Abstract
Choanoflagellates are single-celled aquatic flagellates with a unique morphology consisting of a cell with a single flagellum surrounded by a “collar” of microvilli. They have long interested evolutionary biologists because of their striking resemblance to the collared cells (choanocytes) of sponges. Molecular phylogeny has confirmed a close relationship between choanoflagellates and Metazoa, and the first choanoflagellate genome sequence has recently been published. However, molecular phylogenetic studies within choanoflagellates are still extremely limited. Thus, little is known about choanoflagellate evolution or the exact nature of the relationship between choanoflagellates and Metazoa. We have sequenced four genes from a broad sampling of the morphological diversity of choanoflagellates including most species currently available in culture. Phylogenetic analyses of these sequences, alone and in combination, reject much of the traditional taxonomy of the group. The molecular data also strongly support choanoflagellate monophyly rejecting proposals that Metazoa were derived from a true choanoflagellate ancestor. Mapping of a complementary matrix of morphological and ecological traits onto the phylogeny allows a reinterpretation of choanoflagellate character evolution and predicts the nature of their last common ancestor.
Keywords: evolution, morphology, holozoa, animals, protists
Choanoflagellates are a major group of heterotrophic nanoflagellates, ubiquitously distributed in aquatic environments (1). These single-celled organisms were first described by James-Clark in 1866, who was also the first to note the strong resemblance between the choanoflagellate cell morphology and that of the collared cells (choanocytes) of sponges (Porifera) (2). Based on these similarities, a close relationship between choanoflagellates and Metazoa was long postulated and has now been confirmed (3–9). However, there is still very little molecular data from more than two or three choanoflagellate species. Thus, we have a limited understanding of choanoflagellate phylogeny or how to interpret evolutionary trends within the group. Most importantly, without knowing whether choanoflagellates constitute a monophyletic group, it is difficult to know the relevance of such trends to the early evolution of Metazoa.
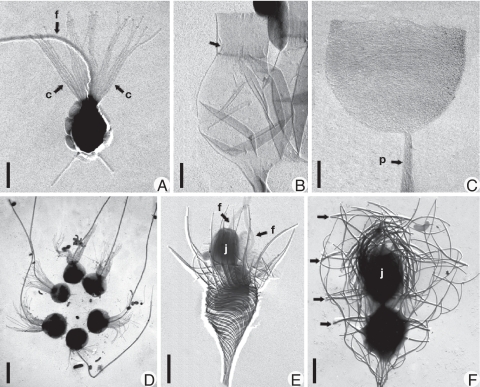
Choanoflagellates are characterized by a distinctive and remarkably uniform cell body (protoplast) morphology. This comprises a spherical to ovoid cell with a single anterior flagellum surrounded by a collar of narrow actin-based microvilli (Fig. 1A) (10). In contrast to the uniformity of the choanoflagellate cell, the morphology of the external covering (periplast) is varied and sometimes striking, ranging from simple organic sheaths to complex silica “cages” up to a 100 μm or more in length.
Fig. 1.
Morphological variation within choanoflagellates. Shadowcast whole mounts of cells or thecae viewed with transmission electron microscopy. (A) Monosiga ovata. (c) collar; f, flagellum. Bar = 2 μm. (B) Salpingoeca urceolata. Empty flask-shaped theca is shown. Arrow denotes inner flange that connects cell (absent) to theca. (Scale bar, 1 μm.) (C) Salpingoeca infusionum. Empty cup-shaped organic theca is shown. (p) peduncle (stalk). (Scale bar, 1 μm.) (D) Salpingoeca amphoridium. Colonial “proterospongia” stage is shown. Note six regularly placed cells held together by fine posterior threads. (Scale bar, 5 μm.) (E) Acanthoeca spectabilis. Immediately after division (nudiform) showing two cells, each with a forwardly directed flagellum (arrows in F) is shown. The juvenile (j) is above the cell remaining in the parent lorica. (Scale bar, 2 μm.) (F) Stephanoeca diplocostata. Tectiform division showing inverted juvenile cell (j) being pushed into an accumulation of costal stripsis shown. Arrows denote transverse (ring) costae. (Scale bar, 2 μm.)
Periplast morphology has formed the basis of the conventional classification of choanoflagellates into three families (11). Members of the Codonosigidae family (Kent 1880) (Fig. 1A) have a thin fibrillar coat, the glycocalyx. This surrounds the cell and may extend posteriorly to join a substantial stalk composed of carbohydrate microfibrils (12). Species of the Salpingoecidae family (Kent 1880) possess a substantial microfibril-based theca. This may be flask- (Fig. 1B), cup- (Fig. 1C) or tube-shaped and is attached to the substratum by a stalk similar to that found in the Codonosigidae family. Members of the Acanthoecidae family (Norris 1965) are characterized by the most distinct periplast morphology. This consists of a complex basket-like lorica constructed in a precise and highly reproducible manner from ribs (costae) composed of rod-shaped silica strips (Fig. 1 E and F) (13). The Acanthoecidae family is further subdivided into nudiform (Fig. 1E) and tectiform (Fig. 1F) species, based on the morphology of the lorica, the stage in the cell cycle when the silica strips are produced, the location at which the strips are stored, and the mode of cell division [supporting information (SI) Text] (14).
