at 235 nm (ε = 23 600 M−1·cm−1) consistent with a Cd(II) thiolate species. This titration yielded a stoichiometry of 1 Cd(II) per 3 peptide monomers, reconfirming the aggregate as a 3-stranded coiled coil, Cd(TRIL12LDL16C)3−. We have used 113Cd NMR to assign the coordination number and environments for Cd(II) bound to these peptide constructs and have found this to be an important tool in interpreting the data for these systems (Olga Iranzo, Tamas Jakusch, Kyung-Hoon Lee, L.H., and V.L.P., unpublished work). Indeed, it was predicted that a 100% 3-coordinate Cd(II) bound to 3 Cys would have a 113Cd NMR chemical shift of 698 ppm (21). Certainly, our value of 697 ppm for 113Cd(TRIL12LDL16C)3− is very close to this optimal value for a CdS3 species. Furthermore, the 111mCd PAC spectrum (Fig. 4B) for this complex confirms that only 1 species is present in solution (on the nanosecond time scale), and that this species has ω0 and η parameters (Table 2) that are predicted using the semiempirical BASIL model for trigonal CdS3 (32). Most important, these NQI parameters are very similar to those observed for 111mCd(TRIL16Pen)3− (21). Therefore, these data are best interpreted as supporting a 100% trigonal planar CdS3 coordination geometry. Furthermore, the high pKa2 of 15.1 for sequestering Cd(II) is consistent with a 3-coordinate CdS3 site. Notably, the values of 697 ppm and 15.1 for the 113Cd NMR chemical shift and pKa2 for binding Cd(II) as Cd(TRIL12LDL16C)3− fall nicely on a correlation established between these 2 parameters [see supporting information (SI) Fig. S1] (Olga Iranzo, Tamas Jakusch, Kyung-Hoon Lee, L.H., and V.L.P., unpublished work). This proves that the high pKa2 associated with binding Cd(II) is clearly due to the coordination number preference of the metal ion and not to the nature of the coordinating group (Pen vs. Cys). We conclude that we have prepared a stable Cd(II)Cys3 species in aqueous solution.
Heterochromic Peptides.
The next goal was to establish whether this approach would allow us to design a single peptide sequence capable of binding 2 equivalents of Cd(II) to 2 very similar Cys sites separated by ≈20 Å. We found that the additional stability provided by an extra heptad repeat in GRAND was important in allowing us to access a heterochromic peptide capable of binding Cd(II) with exclusive coordination geometries. Therefore, our proposed peptide contained our 3-coordinate L12LDL16C segment in a GRAND sequence along with the L26AL30C mutation that has been shown to bind Cd(II) as exclusively 4-coordinate CdS3O was prepared (31). The 2 Cys sites are ≈20 Å apart and are separated by 2 layers of intervening Leu residues that should act to keep these sites independent from one another. The binding of Cd(II) to this construct was again monitored by UV-visible spectroscopy. The absorbance at 230 nm on titrating Cd(II) into a solution of peptide levels off at a 2:3 ratio of Cd(II) to peptide monomer, confirming that (GRANDL12LDL16CL26AL30C)3 is capable of sequestering 2 equivalents of Cd(II). The absorbance at 230 nm of a solution of (GRANDL12LDL16CL26AL30C)3 in the presence of 2 equivalents of Cd(II) was monitored as a function of pH, and although Cd(II) is sequestered at a lower pH than for the analogous TRIL12LDL16C peptide, it occurs at a more basic pH than has been reported for L26AL30C sites (31). However, the pKa2 associated with this site is sensitive to the overall stability of the peptide, and the presence of D-Leu in GRANDL12LDL16CL26AL30C is destabilizing, resulting in a higher pKa2. This leads to a pH profile with the 2 sites displaying closely overlapping titration curves as opposed to the well-resolved profiles observed for GRANDL16PenL26AL30C (31). Once again, the binding of Cd(II) was characterized by 113Cd NMR spectroscopy. The fully loaded peptide, 113Cd2(GRANDL12LDL16CL26AL30C)32−, exhibited 2 distinct and separate peaks for the 2 independent sites at 589 and 690 ppm, indicating these 2 sites are not in chemical exchange with one another. Furthermore, the 589 and 690 ppm chemical shifts are consistent with CdS3O and CdS3 environments, respectively (Olga Iranzo, Tamas Jakusch, Kyung-Hoon Lee, L.H., and V.L.P., unpublished work) (31). These observations demonstrate that the heterochromic diastereopeptide controls the binding of Cd(II) as either 3- or 4-coordinate structures even though the first coordination sphere ligand is cysteine in both sites.
