Abstract
Yeast prions, such as [PSI+], [RNQ+], and [URE3], are heritable elements formed by proteins capable of acquiring self-perpetuating conformations. Their propagation is dependent on fragmentation of the amyloid protein complexes formed to generate the additional seeds necessary for conversion of nascent soluble protein to the prion conformation. We report that, in addition to its known role in [RNQ+] propagation, Sis1, a J-protein cochaperone of Hsp70 Ssa, is also specifically required for propagation of [PSI+] and [URE3]. Whereas both [RNQ+] and [URE3] are cured rapidly upon SIS1 repression, [PSI+] loss is markedly slower. This disparity cannot be explained simply by differences in seed number, as [RNQ+] and [PSI+] are lost with similar kinetics upon inhibition of Hsp104, a remodeling protein required for propagation of all yeast prions. Rather, in the case of [PSI+], our results are consistent with the partial impairment, rather than the complete abolition, of fragmentation of prion complexes upon Sis1 depletion. We suggest that a common set of molecular chaperones, the J-protein Sis1, the Hsp70 Ssa, and the AAA+ ATPase Hsp104, act sequentially in the fragmentation of all yeast prions, but that the threshold of Sis1 activity required for each prion varies.
Keywords: amyloid, Hsp40, Hsp70, molecular chaperone, Sup35
Yeast prions are non-Mendelian inherited elements capable of forming self-perpetuating conformations (1). Of the several prions identified in Saccharomyces cerevisiae, 3 are the best characterized: [PSI+], [RNQ+] (also called [PIN+]), and [URE3], formed by the aggregated states of the cytosolic proteins Sup35, Rnq1, and Ure2, respectively (1). Sup35 is a translation termination factor; Ure2 is a regulator that acts to repress transcription of a set of genes involved in nitrogen catabolism; the function of Rnq1 is unknown. Prion proteins can form different conformational states resulting in prion strains having different heritable traits. For propagation in the cell population, physical transmission of the prion template, often referred to as the propagon or seed, is required to allow conversion of newly synthesized protein to the prion conformation (2).
Somewhat paradoxically, the propagation of yeast prions appears to be inexorably reliant on the function of molecular chaperones, proteins that normally function to prevent protein misfolding (2). Two chaperone systems have been linked to prion propagation: the hexameric AAA+ ATPase Hsp104 and the J-protein (Hsp40):Hsp70 chaperone machinery, with its associated nucleotide exchange factors (3). Hsp104, like its ortholog ClpB, functions in protein remodeling by threading partially folded proteins through its central pore and is stringently required for the propagation of all identified yeast prions (1, 4, 5).
Hsp70s function with their obligate cochaperones, J-proteins, which act to stimulate Hsp70 ATPase activity and stabilize their interaction with client proteins (6). Although J-proteins are very diverse in sequence and structure, they possess a highly conserved J-domain that is responsible for the stimulation of the ATPase activity of Hsp70s. One cytosolic J-protein, Sis1, is required for propagation of [RNQ+] (7). In addition, multiple individual amino acid substitutions in the cytosolic Hsp70s Ssa1/2 that impair propagation of [PSI+] and [URE3] have been identified (3, 8, 9). Participation of Hsp70s in [PSI+] and [URE3] propagation implies an involvement of an unidentified J-protein as well.
The currently favored model for prion propagation posits chaperone-mediated fragmentation of prion complexes to produce sufficient prion seeds to assure consistent transmission of seeds to daughter cells, thus maintaining the prion in the cell population (2, 10–14). Supporting this model, inhibition of Hsp104 activity results in an increase in the size of Sup35 and Rnq1 prion complexes and subsequent prion loss, which has been shown in the case of [PSI+] to be dependent on cell division (10, 11, 15, 16). Additional support for this idea comes from reports of fragmentation of prion fibers in vitro by Hsp104 (17) and the apparent decrease in the number of [PSI+] prion seeds in cells expressing a dominant mutation in the Hsp70 SSA1 gene (8). An increase in the size of Rnq1 polymers, followed by [RNQ+] loss, also occurs upon depletion of Sis1, a partner of Ssa1 (15). Together these data suggest cooperation between the 2 chaperone systems. Such cooperation has precedent, as Hsp104 is known to function in disaggregation of amorphous protein aggregates in conjunction with J-protein:Hsp70 chaperone machinery, with J-protein/Hsp70 and Hsp104 machineries acting sequentially (4).
The yeast cytosol contains 13 J-proteins, 12 of which are thought to function with the Ssa class of Hsp70s (6). To better understand the contribution of J-proteins in prion maintenance, we set out to answer two questions: (i) whether any J-proteins other than Sis1 are required for [RNQ+] maintenance; and (ii) if Sis1, or any other J-protein, is required for propagation of [PSI+] and [URE3]. We found Sis1 to be unique among the cytosolic J-proteins, as no other J-protein was needed for [RNQ+] propagation. Sis1 is required for maintenance of both [PSI+] and [URE3] as well. However, the rates of prion loss upon Sis1 depletion differed among the 3 prions, indicating a similar, but not identical, requirement for molecular chaperone activity.