Speculation on a possible close evolutionary relationship between choanoflagellates and Metazoa was strengthened early on by the discovery of a colonial choanoflagellate, Proterospongia haeckeli, by Saville-Kent in 1880. This taxon superficially resembles a poriferan larva in that it appears to consist of flagellated cells protruding from a matrix bearing internal amoeboid-like cells (11). Although subsequent sightings of this species have not been authenticated, and its original description is somewhat enigmatic, numerous other colonial forms are now known. Thus, choanoflagellates have long been treated in introductory biology texts as a classic example of stepwise evolution of complexity leading to the true multicellularity of Metazoa.
Most molecular phylogenetic trees that include multiple choanoflagellate species have been based on nuclear small subunit, ribosomal gene (SSU rDNA) sequences. These studies have mostly recovered choanoflagellate monophyly but have been hampered by a small sampling of species and some species misidentification (4–5, 7–9). Therefore, to examine major trends in choanoflagellate evolution, we have constructed a taxonomically broad, multigene phylogeny of the group and a complementary matrix of morphological and ecological traits. Phylogenetic analyses of a concatenated four-gene dataset show that the traditional taxonomy of choanoflagellates is flawed, and the evolution of some of their most notable morphological traits is more complex than initially thought.
Results
Molecular Phylogeny of Choanoflagellates.
Large fragments of the nuclear SSU and large subunit (LSU) ribosomal RNA, alpha-tubulin (tubA), and the 90-kDa heat shock protein (hsp90) coding genes were amplified by PCR from total genomic DNA for 16 choanoflagellate species (Table S1). SSU rDNA was sequenced directly from PCR products for all species, from which no evidence of polymorphism was detected. LSU rRNA and both protein genes were amplified in multiple overlapping fragments, all of which matched completely in overlapping regions. Thus, there is no evidence for the existence of paralogs for any of these genes in any of the examined species.
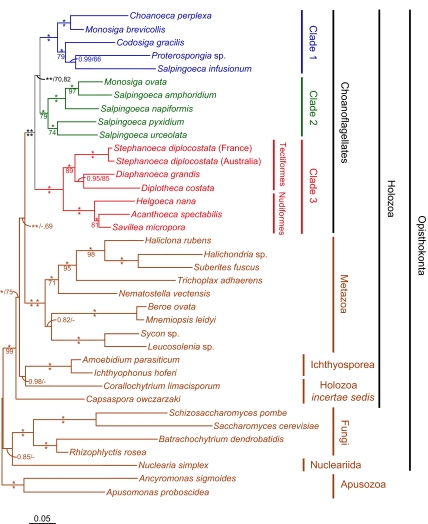
Phylogenetic analyses of each of the four genes independently show similar results for each gene and no strongly supported (bootstrap support >70%) conflicts among them (SI Text and Fig. S1). Therefore, the four genes were combined (concatenated) into a single dataset, along with sequences from representatives of all major opisthokont lineages for which an equivalent combination of sequences was available. These are Fungi, Nucleariida, Ichthyosporea, and Metazoa (7), as well as two holozoan lineages of uncertain affinity, Corallochytrium limacisporum and Capsaspora owczarzaki (7, 9). A close relationship between Opisthokonta and Apusozoa has been well established (15). Therefore, sequences from two apusozoans, Apusomonas proboscidea and Ancyromonas sigmoides, were used to root the opisthokonts in the multigene tree. Additional analyses were also conducted with Corallochytrium limacisporum and Nuclearia simplex deleted, because these were found to form particularly long and unstable branches with these data, and such long branches tend to adversely effect support values, especially for deeper nodes in the tree (Fig. 2) (16).
Fig. 2.
Molecular phylogeny of choanoflagellates based on a concatenated four-gene dataset. The tree shown was derived by Bayesian inference based on a combined tubA, hsp90, SSU, and LSU rDNA nucleotide sequence alignment. Branches are drawn proportional to the number of nucleotide substitutions per site as indicated by the scale bar at the lower left. Branches receiving 1.00 biPP and 100% mlBP support are denoted by an *. biPP and mlBP values are otherwise given above and below branches respectively. Additional values are shown for four important deep branches in the choanoflagellate and Metazoa grouping, indicating support values obtained when the unstable, long-branched taxa Corallochytrium limacisporum and Nuclearia simplex were excluded.