Binding Selectivity.
We next assessed whether the diastereotopic heterochromic peptide, GRANDL12LDL16CL26AL30C, showed selective binding of Cd(II) to 1 of the Cys sites. We had shown with GRANDL16PenL26AL30C that Cd(II) bound at lower pH values to the 4-coordinate site exclusively, and that at higher pH values, where both sites bind Cd(II), the metal still preferred the 4-coordinate environment. Unfortunately, because it was necessary to use penicillamine instead of cysteine in this construct, we could not be certain that the observed selectivity was due solely to the coordination number preference of the metal, or whether other factors, such as inductive effects on the ligating sulfur atoms or access to the site due to steric effects of the bulkier penicillamine, caused the site differentiation. Because Cd2(GRANDL12LDL16CL26AL30C)32− contains only cysteine ligands, our approach allows us to directly assess whether the peptide can discriminate metal complexation based solely on coordination number. This hypothesis was tested by assessing whether binding affinity at the same pH differed between the sites, by performing a titration of Cd(II) into a solution of GRANDL12LDL16CL26AL30C at a pH at which both sites are capable of sequestering Cd(II). From the pH titrations shown in Fig. 3, it appears that the Cd(II) is sequestered at a slightly lower pH to the 4-coordinate L26AL30C site, with the pH profiles for both sites reaching a plateau by pH 9.4. The direct competition of Cys sites in GRANDL12LDL16CL26AL30C for Cd(II) at a pH where they both bind was monitored by 113Cd NMR spectroscopy, which illustrated, under these conditions, that the first equivalent of Cd(II) selectively binds to the 4-coordinate L26AL30C site, and only on further addition of Cd(II) does binding occur to the 3-coordinate L12LDL16C site. This conclusively shows that site selectivity is the result of metal ion coordination geometry preference and appears to be independent of the nature of thiol ligand (Pen vs. Cys). For this selectivity to have been observed, there must be a minimum 10-fold stronger binding to the 4-coordinate site at pH 9.4 over the 3-coordinate site. These experiments provide compelling evidence for 2 key principles governing metalloprotein active site structure. First, we have shown that coordination number can be defined solely by alteration of noncoordinating ligands proximate to the metal center. Second, we prove that a protein can be designed that shows metal-binding selectivity based solely on coordination number control and not by varying the affinity of ligands in the metal first coordination sphere.
Conclusions.
We report here a unique approach for controlling metal ion coordination number in de novo designed metalloproteins by using D-amino acids to reorient side chains. This now opens the door to accessing new constructs that were unattainable using solely L-amino acids, and we anticipate that this approach will allow us to generate challenging metal coordination geometries within a biological construct, that nature is capable of enforcing on a metal that has a preference for a different coordination geometry or ligand environment. These diastereopeptides have led to the successful preparation of a stable trigonal CdS3 bound to L-Cys ligands in aqueous solution, a site likely to be significant in real biomolecules. Furthermore, a single peptide chain has been designed, GRANDL12LDL16CL26AL30C that is capable of independently binding 2 equivalents of Cd(II) to very similar Cys sites, as exclusively 3- and 4-coordinate structures, with Cd(II) displaying selectively for the 4-coordinate, CdS3O, site. This construct allows us to unambiguously establish that such selectivity for very similar Cys sites that differ only in their second-coordination sphere ligands is the direct result of coordination geometry preference of the metal ion and not ligand type. Not only is this a major achievement in the field of de novo metallopeptide design, but one can envision that proteins may control metal ion affinity by controlling coordination number. In particular, this could be essential in metal storage, transport, and transfer, as in metallochaperones. Some sites such as (TRIL16C)3 bind Cd(II) as a mixture of CdS3 and CdS3O, and it seems reasonable that Cd(II) would display a higher affinity for a CdS3O-rich site. Thus, if nature is capable of tuning the CdS3/CdS3O ratio, it may be able to tune the binding affinity of a specific metal ion for that site.