Results
Sis1 Is the Only Cytosolic J-Protein Required for [RNQ+] Maintenance.
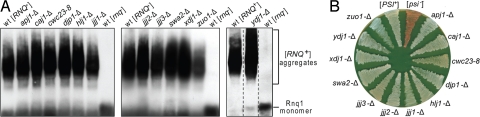
The J-protein Sis1 is required for [RNQ+] maintenance, yet 12 others reside in the cytosol of S. cerevisiae. To determine whether J-proteins other than Sis1 are required for [RNQ+], we tested a set of strains, each carrying a deletion of 1 of the genes encoding cytosolic J-proteins, which we previously constructed from a [RNQ+] parent (18). Before assessing the ability of these deletion strains to maintain [RNQ+], each was serially passaged 10 to 12 times on rich media (≈70–90 generations). Cell extracts were then prepared and analyzed using semidenaturing detergent agarose gel electrophoresis (SDD-AGE), which is able to resolve prion complexes (15). All strains remained [RNQ+], as indicated by the presence of high molecular weight aggregates in all strains (Fig. 1A). We conclude that no cytosolic J-protein other than Sis1 is compulsory for [RNQ+] maintenance.
Fig. 1.
No J-protein other than Sis1 is required for maintenance of [RNQ+] or [PSI+]. [RNQ+] (A) or [PSI+] (B) cells lacking individual J-proteins were passaged for 3 weeks on rich media before analysis. (A) For analysis of [RNQ+], cell lysates were prepared, resolved by SDD-AGE and subjected to immunoblotting using Rnq1-specific antibodies. Dotted lines indicate lanes from different parts of the same gel. Non-specific bands appearing in all lanes at the bottom of the gel have been cropped for clarity. (B) For [PSI+], cells were streaked onto rich medium and grown at 22 °C for color development. Control WT [PSI+] and [psi−] cells were included for comparison. Analysis of extracts from these strains by SDD-AGE was performed (Fig. S1). Additionally, as expected, all [PSI+] strains maintained the ability to grow in the absence of adenine (data not shown).
Sis1, But No Other J-Protein, Is Required for [PSI+] Propagation.
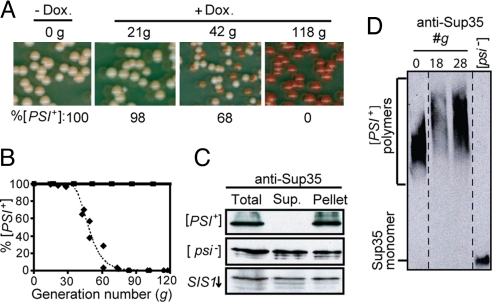
We next wanted to determine the J-protein requirement for [PSI+] propagation. As the J-protein gene deletion strains described earlier were [RNQ+] [psi−], each of the 12 strains were crossed to a [PSI+] strain that also contained a nonsense allele of ADE1, a gene required for adenine synthesis. In the [psi−] state, when soluble Sup35 causes efficient translation termination, colonies are red as a result of the accumulation of a pigment that occurs in the absence of Ade1 function. In [PSI+] cells, read-through of the nonsense codon is enhanced, as most Sup35 is in the aggregated state; thus, colonies are white or light pink. As described earlier, strains were serially passaged on solid media for 3 weeks and the [PSI+] state monitored, both by colony color (Fig. 1B) and by detection of aggregates in semidenaturing agarose gels [supporting information (SI) Fig. S1]. In each case, [PSI+] was maintained. Sis1 is an essential J-protein; therefore, we used a system that had SIS1 under the control of the tetR promoter (TETr), which allows repression of Sis1 synthesis upon addition of the drug doxycycline (15). In the absence of doxycycline, sis1-Δ cells expressing SIS1 from the tetR promoter, sis1-Δ [TETrSIS1], stably maintained [PSI+]. To test whether Sis1 is important for maintenance of [PSI+], drug was added to the culture. Cells were collected at intervals and plated on solid medium to assess the status of [PSI+] based on colony color (Fig. 2A). Repression of SIS1 severely affected [PSI+] propagation. By 49 generations, more than half the colonies were red; by 85 generations, no white colonies were observed, indicating loss of the prion from the population (Fig. 2B).
Fig. 2.
Sis1 is required for [PSI+] propagation. (A and B) Time course of SIS1 repression of [PSI+] cells. Cells were harvested after the indicated number of generations of growth in the presence (diamonds) or absence (squares) of doxycycline and plated onto rich media. The percentage of cells [PSI+] (pink) versus [psi−] (red) was determined (A) and plotted (B). (C) Lysates from sis1-Δ [TETrSIS1] cells, originally [PSI+] before Sis1 repression, prepared 118 generations after addition of doxycycline (SIS1↓), as well as from control [PSI+] and [psi−] cells, were subjected to centrifugation. Equivalent total, supernatant, and pellet fractions were resolved by SDS/PAGE and immunoblot analysis using Sup35-specific antibodies. (D) Lysates from sis1-Δ [TETrSIS1] cells at zero time (0) and 18 or 28 generations after addition of doxycycline and from [psi−] control cells were resolved by SDD-AGE and Sup35 visualized by immunoblot analysis. Dotted lines indicate lanes from different parts of the same gel.