Phylogenetic analysis of 6,415 aligned positions produces a well-resolved tree with strong support for nearly all branches based on Bayesian Inference posterior probabilities (biPP) and maximum likelihood bootstrap percentages (mlBP) (Fig. 2). The tree divides the choanoflagellates into three strongly supported major clades (1.00 biPP, 79–100% mlBP) (Fig. 2). Clades 1 and 2 consist of mixtures of species attributable to the Codonosigidae and Salpingoecidae, whereas Clade 3 corresponds to the traditional Acanthoecidae. The taxonomic mixture of Clades 1 and 2 is highlighted by the fact that the genus Monosiga, including the model organisms Monosiga brevicollis and Monosiga ovata, is split between them (assigned to Clades 1 and 2, respectively). Within Clade 3, the two major morphological types of acanthoeicids, nudiform, and tectiform, are both recovered as strongly supported monophyletic subgroups (1.00 biPP, 89–100% mlBP) (Fig. 2). This is particularly significant with respect to the morphologically disparate nudiform species for it demonstrates that they are an evolutionarily coherent assemblage.
Two colony-forming Proterospongia species are found in Clade 1, although they do not group together, whereas a third colony-forming species, Salpingoeca amphoridium (Fig. 1D), is found in Clade 2. Thus, the potential for developing the colonial “proterospongia” habit is either an ancestral trait of Clades 1 and 2, possibly of choanoflagellates as a whole, or this trait has arisen multiple times independently in the group. This indicates that “Proterospongia” is not a valid taxon. Instead, it is a morphology that probably corresponds to a stage in the life-cycles of sedentary choanoflagellates, as suggested (17). Thus, Proterospongia choanojuncta is simply the colonial phase of Choanoeca perplexa, both species of which have identical sequences for all genes examined (Fig. S1A) (18), and Proterospongia sp. ATCC 50818 is the colonial stage of an unnamed species of Salpingoeca (B.S.C.L., unpublished data).
Both choanoflagellates and Metazoa are strongly supported as monophyletic in the combined gene tree (1.00 biPP, 100% mlBP) (Fig. 2). Thus, there is no indication of either group being paraphyletic with respect to the other, that is, for choanoflagellates having been derived from Metazoa or vice versa. The multigene phylogeny also unites these two groups as sister taxa to the exclusion of all other examined Holozoa (1.00 biPP, <50–69% mlBP). This includes here all known holozoan lineages except for ministeriids, for which only their highly derived SSU rDNA sequences were available (7). A sister-group relationship between Metazoa and choanoflagellates is also robustly recovered by maximum likelihood analyses of much larger multigene phylogenies (8, 9).
Analyses of the concatenated four-gene dataset place Clades 1 and 2 together to the exclusion of Clade 3, with strong posterior probability but only moderate bootstrap support (1.00 biPP, 70–82% mlBP) (Fig. 2). Thus, the position of the root of the choanoflagellates is not confidently assigned at this time. However, the grouping of Clades 1 and 2 is also seen in most of our single-gene trees (Fig. S1), and none of these trees shows strong support for any alternative root. If correct, this division would split the choanoflagellates into species with either wholly organic (Clades 1 and 2) or silica-based periplasts (Clade 3) and indicate that the lorica was an early invention in choanoflagellate evolution. The shallow depth of this node, that is the short branch uniting Clades 1 and 2 in the multigene tree (Fig. 2), suggests that considerably more data will be required to resolve this issue.
Phylogenetic Trait Mapping.
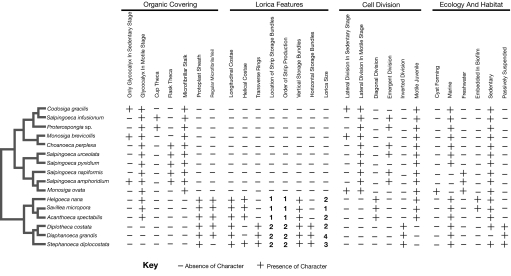
A total of 27 morphological and ecological characters were scored for all choanoflagellate species in the molecular tree (Fig. 3, Table S2, and Table S3). Detailed descriptions of characters and states are given in the SI Text along with a short tutorial on choanoflagellate morphology, systematics, and life-history.
Fig. 3.
Matrix of morphological and ecological characters for choanoflagellates. Characters (27) were scored for all taxa for which information was available either from the literature or our own observations. Species are arranged in order corresponding to the molecular phylogeny (Fig. 2) as indicated by the simplified schematic to the left of the matrix. Accumulation of costal strips: (1) around surface of juvenile; (2) inside the top of the parent collar. Order of costal strip production: (1) longitudinal first, helical second; (2) transverse first, longitudinal second. Lorica size: (1) <8 μm; (2) 9–12 μm; (3) 12–15 μm; and (4) >15 μm.
Organic Covering.
All choanoflagellates have some form of organic covering, and all species except for tectiform loricates have a motile stage with only a glycocalyx, which is obscured when cells become sedentary and form a theca or lorica. The most widely noted organic covering in choanoflagellates is the theca, and this character is central to traditional choanoflagellate taxonomy as a defining feature of one of the three major divisions (Salpingoecidae). However, the phylogeny indicates that the theca is older than originally thought and was present, at the latest, in the last common ancestor of Clades 1 and 2. Thus, all members of Clades 1 and 2 possess a theca except for Codosiga gracilis, Monosiga brevicollis, and Monosiga ovata (19), and these thecae appear to have a similar structure in both groups (B.S.C.L., unpublished data).