Experimental Procedures
Peptide Synthesis and Purification.
Both TRI and GRAND peptides (sequences given in Table 1) were synthesized on an Applied Biosystems 433A peptide synthesizer using standard protocols (33) and purified and characterized as reported (14). Stock solutions of the apopeptides were prepared in doubly distilled water, and their concentrations determined by calculating the thiol concentration using a published assay with 4,4′-dipyridyl disulphide (34). All solutions were degassed with argon before use so as to prevent oxidation of peptide and formation of disulfide bonds.
CD Spectroscopy.
CD spectra were of 40 μM peptide solutions in 50 mM Tris buffer, pH 8.5, were recorded in a 0.1-cm quartz cuvette on an Aviv 62DS spectrometer from 280 to 200 nm at 298 K. The observed ellipticity in millidegrees has been converted to molar ellipticity, [θ], and is reported in units of deg·dmol−1·cm2.
UV-Visible (UV-Vis) Spectroscopy.
Cd(II) into peptide titrations were performed on a Cary 100 Bio UV-vis spectrometer in 1-cm quartz cuvette. Aliquots of 4.47-Mm CdCl2 were added to a 30-μM peptide solution in 50 mM Tris at pH 8.5. Difference spectra were obtained by subtracting the background spectrum of peptide (30 μM in 50 mM Tris, pH 8.5) in the absence of metal.
pH Titrations.
All UV-vis pH titrations were performed at ambient temperature on an Ocean Optics SD 2000 fiber-optic spectrometer and the pH measured using a mini-glass combination pH electrode coupled to a Fisher Accumet model 805 pH meter. Titrations were performed by adding small aliquots of concentrated solutions of KOH and HCl and the absorbance at λmax (230/235 nm) monitored as a function of pH. Reverse titrations with HCl were performed to ensure reversibility of the process. Experimental data for TRIL12LDL16C were fit to a model published for the simultaneous release of 2 protons on binding Cd(II) to 3 thiolates (16).
PAC Spectroscopy.
PAC spectroscopy and sample preparation were performed as described (35, 36); however, a typing error of “10–40 mL” should be “10–40 μL,” and the current experiment with 111mCd and TRIL12LDL16C was carried out in a 1:12 ratio, at pH 9.1, 1 °C, and in 55% wt/wt sucrose to further reduce the Brownian tumbling of the molecules. Finally, all fits were carried out with 300 data points, disregarding the 5 first points due to systematic errors in these.
113Cd NMR Spectroscopy.
All spectra were collected at room temperature on a Varian Inova 500 spectrometer (110.92 MHz for 113Cd) equipped with a 5-mm broadband probe and externally referenced to a 0.1 M Cd(ClO4)2 solution in D2O. A spectral with of 847 ppm (93,897 Hz) was sampled using a 5-μs 90° pulse and 0.05-s acquisition time, with no delay between scans. Samples were prepared by dissolving 25–35 mg of lyophilized and degassed peptide in 500 μL of 15% D2O/H2O under a flow of argon. Peptide concentration was determined as described by using an assay with 4,4′-dipyridyl disulphide (34) and the desired amount of a 250 mM 113Cd(NO3)2 solution (prepared from 95% isotopically enriched 113CdO obtained from Oak Ridge National Laboratory) added. The pH of the solution was measured and adjusted to ≈9.4 by addition of concentrated solutions of HCl or KOH and the pH measured both before and after recording the spectrum. Attempts were made to maintain an argon environment; however, the solutions did come into contact with the air on recording and adjusting the pH. The data were processed by using MestRe-C (37) and the free induction decays zero-filled and treated with an exponential function with a line-broadening value of 100 Hz before Fourier transformation.
Supplementary Material
Acknowledgments.
V.L.P. thanks the National Institutes of Health for support of this research (Grant R01 ES0 12236). L.H. thanks the Danish Natural Science Research Council for support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806792105/DCSupplemental.