To confirm loss of [PSI+], we carried out 2 biochemical and 2 biological assays. Aggregation of Sup35 was monitored by centrifugation of cell lysates. In control [PSI+] lysates, all Sup35 was found in the pellet fraction; in [psi−] lysates, approximately 50% was soluble. Analysis of lysates made from cells ≈100 generations after SIS1 repression revealed that ≈50% of Sup35 was soluble, as with [psi−] lysates (Fig. 2C). We also analyzed lysates using SDD-AGE. Curing of [RNQ+] upon Hsp104 inhibition or Sis1 depletion is preceded by an increase in the size of prion complexes, as is the curing of [PSI+] upon Hsp104 inhibition (16). The size of Sup35 complexes increased in size after repression of SIS1 (Fig. 2D), consistent with a role of Sis1 in [PSI+] prion fragmentation along with Hsp104. In addition, we conducted 2 biological assays to determine the ability of cells to transmit the prion either through cytoplasmic contact (i.e., cytoduction) or through cotransformation of cell lysates with an essential plasmid (i.e., lysate transformation). Cells were tested before and after Sis1 repression; before Sis1 depletion, 100% of cytoductants and 85% of transformants isolated were [PSI+], whereas, after Sis1 depletion, 100% of cells isolated were [psi−] based on either assay (Table S1).
The results described herein demonstrate that [PSI+] is lost when Sis1 is depleted. However, as modest overexpression of Hsp104 is known to destabilize [PSI+] (19), we tested the level of Hsp104 after SIS1 repression. Hsp104 levels were slightly elevated during the timeframe of prion loss in cells following SIS1 repression compared with cells that have normal levels of the protein (Fig. S2A). Therefore we directly tested the involvement of Hsp104 in prion loss by substituting a single amino acid variant, Hsp104T160M, for WT Hsp104. Hsp104T160M is competent to maintain [PSI+], but incapable of driving prion loss when overexpressed (20). [PSI+] was lost when Sis1 was depleted, regardless of whether WT Hsp104 or Hsp104T160M was expressed (Fig. S2B). In sum, we conclude that Sis1, but not other cytosolic J-proteins, is required for maintenance of [PSI+].
Loss of [PSI+] Is Significantly Slower than Loss of [RNQ+] upon SIS1 Repression.
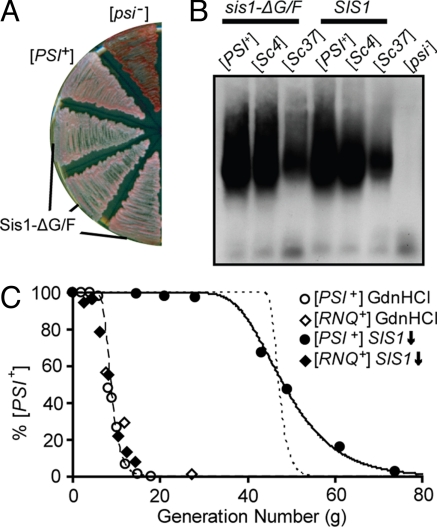
Recently we reported the use of sis1-Δ [TETrSIS1] to analyze the loss of the [RNQ+] prion upon SIS1 repression (15). The loss of [RNQ+] was dramatically faster than loss of [PSI+]: approximately 50% loss in 10 generations, compared with 50 generations. This difference in rate of loss of [RNQ+] and [PSI+] upon Sis1 depletion suggested to us that the 2 prions may have different Sis1 requirements for propagation. To test this idea further, we made use of a Sis1 mutant protein, Sis1-ΔG/F, which lacks the 51-aa glycine/phenylalanine-rich region (residues 71–121) adjacent to the J-domain. Sis1-ΔG/F supports cell viability but cannot maintain [RNQ+] (7). [PSI+] strains that have a chromosomal deletion of SIS1 but express Sis1-ΔG/F from a low-copy plasmid were obtained and passaged on solid media for 3 weeks. [PSI+] was maintained, as indicated both by the light pink colony color (Fig. 3A) and by the presence of large aggregates in cell lysates (Fig. 3B, first and fourth lanes).
Fig. 3.
The stringency of the Sis1 requirement for [RNQ+] and [PSI+] differ. (A) Sis1-ΔG/F supports [PSI+]. sis1-ΔG/F cells were passaged for 3 weeks and then streaked onto rich media along with control WT [PSI+] and [psi−] cells. (B) Maintenance of Sc4 (strong) and Sc37 (weak) [PSI+] strains by Sis1-ΔG/F. Cell lysates were prepared from sis1-ΔG/F cells described in A and sis1-ΔG/F harboring [PSI+] strains Sc4 or Sc37, as indicated. Lysates were resolved by SDD-AGE and Sup35 detected by immunoblot analysis. (C) Kinetics of curing [RNQ+] (open and filled diamonds) and [PSI+] (open and filled circles) by GdnHCl treatment (open diamonds and circles) or SIS1 repression (filled diamonds and circles). Kinetic modeling was performed as described in SI Methods. The dashed line represents an assumption of cells initially having 265 seeds/cell and no fragmentation of seeds occurs. The dotted line represents kinetic modeling of 1014 seeds/cell and no fragmentation. The solid line represents 265 seeds/cell, and 77% of seeds are fragmented each generation.