Evolution of the theca is further complicated by the fact that it occurs in multiple forms, most commonly as a flask consisting of three layers (20) or a simple cup consisting of one (B.S.C.L., unpublished data). The sporadic distribution of these forms in Clade 1 indicates that the theca, even when present, must have been modified on multiple occasions. Either the flask was reduced to a cup several times, or the relatively simple single-layered cup was invented at least twice independently. The latter possibility is suggested by the fact that not all aspects of the cups in Clade 1 are homologous (B.S.C.L., unpublished data). Thus, the molecular phylogeny indicates that the choanoflagellate theca has had a complex evolutionary history, including multiple independent losses and modifications.
Lorica.
The silica lorica is a striking and unique feature, and the phylogeny indicates that it has evolved only once (Fig. 2). All loricae are based on two layers of rib-like arrays of silica strips (costae); an outer, usually longitudinal, costal layer is held in place by an inner layer of helical and/or transverse (ring) costae (21). All costae are built from prefabricated strips that are accumulated in bundles on the cell surface. Once a full set of strips has been accumulated, the lorica is then assembled in a series of rapid coordinated movements. During this spectacular feat of nanoconstruction, individual longitudinal and helical costae are assembled from single bundles of costal strips (accumulated in a vertical orientation on the surface of the juvenile cell), whereas each transverse costa (ring) is assembled from portions of several different bundles (accumulated in a horizontal orientation on the juvenile cell) (22). Because helical costae are found in all nudiform and some tectiform species, whereas only tectiform species have rings (Fig. 1F) (22, 23), it appears that longitudinal and helical costae are the ancestral state from which transverse (ring) costae were derived.
Cell Division.
Differences in periplast morphology have important consequences for cell division in choanoflagellates, which varies considerably across the group. Lateral cell division is found in species with a glycocalyx, which is a thin and expandable structure (12). However, this cannot occur in species with the thicker, more rigid theca (20). Therefore, cells of non-thecate members of Clades 1 and 2 divide laterally, whereas in thecate species, dividing cells first become amoeboid before emerging from the anterior aperture and dividing outside the periplast (20). Thus, the last common ancestor of Clades 1 and 2 appears to have used emergent cell division, consistent with it having possessed a theca.
Cell division in loricate species is further complicated by the fact that the daughter cell lorica must be produced after the new cell emerges from the parent lorica. Two fundamentally different solutions to this are found. In nudiform taxa, the motile daughter cell swims away, becomes sedentary, accumulates a complete set of strips, and then produces a lorica (Fig. 1E) (22, 25). In tectiform species, the daughter cell (the juvenile) inverts and exits the parent lorica backwards, picking up a complete set of newly synthesized costal strips as it leaves (Fig. 1F) (22). These strips are then quickly assembled into a lorica so that the tectiform juvenile is in possession of a fully formed lorica soon after separating from the parent cell.
Ecology and Habitat.
The majority of described species of choanoflagellates are marine, but there are also over 50 described freshwater species. All freshwater species sampled here form a single subgroup in Clade 2 (Fig. S1A). However, the overall range in morphology of freshwater species is similar to that of marine species. Thus, it is unlikely that all freshwater species belong to a single clade. Rather, there were probably multiple invasions of fresh water by choanoflagellates, but sampling of further taxa would be needed to test this. Loricate species, which are exclusively marine, show the greatest ecological diversity ranging from microbial biofilms to numerous different microniches in the water column (26).
Discussion
A robust phylogeny of choanoflagellates allows a reevaluation of the group's position within Holozoa. All individual analyses of the four different genes indicate that choanoflagellates are a monophyletic group (Fig. S1), and analyses of these genes combined indicate that this monophyletic group is the sister group to Metazoa (Fig. 2). We find no evidence to indicate that choanoflagellates could be paraphyletic with respect to Metazoa or vice versa. This is despite examining a wide diversity of choanoflagellate morphologies and a broad sampling of Metazoa with representatives of all early branches for which sequence data are currently available. These include two branches of Porifera, Demospongiae (Haliclona, Halichondria, and Suberites), and Calcarea (Sycon and Leucosolenia), as well as Ctenophora (Beroe and Mnemiopsis), Cnidaria (Nematostella), and Placozoa (Trichoplax) (Fig. 2). Thus, there is no evidence that Metazoa are derived from a choanoflagellate ancestor or that any division of choanoflagellates has an exclusive relationship with Metazoa. This is particularly relevant with regard to “Proterospongia”, with its resemblance to a simple poriferan body plan. Instead, the molecular phylogeny indicates that the ability in choanoflagellates to form colonies evolved early in the group. In fact, it cannot be ruled out that this was ancestral trait or even one shared with the ancestor of Metazoa.