References
- 1.Holm RH, Kennepohl P, Solomon EI. Structural and functional aspects of metal sites in biology. Chem Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 2.Special Edition on Protein Design. Chem Rev. 2001;101:3025–3232. [Google Scholar]
- 3.Forum on Biomolecular Design in Inorg Chem. Inorg Chem. 2005;46:9927–10402. [Google Scholar]
- 4.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. De novo design and structural characterization of proteins and metalloproteins. Annu Rev Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 5.Iranzo O, Ghosh D, Pecoraro VL. Assessing the integrity of designed homomeric parallel three-stranded coiled coils in the presence of metal ions. Inorg Chem. 2006;45:9959–9973. doi: 10.1021/ic061183e. [DOI] [PubMed] [Google Scholar]
- 6.Bryson JW, et al. Protein design: A hierarchic approach. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 7.Summers MF. 113Cd NMR spectroscopy of coordination compounds and proteins. Coord Chem Rev. 1988;86:43–134. [Google Scholar]
- 8.Armitage IM, Pajer RT, Uiterkamp AJMS, Chlebowski JF, Coleman JE. Cadmium-113 Fourier transform nuclear magnetic resonance of cadmium(II) carbonic anhydrases and cadmium(II) alkaline phosphatase. J Am Chem Soc. 1976;98:5710–5712. doi: 10.1021/ja00434a058. [DOI] [PubMed] [Google Scholar]
- 9.Hecht MH. De novo design of β-sheet proteins. Proc Natl Acad Sci USA. 1994;91:8729–8730. doi: 10.1073/pnas.91.19.8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kortemme T, Ramírez-Alvarado M, Serrano L. Design of a 20-amino acid, three-stranded β-sheet protein. Science. 1998;281:253–256. doi: 10.1126/science.281.5374.253. [DOI] [PubMed] [Google Scholar]
- 11.Silverman JA, Balakrishnan R, Harbury PB. Reverse engineering the (β/α)8 barrel fold. Proc Natl Acad Sci USA. 2001;98:3092–3097. doi: 10.1073/pnas.041613598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dieckmann GR, et al. De novo design of mercury-binding two- and three-helical bundles. J Am Chem Soc. 1997;119:6195–6196. [Google Scholar]
- 13.Dieckmann GR, et al. The role of protonation and metal chelation preferences in defining the properties of mercury-binding coiled coils. J Mol Biol. 1998;280:897–912. doi: 10.1006/jmbi.1998.1891. [DOI] [PubMed] [Google Scholar]
- 14.Farrer BT, Harris NP, Balchus KE, Pecoraro VL. Thermodynamic model for the stabilization of trigonal thiolato mercury(II) in designed three-stranded coiled coils. Biochemistry. 2001;40:14696–14705. doi: 10.1021/bi015649a. [DOI] [PubMed] [Google Scholar]
- 15.Farrer BT, McClure CP, Penner-Hahn JE, Pecoraro VL. Arsenic(III)-cysteine interactions stabilize three-helix bundles in aqueous solution. Inorg Chem. 2000;39:5422–5423. doi: 10.1021/ic0010149. [DOI] [PubMed] [Google Scholar]
- 16.Matzapetakis M, Ghosh D, Weng T-C, Penner-Hahn JE, Pecoraro VL. Peptidic models for the binding of Pb(II), Bi(III) and Cd(II) to mononuclear thiolate binding sites. J Biol Inorg Chem. 2006;11:876–890. doi: 10.1007/s00775-006-0140-7. [DOI] [PubMed] [Google Scholar]
- 17.Farrer BT, Pecoraro VL. Hg(II) binding to a weakly associated colied coil nucleates an encoded metalloprotein fold: A kinetic analysis. Proc Natl Acad Sci USA. 2003;100:3760–3765. doi: 10.1073/pnas.0336055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iranzo O, Thulstrup PW, Ryu S, Hemmingsen L, Pecoraro VL. The application of 199 Hg of NMR and 199 mHg perturbed angular correlation (PAC) spectroscopy to define the biological chemistry of HgII: A case study with designed two- and three-stranded coiled coils. Chem Eur J. 2007;13:9178–9190. doi: 10.1002/chem.200701208. [DOI] [PubMed] [Google Scholar]