The ability of Sis1-ΔG/F to support [PSI+], together with the differences in curing rates, is consistent with the idea that Sis1 is more stringently required by [RNQ+] than by [PSI+]. However, we also considered whether the particular conformational state, rather than the primary sequence of the prion protein, was responsible for the difference observed. To address this issue, we made use of the 2 defined [PSI+] strains, Sc4 (strong) and Sc37 (weak), which were generated by transformation of Sup35 fibers formed at 4 °C and 37 °C, respectively (21). Both the Sc4 and Sc37 [PSI+] strains were cured by Sis1 depletion (data not shown). Additionally, both were maintained in sis1-ΔG/F cells through many generations of growth (Fig. 3B). In sum, although Sis1 is required for the propagation of both [RNQ+] and [PSI+], a difference in the nature of this requirement by the 2 prions is evident.
Because both prions also require Hsp104 for propagation, we compared the loss of [PSI+] with that of [RNQ+] upon inhibition of Hsp104 by treatment with guanidinium hydrochloride (GdnHCl). Consistent with previous results from other strain backgrounds (13), [PSI+] was cured rapidly, with approximately 50% loss occurring in 8 generations, and complete loss occurring within 15 generations (Fig. 3C). This rate of loss of [PSI+] is very similar to that of [RNQ+] upon GdnHCl treatment (15). We conclude that the kinetics of loss of the 2 prions are very similar when Hsp104 is inhibited, but not when Sis1 is depleted.
In considering the loss of [PSI+] upon Sis1 depletion, we noted two things: (i) the lag time before prion loss becomes detectable was considerably longer than observed for Hsp104 inhibition, and (ii) once loss became detectable, it proceeded slowly over approximately 30 generations. These kinetics were surprising, as the “propagon/seed model” of prion loss posits that, upon complete inhibition of new seed formation, the prion is retained at high levels in the population until the number of seeds are reduced by dilution; however, at this time, loss occurs very rapidly, with the length of the lag being dependent upon the number of seeds present before inhibition (11, 13, 22). The rapid loss of both [PSI+] and [RNQ+] upon Hsp104 inhibition and [RNQ+] upon Sis1 depletion are consistent with this model (Fig. 3C, dashed line), whereas the loss of [PSI+] upon Sis1 depletion is not. Increasing the number of seeds to account for the increased lag time resulted in unreasonable seed numbers (1014 seeds/cell) and still failed to fit the data (Fig. 3C, dotted line). We reasoned that relaxing the assumption of complete inhibition might improve the fit (see Materials and Methods for details). The seed number was held constant at 265 seeds/cell—the number calculated from the results obtained when Hsp104 is inhibited—while the efficiency of fragmentation per generation was allowed to vary between 0% and 100%. An excellent fit (R2 > 0.99) was obtained when, on average, 77% of seeds (as opposed to 0%) were fragmented once per generation over the course of the experiment (Fig. 3C, solid line). Thus, the kinetic data indicate that [PSI+] prion fragmentation is impaired, rather than abolished, when Sis1 is repressed, resulting in the eventual depletion of seeds and the loss of [PSI+] from the cell population.
Sis1 Is Required for [URE3] Propagation.
Because Sis1 is required for propagation of both [RNQ+] and [PSI+], we asked whether it also plays a role in [URE3] propagation. For these tests, we made use of a red/white colony color assay for discrimination between cells that possess or lack the prion ([URE3] or [ure-o], respectively). Like the assay for [PSI+], this assay is based on the accumulation of the red pigmented intermediate in the adenine biosynthesis pathway (23, 24). Because Ure2 acts as a transcriptional suppressor in nitrogen metabolism, the ADE2 gene was placed under the control of a Ure2 target promoter, DAL5. When Ure2 is sequestered in [URE3] aggregates, DAL5 is active, rendering [URE3] colonies white and [ure-o] colonies red. [URE3] is less mitotically stable than either [RNQ+] or [PSI+], but can be stably maintained by growth on poor nitrogen sources such as proline (25).
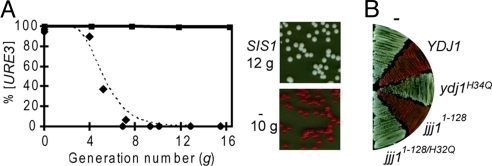
To test for a requirement for Sis1 in [URE3] propagation, a sis1-Δ [TETrSIS1] [URE3] strain with ADE2 under the control of the DAL5 promoter was constructed. Cells were transferred to proline-based medium containing doxycycline to repress SIS1 expression and samples removed for plating onto rich medium for determination of colony color. [URE3] was rapidly lost; only red colonies were observed after 8 generations of growth in doxycycline (Fig. 4A). As a control, sis1-Δ [TETrSIS1] [URE3] cells carrying a second plasmid expressing SIS1 from the endogenous promoter were treated with drug. [URE3] was stably maintained in these cells, as 100% of cells plated after 16 generations formed white colonies; however, when the additional SIS1 plasmid was not present, sis1-Δ [TETrSIS1] [URE3] cells slowly lost the prion in the absence of drug, with ≈25% of the cells being [ure-o] at 10 generations and 45% at 20 generations, indicating that [URE3] is highly sensitive to the dosage of Sis1.