The phylogeny also allows a reexamination of the long-established morphological taxonomy of choanoflagellates. We find that all examined species fall cleanly into one of three well-supported clades, only one of which (Clade 3) corresponds to a traditional taxon (Acanthoecidae) (Fig. 2). Clades 1 and 2 are mixtures of thecate and non-thecate taxa, whereas Clade 3 consists entirely of species that possess a silica-based lorica. Homology of the thecae found in Clades 1 and 2 indicates that this structure arose once and early in choanoflagellate evolution, at least before the last common ancestor of Clades 1 and 2. The silica lorica also appears to have been an early invention, and because the root of the tree remains equivocal (Fig. 2), the question of whether the last common ancestor of all extant choanoflagellates possessed a theca or a lorica remains open.
The confusion in traditional choanoflagellate taxonomy arises in part from the fact that the theca, used to define one of the proposed major divisions of the group, has a much more complex history than originally thought. This structure appears to have arisen early in choanoflagellate evolution rather than in an ancestor of the traditional Salpingoecidae. Mapping of this character onto the new phylogeny also indicates that, in addition to multiple independent losses, this structure has been subject to a number of further modifications. These include either multiple reversions or parallel independent inventions of a cup-shaped variant and at least one origin of a tube-shaped variant not examined here.
Interpretation of morphological and ecological traits (Fig. 3) in light of the phylogeny (Fig. 2) allows us to make some predictions about the nature of the last common ancestor of choanoflagellates. This was probably a sedentary marine organism, which on division, produced a motile daughter cell with a glycocalyx. This organic covering would have been retained in the sedentary phase, in most cases becoming obscured by either a theca or a lorica. In fact, an organic covering can still be observed in all examined species (B.S.C.L., unpublished data) including those with a lorica (where it takes the form of a sheath around the protoplast) (B.S.C.L., unpublished data). The primary function of this structure is to secure the cell to a substratum, which in the case of the loricate species, is the lorica itself.
The last common ancestor of choanoflagellates was also probably a marine organism, a habit that most species have retained. The current phylogeny further indicates a single invasion of the marine planktonic environment within choanoflagellates. This was achieved by tectiform loricates via a modification of the lorica, reducing the thicker helical costae to fewer and simpler rings, thus reducing overall weight and retaining rigidity. In addition, changes in tectiform cell division allowed the cell to possess a lorica nearly throughout its entire life cycle (26). This strategy appears to have been highly successful because over 150 species of tectiform loricates are known versus only 5–6 nudiform species (23).
The molecular phylogeny (Fig. 2) includes nearly every choanoflagellate species currently available in culture. However, this is still only 16 species, whereas over 244 species have been described to date. Of these species, ≈128 are tectiform loricate taxa. For these 128 loricate taxa, the morphologically diverse selection of species examined here clearly form a monophyletic group (Fig. 2), so we do not expect that any of the remaining loricate taxa represent additional deep clades. The remaining ≈116 non-loricate taxa are more problematic as they are almost entirely differentiated by the size and shape of their periplasts, which the data presented (Figs. 2 and 3) show to be unreliable taxonomic characters. Thus, it cannot be ruled out that there are additional major divisions of choanoflagellates not detected in the present study.
The phylogeny places choanoflagellates as the sister group to Metazoa to the exclusion of all other sampled holozoan taxa with strong posterior probability and moderate bootstrap support (Fig. 2). Thus, our study and others indicate that choanoflagellates are sister to Metazoa. The only possibly closer sister taxa to the Metazoa are the ministeriids, solely represented in culture collections by Ministeria vibrans (5, 7). However, sequences from this taxon tend to produce long, weakly supported branches in phylogenetic trees, making their placement uncertain (5, 7). Furthermore, a recent phylogeny based on 78 protein sequences strongly placed this Ministeria vibrans together with Capsaspora owczarzaki as a sister group to an exclusive and strongly supported choanoflagellate + Metazoa clade (27). Thus, although the branching order among the deeper lineages within Holozoa remains controversial, there is now a substantial body of evidence supporting a choanoflagellate + Metazoa clade (Fig. 2) (8, 9, 27).
These data indicate that the order Choanoflagellida requires considerable taxonomic revision. Codonosigidae is clearly shown to be polyphyletic, and Salpingoecidae is paraphyletic. However, the status of the family Acanthoecidae should remain unchallenged. In addition, because of their enhanced relevance, we recommend that the terms nudiform and tectiform should be given official taxonomic recognition. To retain the terminology without contravening the International Code of Zoological Nomenclature guidelines (28), these two groups should be given intermediate-rank status and known by the informal family-group names of Nudiformes and Tectiformes, respectively (Fig. 2). Within Clades 1 and 2, the genera Proterospongia and Monosiga are clearly polyphyletic, whereas Salpingoeca is paraphyletic. The described species within these genera should be reallocated on the basis of more precisely circumscribed diagnoses into existing or new genera.