- 19.Touw DS, Nordman CE, Stuckey JE, Pecoraro VL. Identifying important structural characteristics of arsenic resistance proteins by using designed 3-stranded coiled coils. Proc Natl Acad Sci USA. 2007;104:11969–11974. doi: 10.1073/pnas.0701979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K-H, Matzapetakis M, Mitra S, Marsh ENG, Pecoraro VL. Control of metal coordination number in de novo designed peptides through subtle sequence modifications. J Am Chem Soc. 2004;126:9178–9179. doi: 10.1021/ja048839s. [DOI] [PubMed] [Google Scholar]
- 21.Lee K-H, Cabello C, Hemmingsen L, Marsh ENG, Pecoraro VL. Using nonnatural amino acids to control metal-coordination number in three-stranded coiled coils. Angew Chem Int Ed. 2006;45:2864–2868. doi: 10.1002/anie.200504548. [DOI] [PubMed] [Google Scholar]
- 22.Mahalakshmi R, Balaram P. In: D-Amino Acids: A New Frontier in Amino Acid and Protein Research. Konno R, Fisher GH, Brueckner H, d'Aniello A, Fujii N, Homma H, editors. NY: Nova Science; 2006. pp. 415–430. [Google Scholar]
- 23.Aravinda S, Shamala N, Roy RS, Balaram P. Non-protein amino acids in peptide design. Proc Ind Acad Sci. 2003;115:373–400. [Google Scholar]
- 24.Zawadzke LE, Berg JM. A racemic protein. J Am Chem Soc. 1992;114:4002–4003. [Google Scholar]
- 25.Haack T, González MJ, Sánchez Y, Giralt E. D-amino acids in protein de novo design. II. Protein-diastereomerism versus protein-enantiomerism. Lett Pept Sci. 1997;4:377–386. [Google Scholar]
- 26.Fairman R, Anthony-Cahill SJ, DeGrado WF. The helix-forming propensity of D-alanine in a right-handed α-helix. J Am Chem Soc. 1992;114:5458–5459. [Google Scholar]
- 27.Karle IL, Gopi HN, Balaram P. Crystal structure of a hydrophobic 19-residue peptide helix containing three centrally located D amino acids. Proc Natl Acad Sci USA. 2003;100:13946–13951. doi: 10.1073/pnas.2336106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aravinda S, Shamala N, Desirajub S, Balaram P. A right handed peptide helix containing a central double D-amino acid segment. Chem Comm. 2002:2454–2455. doi: 10.1039/b207960g. [DOI] [PubMed] [Google Scholar]
- 29.Sia SK, Kim PS. A designed protein with packing between left-handed and right-handed helices. Biochemistry. 2001;40:8981–8989. doi: 10.1021/bi010725v. [DOI] [PubMed] [Google Scholar]
- 30.Van Regenmortel MHV, Muller S. D-peptides as immunogens and diagnostic reagents. Curr Opin Biotechnol. 1998;9:377–382. doi: 10.1016/s0958-1669(98)80011-6. [DOI] [PubMed] [Google Scholar]
- 31.Iranzo O, Cabello C, Pecoraro VL. Heterochromia in designed metallopeptides: Geometry-selective binding of CdII in a de novo peptide. Angew Chem Int Ed. 2007;46:6688–6691. doi: 10.1002/anie.200701729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer R, Jensen SJ, Schmidt-Nielsen B. The angular overlap model applied to the calculation of nuclear quadrupole interactions. Hyp Int. 1988;39:203–234. [Google Scholar]
- 33.Chan WC, White PD. In: Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Hames BD, editor. New York: Oxford Univ Press; 2000. p. 346. [Google Scholar]
- 34.Mantle M, Stewart G, Zayas G, King M. The disulphide-bond content and rheological properties of intestinal mucins from normal subjects and patients with cystic fibrosis. Biochem J. 1990;266:597–604. [PMC free article] [PubMed] [Google Scholar]
- 35.Hemmingsen L, et al. Cd-substituted horse liver alcohol dehydrogenase: Protein conformation and metal site coordination geometry in binary and ternary inhibitor complexes. Eur J Biochem. 1996;241:546–551. doi: 10.1111/j.1432-1033.1996.00546.x. [DOI] [PubMed] [Google Scholar]
- 36.Matzapetakis M, et al. Comparison of the binding of cadmium(II), mercury(II), and arsenic(III) to the de novo designed peptides TRI L12C and TRI L16C. J Am Chem Soc. 2002;124:8042–8054. doi: 10.1021/ja017520u. [DOI] [PubMed] [Google Scholar]
- 37.Cobras C, Cruces J, Sardina FJ. Mestre C, Version 2.3. Santiago de Compostela, Spain: Universidad de Santiago de Compostela; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.