Fig. 4.
Loss of [URE3] upon Sis1 depletion. (A) sis1-Δ [TETrSIS1] [URE3] carrying either an extra copy of SIS1 under control of its own promoter in a low copy plasmid (squares) or an empty vector (diamonds). Cells were harvested after the indicated number of generations in the presence of doxycycline and plated onto rich media. Left: The percentage of [URE3] (white) versus [ure-o] (red) colonies was determined and plotted. The dotted line represents kinetic modeling of 25 seeds/cell and no fragmentation. Right: Representative sample of sis1-Δ (TETrSIS1) [URE3] cells, which have an additional copy of SIS1 or an empty vector (−), taken at 12 and 10 generations, respectively. (B) [URE3] cells were transformed with plasmids bearing indicated overexpression plasmids with the GPD promoter and passaged on rich media. No insert (−); WT YDJ1 (YDJ1); YDJ1 with a mutation in J-domain (ydj1H34Q); 128 residue J-domain fragment of JJJ1 (jjj11-128); jjj11-128 fragment with mutation altering the J-domain (jjj11-128/H32Q).
As with [RNQ+] and [PSI+], data we obtained from doxycycline-treated sis1-Δ [TETrSIS1] [URE3] cells were fit to a kinetic model. [URE3] loss upon Sis1 depletion conformed to a model in which cells have ≈25 seeds/cell and no fragmentation occurs after Sis1 repression (Fig. 4A). This number is consistent with the previously reported value of ≈20 seeds/cell, a number obtained by inhibition of Hsp104 upon addition of GdnHCl (26). Thus, the rate of [URE3] loss upon Sis1 depletion indicates that Sis1, like Hsp104, is required for the fragmentation of [URE3] prion complexes.
Overexpression of a J-Domain Specifically Destabilizes [URE3].
The results described here indicate an important role of Sis1 in [URE3] propagation. We were interested in whether Ydj1, the most abundant cytosolic J-protein, also played a role, because it has been reported that its overexpression causes [URE3] loss (27). Therefore, we crossed a [URE3] and a ydj1-Δ strain and obtained ydj1-Δ [URE3] haploids, suggesting that Ydj1 is not critical for [URE3] maintenance. To more rigorously test this idea, cells were passaged for an additional 3 weeks; even after this interval, colonies remained white, indicating long-term maintenance of [URE3] in the absence of Ydj1. We wanted to corroborate that the white colony color indicated the presence of [URE3] rather than an indirect effect of the lack of Ydj1 on some other aspect of cell physiology. Therefore, we tested whether inhibition of Hsp104, which is known to be required for [URE3] propagation, resulted in red pigment accumulation. After treatment with GdnHCl, ydj1-Δ cells formed red colonies (data not shown), indicating that the prion was present before Hsp104 inhibition. Thus, we conclude that Ydj1 is not required for propagation of [URE3].
Because we did not detect an essential role for Ydj1 in the maintenance of [URE3], we wanted to examine more closely the effects of Ydj1 overexpression. First, we confirmed the loss of [URE3] in our system by overexpressing Ydj1 from the strong glucose-6-phosphate dehydrogenase (GPD) promoter (Fig. 4B). Second, we tested the effect on [URE3] propagation of GPD promoter driven expression of 8 other J-proteins (Apj1, Cwc23, Jjj1, Jjj2, Jjj3, Sis1, Swa2, and Zuo1). None of these constructs affected [URE3] stability. We also tested the effect of these constructs, as well as the YDJ1 construct, on the propagation of [PSI+] and [RNQ+]. No effect was observed with any construct on either [PSI+] or [RNQ+] propagation.
We reasoned that this apparent specificity of Ydj1 in regard to [URE3] stability was because Ydj1 is specifically involved in [URE3] propagation or was misleading because other J-proteins were either not expressed at sufficiently high levels or were sequestered in the cytosol such that they could not interact with prion complexes. To differentiate among these possibilities, we first asked if a functional J-domain of Ydj1 was required for the effect. We made use of an alteration in the highly conserved HPD motif, His34→Gln, which renders the J-domain of Ydj1 nonfunctional in vivo and incompetent for Hsp70 stimulation (28). Overexpression of the Ydj1H34Q variant did not result in [URE3] curing (Fig. 4B), indicating a requirement for a functional J-domain and thus an interaction with Hsp70. Next we asked if other regions of Ydj1 are required for destabilization of [URE3]. The N-terminal 134-aa fragment containing only the J-domain and adjacent G/F region, but lacking the client peptide binding and dimerization domains (Ydj11–134), was tested. Destabilization of [URE3] was observed (data not shown). Finally, we asked if the overexpression of a J-domain from another protein was sufficient. We tested a small, 128-aa fragment of the J-protein Jjj1 (Jjj11–128), which has only the J-domain in common with Ydj1, along with its corresponding inactive HPD motif mutant Jjj11–128/H32Q (18). Overexpression of Jjj11–128, but not Jjj11–128/H32Q, destabilized [URE3] (Fig. 4B), even though the full-length protein, which associates with the pre60S ribosomal subunits in the cytosol, did not (data not shown). We conclude that the effect of Ydj1 overexpression is not specific to Ydj1 itself, but rather results from the more generic effect of higher levels of a J-domain. However, when either of the J-domain fragments Ydj11–134 or Jjj11–128 were overexpressed in a [RNQ+] or [PSI+] strain, no disruption in the maintenance of either prion was observed; thus, these results reveal another difference in chaperone sensitivity among these 3 prions.