The data presented here indicate that choanoflagellates are an important key to understanding the starting material from which Metazoa evolved. However, hypotheses proposing that either group is derived from the other (reviewed in ref. 29) are strongly rejected by the molecular phylogeny (Fig. 2). It appears instead that both groups are descended from a common marine protistan ancestor. From this ancestor, the Metazoa evolved into truly multicellular organisms, whereas the choanoflagellates have maintained a predominantly solitary existence, albeit with a widespread ability to form colonial stages in their life cycle. This has proved a highly successful strategy. From a sedentary marine ancestor, choanoflagellates have invaded habitats as diverse as bacterial biofilms, the marine water column, and freshwaters. In the process, they have become one of the preeminent groups of heterotrophic protists in aquatic environments.
Materials and Methods
Cell Culture and DNA Extraction.
All choanoflagellate species were maintained in stock cultures of 20 ml at 15°C. Salpingoeca amphoridium medium was comprised of 18 ml of sterile distilled water and 1 ml of cereal grass infusion (0.5% cereal grass powder). Monosiga ovata medium was comprised of five parts Pratt's medium (23, 30) to one part of cereal grass infusion. All other species were maintained in the following seawater cultures: 19 ml of sterile seawater and one rice grain - Diaphanoeca grandis (ATCC 50111), Helgoeca nana (= Acanthoecopsis unguiculata ATCC 50073), Salpingoeca pyxidium (ATCC 50929), Salpingoeca urceolata (ATCC 50560), and Savillea micropora; 18.9 ml of sterile seawater and 0.1 ml of peptone/yeast solution (0.4% proteose peptone; 0.08% yeast extract in distilled water) - Acanthoeca spectabilis (ATCC PRA-103), Choanoeca perplexa (ATCC 50453), Codosiga gracilis (ATCC 50454), Diplotheca costata, and Stephanoeca diplocostata (ATCC 50454); 18 ml of seawater and 1 ml of cereal grass infusion - Monosiga brevicollis (ATCC 50154), Proterospongia sp. (ATCC 50818), and Salpingoeca infusionum (ATCC 50559). DNA extractions were performed on 40-ml cultures. Cells were centrifuged at 2,700 × g for 40 min and DNA was extracted by using a NaCl/ethanol protocol (31).
Molecular phylogeny (Fig. 2 and Fig. S1) shows that ATCC Culture 50073 (deposited as Acanthoecopsis unguiculata) was misidentified and is actually a nudiform choanoflagellate. We have recently named this species as Helgoeca nana (25). The SSU rDNA molecular phylogeny (Fig. S1A) also shows that GenBank sequence AF272000 is misattributed to a tectiform taxon Calliacantha sp. CEE-2003 and is therefore herein referred to as “AF272000”.
PCR Amplification, Cloning, and DNA Sequencing.
All PCR amplifications used total genomic DNA. PCR protocols consisted of 2 min denaturing at 94°C, followed by 30 cycles of 30 seconds denaturing at 94°C, varying annealing temperatures, and 1 min extension at 72°C, with a final finishing step of 10 min at 72°C. PCR products were analyzed on 1% agarose gels and purified by using the QIAquick gel extraction kit (Qiagen). PCR and sequencing strategies for individual genes are detailed in the SI Text.
Phylogenetic Analysis.
Multiple sequence alignments were created for each gene separately, and each individual alignment was subjected to an initial phylogenetic analysis. This was to facilitate the identification of potential paralogs and to determine whether all genes had phylogenetically compatible histories (no strongly supported conflicting clades) and were therefore suitable for concatenation. In all cases, no evidence of paralogy or incompatible histories was observed. Because no LSU sequence was available for Leucosolenia sp. [National Center for Biotechnology Information (NCBI) Taxonomy ID86013], the LSU sequence from Leucosolenia sp. MMM-2001 (NCBI Taxonomy ID154963) was used instead.
Multiple Sequence Alignment.
Alignments were created separately for each gene by using ClustalX (32), including all available choanoflagellate sequences plus outgroup sequences. Additional downloaded sequences are detailed in the SI Text. Each alignment was then modified by eye to minimize insertion-deletion events, and regions that could not be unambiguously aligned among all sequences were excluded from the phylogenetic analyses. The individual alignments and a concatenated alignment created from all four genes were then examined with Modeltest (version 3.7) (33), which indicated in each case that GTR+I+γ (34) was the most appropriate nucleotide substitution model. All alignments are available from M.C. or S.L.B. on request.
Bayesian Inference.
Phylogenetic trees were estimated with Bayesian inference by using MrBayes (version 3.1.1) (35). All parameters for the GTR+I+γ model were estimated by the program from BioNJ starting trees. Tree searches consisted of 2 parallel sets of 4 chains (3 heated and 1 cold) run for 250,000 generations for the concatenated dataset and 1,000,000 generations for individual gene analyses. This number of generations was sufficient to reach convergence for these 2 datasets as measured by split frequencies ≤0.01. Trees were sampled every 10th generation, and the first 25% of the sampled trees were discarded as burn in.