Discussion
The J-protein Sis1 functions in the propagation of the 3 well known yeast prions, [PSI+], [RNQ+], and [URE3], suggesting a common mechanism of prion propagation. Data are most consistent with a model in which Sis1 functions with the Hsp70 Ssa, cooperating with Hsp104 to facilitate the fragmentation of prion complexes.
Specific Sis1 Requirement in Prion Propagation.
Of the 13 J-proteins of the yeast cytosol, Sis1 is the only one required for the propagation of [PSI+] and [RNQ+]. Sis1 is also crucial for the propagation of [URE3]. Most surprising to us was the failure to detect a specific role in prion maintenance for the most abundant J-protein of the cytosol, Ydj1, which is known to be involved in many cellular processes (6). Ydj1 has also been shown to bind Ure2 in vitro (29), and its overexpression affects the stability of [URE3] and some [RNQ+] variants (27, 30). However, the results reported here demonstrate that overexpression of a small, functional J-domain-containing fragment from Ydj1, or from another J-protein having no sequence similarity with Ydj1 besides the J-domain, is sufficient to destabilize [URE3]. Thus, the effect is not specific for Ydj1 and may be a result of competition for Hsp70 by the overexpressed J-domain, or of increased stimulation of the ATPase activity of Hsp70. A similar effect was not observed for [RNQ+] or [PSI+] upon overexpression of any J-protein or J-domain construct, however, indicating that [URE3] is considerably more sensitive to the alterations in chaperone activity that result from excessive J-domain expression.
The requirement for Sis1 for propagation of 3 prions points to a common molecular mechanism for prion propagation. In addition, both Sup35 and Rnq1 polymers increased in size upon depletion of Sis1 (15). Similarly, an increase in size of polymers is found upon inhibition of Hsp104 (15, 16). Together, these data support the idea that Sis1 acts as the specific J-protein partner of Ssa and collaborates with Hsp104 in fragmentation of prion complexes, generating the seeds necessary for propagation. The idea that Sis1:Ssa functions before action of Hsp104 is supported by several observations. First, both Sis1 and Ssa (but not Hsp104) are stably associated with prion aggregates (7, 31). Second, in both the bacterial and eukaryotic systems, the J-protein:Hsp70 system functions before the AAA+ ATPase ClpB or Hsp104 in dissolution of non-prion amorphous protein aggregates (4, 14). Third, [PSI+] propagation requires transfer of the Sup35 protein through the central pore of Hsp104, as is required for dissolution of amorphous aggregates by both ClpB and Hsp104 (4, 14).
Stringency of the Requirements for Chaperone Function in [PSI+], [RNQ+], and [URE3] Propagation.
Despite the requirement of Sis1 for the propagation of [PSI+], [RNQ+], and [URE3], the rate of loss of [RNQ+] and [URE3] was much more rapid than [PSI+]. This disparity cannot be attributed to differences in seed number, as the kinetics of loss of the 3 prions were similar when Hsp104 was inhibited, with estimates of 25, 100, and 250 seeds for [URE3], [RNQ+], and [PSI+], respectively. Similar numbers were obtained for loss of [URE3] and [RNQ+] upon depletion of Sis1. However, such a calculation for [PSI+] loss upon Sis1 repression estimates >1014 seeds (Fig. 3C), a number that is unquestionably infeasible, as there are only 105 Sup35 molecules per cell (32). Thus, unlike the situation with [URE3] and [RNQ+], our data for [PSI+] are not consistent with a model of complete inhibition of fragmentation upon Sis1 repression. In contrast, our data fit well to a model in which seed fragmentation is only partially impaired, rather than abolished.
Several factors could account for the partial inhibition of fragmentation upon Sis1 depletion in the case of [PSI+]: (i) another of the 12 J-proteins of the cytosol could partially substitute for Sis1 in [PSI+] fragmentation; (ii) the residual amount of Sis1 required to maintain cell viability may allow the fragmentation of [PSI+], but not [RNQ+] or [URE3], prion complexes; or (iii) low levels of [PSI+] fragmentation could occur even in the absence of Sis1 or any other J-protein. In this vein, it has been reported that Hsp104 is capable of fragmenting Sup35 fibers in vitro in the absence of any other components (17). Thus, in the case of [PSI+], we suggest that the Hsp70/J-protein machinery may accelerate prion fragmentation, allowing it to keep pace with cell division, but may not be absolutely required for each fragmentation event.
Regardless of which of the aforementioned explanations is correct, our observations suggest a disparity between [PSI+] and both [RNQ+] and [URE3], suggesting fundamental differences in prion aggregate structures that remain uncharacterized. However, the observation that both weak and strong [PSI+] strains are maintained by Sis1-ΔG/F, whereas [RNQ+] is not, indicates that this difference in chaperone requirement is inherent to the different prions themselves rather than strain-specific conformations. Despite having dissimilar sequences, the prion-forming domains of [PSI+], [RNQ+], and [URE3] are all capable of assuming a similar amyloid structure, and thus sequence-rooted variations of this core structure may be responsible. Alternatively, non-prion-forming domains may either facilitate or obviate chaperone interactions with aggregates through direct interaction or steric hindrance. Thus, further analysis is needed to reveal the factors underlying the disparate chaperone requirements among yeast prions reported here.
In sum, we have demonstrated a specific requirement for the J-protein Sis1 in the maintenance of 3 yeast prions, and we think it is likely that it is required for all yeast prions. Although the chaperone machinery that maintains all yeast prions may be shared, the amount of activity may vary, resulting in unequal stringency of the requirement, as reported here for Sis1.
Materials and Methods
Yeast Strains.
Saccharomyces cerevisiae W303-derived haploid strains were used throughout. Most were derived from PJ513a: [RNQ+] [psi−] MAT a trp1-1 ura3-1 leu2-3,112 his3-11,15 ade2-1 can1-100 GAL2 met2-1 lys2-2 (18). Strains that had SIS1 under the control of the tetracycline repressible (TETr) promoter (sis1-Δ::LEU2 [TETrSIS1]), as well as strains bearing individual deletions of J-protein genes were described previously (15, 18), with the exception of the cwc23-8 allele, which will be described elsewhere. W303 YJW616 [PSI+] [rnq−], [PSISc4+] [rnq−], and [PSISc37+] [rnq−] strains that had ade1-14 (UGA) were gifts from Jonathan Weissman (San Francisco, CA) (33). To generate [PSI+] [rnq−] ade1-14, ADE2 haploids with J-protein gene deletions, YJW616 was crossed with individual deletion strains, followed by sporulation and tetrad dissection.
W303 strains CC30 and Sc[URE3] were obtained from Christophe Cullin (L'Institut de Biochimie et Génétique Cellulaires, Bordeaux, France) (24, 34). The ura2-Δ::HIS3 locus in CC30 was replaced by the ura2-Δ::kanMX4 cassette derived from BY ura2-Δ::kanMX4 strain of yeast knockout library (Open Biosystems) (35), and the resulting strain was crossed to Sc[URE3] to obtain a [URE3] ura2Δ::kanMX4 URA3 dal5-Δ::PDAL5-ADE2 haploid. Subsequent mating to J-protein-deletion strains described earlier and dissection yielded the desired haploids. Δhsp104 strains used in this investigation are described in SI Methods. For use as controls, [rnq−], [psi−], and [ure-o] strains were obtained by growth of the appropriate strain on liquid media containing 3 mM GdnHCl for 2 days.
Plasmids.
The plasmid bearing TETrSIS1 was described previously (15). Hsp104 plasmids pJ309 and pD816 (20), have HSP104 and hsp104-T160M, respectively, under the native promoter. Unless otherwise indicated, all others used in this study were based on the pRS plasmid series (36). J-protein overexpression 2μ-plasmids bearing either full-length or J-domain-containing fragments under the GPD promoter were previously described (18). The plasmids [TRP1-YDJ1] and [TRP1-ydj1-H32Q] were created by first cloning a genomic copy of YDJ1 into the CEN-based plasmid pRS414-GPD and then introducing the H34→Q mutation by site-directed mutagenesis.
Biochemical Analysis of Prion Particle Size.
SDD-AGE was performed as described elsewhere (15, 37) with modifications. Aliquots of pre-cleared lysates were incubated in non-reducing sample buffer for 7 min at 25 °C, and resolved in a 1.5% Tris-glycine (0.1% SDS) agarose gel (SeaKem Gold PFGE agarose). Protein was transferred to a nitrocellulose membrane at 1A for 1 h at 25 °C in a Tris-glycine/methanol buffer and probed with antibodies specific for Rnq1 or Sup35. The centrifugation assay was performed essentially as described previously (15). Pre-cleared lysates were centrifuged at 80,000 rpm (Beckman rotor TLA120.1) for 30 min. Supernatant and pellet fractions were isolated and resolved by standard SDS/PAGE and immunoblot analysis.
Assays for Prion Loss.