For the protein-coding genes, first and second codon positions were modeled together whereas third-codon positions were modeled separately. For the concatenated dataset, the data were divided into three partitions: 1) ribosomal DNA genes, 2) first-and second-codon positions, and 3) third-codon positions. Analyses of the concatenated data with all third-codon positions deleted showed no change in topology with the exception of Clade 3 (data not shown). Here, in the absence of third-codon positions, the tectiform species Diplotheca costata appears as the earliest branch in the clade, thus, depicting nudiforms as derived from within tectiforms. However, paraphyly of tectiform choanoflagellates is strongly rejected by parsimony criteria, which require an additional 5 steps (out of a total of 42) to fit the morphological data (Fig. 3) onto this alternative topology.
To rule out the possibility that third positions are mutationaly saturated within Clade 3, pairwise distances at first- and second-codon positions were plotted against those at third positions (36) with raw (p) measured by using PAUP* 4.0b10 (37). These analyses show no indication of third-position saturation in Clade 3. For tubA, a linear trend was observed in the plot (y = 10.5x + 0.1771, R2 = 0.6277; where y = pairwise distances at first and second positions and x = pairwise distances at third positions). Helgoeca nana formed a set of outlier pairwise distances in the hsp90 plot. In comparisons involving Helgoeca nana with all other species, and comparisons involving all species excluding Helgoeca nana, linear trends were observed in both plots (y = 6.9066x − 1.2784, R2 = 0.9638 and y = 2.169x + 0.2466, R2 = 0.7815, respectively). Thus, concatenated analyses including third-codon positions (Fig. 2) are likely to give the most accurate resolution of Clade 3 and have no apparent affect on any other part of the tree.
Maximum Likelihood.
Maximum likelihood trees were produced by using the program RAxML (version 7.3) (38) with 1,000 bootstrap replicates by using the GTRCAT model. All model parameters were estimated by the program from its own maximum parsimony starting trees. The alignment was divided into the same three partitions as used for the Bayesian analysis.
Supplementary Material
Acknowledgments.
We thank B. Hall for help with sequencing, S. Innes for information on Helgoeca nana, J. Kent for image preparation, A. Polaszek for nomenclatural advice, and L. Medlin (Alfred Wegener Institute, Bremerhaven) for help with SSU rDNA amplification. We also thank P. Holland (Oxford University) for providing the culture of Monosiga ovata, T. Cavalier-Smith (Oxford University) for genomic DNA from Corallochytrium limacisporum, and R. Lichtwardt (University of Kansas) for genomic DNA from Amoebidium parasiticum. We thank the Joint Genome Institute, the Broad Institute, and StellaBase for the use of unpublished sequence data. This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/C514990/1 (to S.L.B. and B.S.C.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data Deposition: Sequences reported in this paper have been deposited in the GenBank database (accession numbers EU011922–EU011972).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801667105/DCSupplemental.
References
- 1.Leadbeater BSC, Thomsen HA. Order Choanoflagellida. In: Lee JJ, Leedale GF, Bradbury P, editors. The Illustrated Guide to The Protozoa. 2nd Ed. Vol 1. Lawrence, Kansas: Society of Protozoologists; 2000. pp. 14–38. [Google Scholar]
- 2.James-Clark H. Conclusive proofs on the animality of the ciliate sponges, and their affinities with the Infusoria Flagellata. Am J Sci Ser. 2. 1866;42:320–325. [Google Scholar]
- 3.Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the Metazoa: An evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 4.Amaral-Zettler LA, Nerad TA, O'Kelly CJ, Sogin ML. The nucleariid amoebae: More protists at the animal-fungal boundary. J Eukaryot Microbiol. 2001;48:293–297. doi: 10.1111/j.1550-7408.2001.tb00317.x. [DOI] [PubMed] [Google Scholar]
- 5.Cavalier-Smith T, Chao EEY. Phylogeny of Choanozoa, Apusozoa, and other protozoa and early eukaryotic megaevolution. J Mol Evol. 2003;56:540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- 6.Lang BF, O'Kelly C, Nerad TA, Gray MW, Burger G. The closest unicellular relatives of Animals. Curr Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 7.Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol Biol Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- 8.Jiménez-Guri E, Philippe H, Okamura B, Holland PWH. Buddenbrockia is a cnidarian worm. Science. 2007;317:116–118. doi: 10.1126/science.1142024. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of Metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- 10.Karpov SA, Leadbeater BSC. Cytoskeleton structure and composition in choanoflagellates. J Eukaryot Microbiol. 1998;45:361–367. [Google Scholar]