Time course experiments for [PSI+] or [URE3] curing were executed as previously reported for [RNQ+] (15) with some modifications. Cell cultures were maintained in exponential growth phase by continual subculturing in the presence of either 5 μg/ml doxycycline (Sigma) or 3 mM GdnHCl when indicated. [PSI+] curing experiments used either rich or synthetic glucose-based media when necessary to maintain non-essential plasmids with auxotrophic markers. Sis1-depleted cells remained viable for at least 120 generations and typically doubled approximately every 2.5 h. Potential growth advantages resulting from differential adenine production in these strains were minimized in cultures by supplementing rich media with additional adenine (40 mg/L). [URE3] curing experiments used proline-based synthetic media lacking ammonia to avoid counter-selection against [URE3], which can occur on rich media as a result of the inability of these cells to suppress the uptake of poor nitrogen sources (25).
Cytoduction assays were conducted as previously described using a [psi−] recipient strain (15); the presence of [PSI+] in resulting cytoductants was determined by SDD-AGE. Yeast lysate transformations were carried out by cotransforming spheroplasts of [psi−] cells with cell extracts of the test strain containing equivalent amounts of total protein, and a URA3-containing plasmid (38). Transformants were selected on medium lacking uracil and patched onto rich media to analyze prion status based on colony color. Loss of [URE3] was confirmed by mating experimental strains to a [ure-o] strain. Following sporulation and tetrad dissection, the presence of the [URE3] prion in the parental strain was inferred from the 4:0 inheritance of adenine prototrophy (white colony color), which is curable by GdnHCl treatment, whereas [ure-o] strains give rise to only red colonies.
Kinetic Analyses of Prion Curing.
For the purposes of kinetic analyses, the curing of [PSI+] and [URE3] were monitored by colony color counting assays (22, 26), with sectored colonies being counted as [PSI+] or [URE3] (13). No analogous assay is available to monitor [RNQ+]. Therefore, estimates of [RNQ+]/[rnq−] population numbers were made from our previously published Rnq1-GFP cell counting data, assuming “small dot” and “large dot” phenotypes represent [RNQ+] cells and “diffuse” and “diffuse + dot” phenotypes represent [rnq−] cells (15). Kinetic models used to fit prion curing data are discussed in depth in the SI Methods.
Supplementary Material
Acknowledgments.
We thank Mick Tuite for Sup35 antibodies; Daniel Masison (National Institute of Diabetes, Digestive, and Kidney Disease, Bethesda, MD) for Hsp104 plasmids; Jonathan Weissman for [PSI+] strains; Susan Lindquist (Whitehead Institute for Biomedical Research, Cambridge, MA) for the hsp104-Δ and Hsp104 antibodies; Christophe Cullin (L'Institut de Biochimie et Génétique Cellulaires) for [URE3] and [ure-o] strains; and members of the Craig laboratory for thoughtful comments on the manuscript. This work was supported by National Institutes of Health Grant GM53655 and the USDA Cooperative State Research, Education and Extension Service (CSREES) project WISO4769 (to E.A.C.); Human Frontier Science Program Long-Term Fellowship (T.H.); and a National Institutes of Health National Research Service Award postdoctoral fellowship (J.K.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808934105/DCSupplemental.
References
- 1.Wickner RB, et al. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones GW, Tuite MF. Chaperoning prions: The cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–832. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- 3.Perrett S, Jones GW. Insights into the mechanism of prion propagation. Curr Opin Struct Biol. 2008;18:52–59. doi: 10.1016/j.sbi.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Mogk A, et al. Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem Soc Trans. 2008;36:120–125. doi: 10.1042/BST0360120. [DOI] [PubMed] [Google Scholar]
- 5.Du Z, et al. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig EA, et al. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- 7.Sondheimer N, et al. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung G, et al. A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts BT, Moriyama H, Wickner RB. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast. 2004;21:107–117. doi: 10.1002/yea.1062. [DOI] [PubMed] [Google Scholar]
- 10.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:e24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrne LJ, et al. Cell division is essential for elimination of the yeast [PSI+] prion by guanidine hydrochloride. Proc Natl Acad Sci USA. 2007;104:11688–11693. doi: 10.1073/pnas.0701392104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paushkin SV, et al. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 13.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 15.Aron R, et al. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kryndushkin DS, et al. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 17.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chernoff YO, et al. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol CellBiol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka M, et al. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 22.Eaglestone SS, et al. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–7046. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talarek N, et al. The [URE3] prion is not conserved among Saccharomyces species. Genetics. 2005;171:23–34. doi: 10.1534/genetics.105.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shewmaker F, et al. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics. 2007;176:1557–1565. doi: 10.1534/genetics.107.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ripaud L, Maillet L, Cullin C. The mechanisms of [URE3] prion elimination demonstrate that large aggregates of Ure2p are dead-end products. EMBO J. 2003;22:5251–5259. doi: 10.1093/emboj/cdg488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai J, Douglas MG. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 29.Lian HY, et al. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J Biol Chem. 2007;282:11931–11940. doi: 10.1074/jbc.M606856200. [DOI] [PubMed] [Google Scholar]
- 30.Bradley ME, et al. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99(suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagriantsev SN, et al. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 33.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-Bellot E, Guillemet E, Cullin C. The yeast prion [URE3] can be greatly induced by a functional mutated URE2 allele. EMBO J. 2000;19:3215–3522. doi: 10.1093/emboj/19.13.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 36.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 37.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- 38.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1125. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.