- 11.Kent SW. A Manual of the Infusoria. Vols 1–3. London: D. Bogue; 1882. p. 913. [Google Scholar]
- 12.Leadbeater BSC, Morton C. A microscopical study of a marine species of Codosiga James-Clark (Choanoflagellata) with special reference to the ingestion of bacteria. Biol J Linn Soc. 1974;6:337–347. [Google Scholar]
- 13.Leadbeater BSC. Developmental studies on the loricate choanoflagellate Stephanoeca diplocostata Ellis. VII. Dynamics of costal strip accumulation and lorica assembly. Europ J Protistol. 1994;30:111–124. [Google Scholar]
- 14.Manton I, Bremer G, Oates K. Problems of structure and biology in a large collared flagellate (Diaphanoeca grandis Ellis) from arctic seas. Proc Roy Soc Lond B. 1981;213:15–26. [Google Scholar]
- 15.Kim E, Simpson AGB, Graham LE. Evolutionary relationships of Apusomonads inferred from taxon-rich analyses of 6 nuclear encoded genes. Mol Biol Evol. 2006;23:2455–2466. doi: 10.1093/molbev/msl120. [DOI] [PubMed] [Google Scholar]
- 16.Hendy MD, Penny D. A framework for the quantitative study of evolutionary trees. Syst Zool. 1989;38:297–309. [Google Scholar]
- 17.Leadbeater BSC. Life-history and ultrastructure of a new marine species of Proterospongia (Choanoflagellida) J Mar Biol Assoc UK. 1983;63:135–160. [Google Scholar]
- 18.Medina M, et al. Phylogeny of Opisthokonta and the evolution of multicellularity and complexity in Fungi and Metazoa. Int J Astrobiol. 2003;2:203–211. [Google Scholar]
- 19.Leadbeater BSC, Karpov SA. Cyst formation in a freshwater strain of choanoflagellate Desmarella moniliformis Kent. J Eukaryot Microbiol. 2000;47:433–439. doi: 10.1111/j.1550-7408.2000.tb00071.x. [DOI] [PubMed] [Google Scholar]
- 20.Leadbeater BSC. Observations on the life-history and ultrastructure of the marine choanoflagellate Choanoeca perplexa Ellis. J Mar Biol Assoc UK. 1977;57:285–301. [Google Scholar]
- 21.Thomsen HE, Buck KR, Coale SL, Garrison DL, Gowing MM. Loricate choanoflagellates (Acanthoecidae, Choanoflagellida) from the Weddell Sea, Antarctica. Zool Scripta. 1990;19:367–387. [Google Scholar]
- 22.Leadbeater BSC. Developmental studies on the loricate choanoflagellate Stephanoeca diplocostata Ellis. VIII. Nuclear division and cytokinesis. Europ J Protistol. 1994;30:171–183. [Google Scholar]
- 23.Leadbeater BSC. Choanoflagellate lorica construction and assembly: The nudiform condition. I. Savillea Species Protist. 2008;159:259–268. doi: 10.1016/j.protis.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Karpov SA, Leadbeater BSC. Cell and nuclear division in a freshwater choanoflagellate. Monosiga ovata Kent Europ J Protistol. 1997;33:323–334. [Google Scholar]
- 25.Leadbeater BSC, Hassan R, Nelson M, Carr M, Baldauf SL. A new genus, Helgoeca gen nov, for a nudiform choanoflagellate. Europ J Protistol. 2008;44:227–237. doi: 10.1016/j.ejop.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Leakey RG, Leadbeater BSC, Mitchell E, McCready SM, Murray AA. The abundance and biomass of choanoflagellates and other nanoflagellates in waters of contrasting temperature to the north-west of South Georgia in the Southern Ocean. Europ J Protistol. 2002;38:333–350. [Google Scholar]
- 27.Shalchian-Tabrizi K, et al. Multigene phylogeny of Choanozoa and the origin of animals. PLoS ONE. 2008;3:e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ride WDL, et al., editors. International Code of Zoological Nomenclature. 4th Ed. London: International Trust for Zoological Nomenclature; 1999. [Google Scholar]
- 29.Maldonado M. Choanoflagellates, choanocytes, and animal multicellularity. Invertebr Biol. 2004;123:1–22. [Google Scholar]
- 30.Pratt JM. Transcription and Translation: A Practical Approach. Oxford: IRL Press; 1984. pp. 179–209. [Google Scholar]
- 31.Carr M, Cotton S, Földvári M, Kotrba M. A description of a new species of Diasemopsis (Diptera, Diopsidae) from the Comoro Islands with morphological, molecular and allometric data. Zootaxa. 2006;1211:1–19. [Google Scholar]
- 32.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez F, Oliver JL, Marín A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- 35.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 36.Baker RH, Wilkinson GS, DeSalle R. Phylogenetic utility of different types of molecular data used to infer evolutionary relationships among stalk-eyed flies (Diopsidae) Syst Biol. 2001;50:87–105. [PubMed] [Google Scholar]
- 37.Swofford DL. Sunderland, Massachusetts: Sinauer Associates; 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Version 4. [Google Scholar]
- 38.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.