Abstract
Synthetic lethal genetic interaction networks define genes that work together to control essential functions and have been studied extensively in Saccharomyces cerevisiae using the synthetic genetic array (SGA) analysis technique (ScSGA). The extent to which synthetic lethal or other genetic interaction networks are conserved between species remains uncertain. To address this question, we compared literature-curated and experimentally derived genetic interaction networks for two distantly related yeasts, Schizosaccharomyces pombe and S. cerevisiae. We find that 23% of interactions in a novel, high-quality S. pombe literature-curated network are conserved in the existing S. cerevisiae network. Next, we developed a method, called S. pombe SGA analysis (SpSGA), enabling rapid, high-throughput isolation of genetic interactions in this species. Direct comparison by SpSGA and ScSGA of ∼220 genes involved in DNA replication, the DNA damage response, chromatin remodeling, intracellular transport, and other processes revealed that ∼29% of genetic interactions are common to both species, with the remainder exhibiting unique, species-specific patterns of genetic connectivity. We define a conserved yeast network (CYN) composed of 106 genes and 144 interactions and suggest that this network may help understand the shared biology of diverse eukaryotic species.
Keywords: comparative genomics, Saccharomyces cerevisiae, Schizosaccharomyces pombe, synthetic genetic array
A better understanding of genetic interactions may illuminate diverse aspects of biology and improve the diagnosis and treatment of complex human diseases (1–3). It remains technically difficult to obtain genetic interaction data for metazoans; therefore, the ability to accurately predict genetic interactions in these organisms using data obtained from experimentally tractable model systems, such as yeast, could be especially useful (4). The major goal of this study was to compare synthetic lethal genetic interaction networks in two distantly related eukaryotic organisms to assess the degree to which these networks are conserved and rewired.
To date, medium-to-large-scale synthetic lethal genetic interaction networks have been generated for simple eukaryotes such as Saccharomyces cerevisiae (5–7) and Caenorhabditis elegans (8, 9). Comparative analysis of these networks revealed that synthetic lethal genetic interactions between orthologous nonessential gene pairs may not be conserved (8, 10). However, three factors potentially confound these results. First, studies in S. cerevisiae have used ORF deletion libraries (11), ensuring that a given gene product is eliminated completely. The C. elegans studies used RNA interference (RNAi)-based methods to inactivate one or both genes, which can result in a variable, gene-specific and incomplete knockdown of gene expression (12, 13). Second, genetic interactions identified in a single-celled organism like S. cerevisiae could, for numerous reasons, be difficult or impossible to detect in a multicellular organism like C. elegans, especially if the phenotype in S. cerevisiae is subtle and only detected accurately by quantitative analysis (14). Third, studies in S. cerevisiae have identified synthetic lethal interactions by measuring colony size. This phenotypic measure may not correlate with the phenotypic measurements used in the studies of C. elegans—namely, animal fecundity and overall growth rate. We hypothesized that a comparative analysis of genetic interactions in S. pombe and S. cerevisiae, two distantly related single-celled fungi, using analogous ORF deletion libraries and scoring techniques, would mitigate these three confounding factors, enabling us to identify conserved as well as species-specific genetic interactions.
S. pombe and S. cerevisiae share substantial gene content, with ≈75% of S. pombe genes having one or more recognizable orthologs in S. cerevisiae. Nevertheless, these species are separated by as much as 1 billion years of evolution (15) and therefore, not surprisingly, the biology of these two organisms is dissimilar in several important ways. For example, S. pombe cells divide by medial fission, a process analogous to the division of cells in many metazoan organisms, whereas S. cerevisiae cells divide by budding. Furthermore, in S. pombe, mating and meiosis are tightly coupled, such that only the zygote is ever a diploid, and even here only transiently. By contrast, in S. cerevisisae, mating and meiosis are not coupled, and this organism prefers the diploid state. Species-specific gene gains and losses are also apparent. Specifically, genes required for functional complexes involved in pre-mRNA splicing, RNAi-mediated heterochromatin silencing, and signalosome function are present in S. pombe (and other metazoan organisms, including humans) but lost in S. cerevisiae (16, 17). The structure of the centromeres in S. pombe is also considerably more complex and metazoan-like in comparison to the relatively simple centromeres of S. cerevisiae (18). Genome-wide microarray, protein localization, and proteomic analyses suggest moderate conservation between the expression, accumulation, and subcellular localization of orthologous gene products and proteins in these two yeasts (19–21). Similarly, a pilot analysis of 85 S. pombe gene deletions found that only 66% of the essential genes identified in S. pombe are also essential in S. cerevisiae (22). Together, these results suggest that rewiring of genetic interaction networks between these two species has occurred and should reflect the different biology of these two species. However, the extent to which this is the case remains unclear.
Here, we create both literature-curated and experimental datasets to help define a synthetic lethal genetic interaction network for S. pombe. Using similar SGA techniques in each species, we compared an S. pombe network to the equivalent S. cerevisiae network and found experimental evidence for conservation on the level of 29% of synthetic lethal (SL) or synthetic sick (SS) genetic interactions tested. Thus, despite substantial rewiring of the genetic interaction networks in each species, there is also a significant conserved core network, shared across hundreds of millions of years of evolution.
Results and Discussion
A High-Confidence Literature-Curated Genetic Interaction Network for S. pombe.
To facilitate the comparison of genetic interaction networks between S. pombe and S. cerevisiae, we first identified comparable high-quality literature-curated genetic interaction datasets for both species. For S. pombe, we curated 1974 published manuscripts and identified a set of 2922 unique genetic interactions, the SpIOB dataset. We also obtained a smaller, independently curated set of 1310 unique genetic interactions for S. pombe from BioGRID (23), the SpBioGRID dataset. In total, 17% (857/4996) of all S. pombe genes have at least one literature-curated annotation in these two datasets. Comparison of the SpIOB and SpBioGRID datasets reveals 2252 unique SpIOB interactions, 640 unique SpBioGRID interactions, and 670 interactions in common [supporting information (SI) Table S1]. These 670 overlapping interactions, identified by two independent efforts, result in a single high-confidence literature-curated network, referred to as SpGI-Overlap. For S. cerevisiae, we derived from BioGRID an analogous, but much more extensive, literature-curated network containing 18,109 genetic interactions, the ScBioGRID dataset (Table S1).
The SpGI-Overlap network contains 273 genetic interactions annotated as SL/SS where both genes have an identifiable sequence ortholog in the ScBioGRID network. We find that 62/273 (23%) of these known S. pombe SL/SS interactions are conserved in S. cerevisiae, significantly more than expected by chance (10,000 randomized networks, P < 0.0001; Table 1, and see Table S2, which lists all SL/SS genetic interactions reported in this study). We also find that 18/170 (11%) and 9/123 (7%) interactions annotated as phenotypic enhancement or phenotypic suppression, respectively, are conserved (Table 1). Thus, these types of interactions are either more poorly conserved than SL/SS interactions or simply more poorly annotated. Regardless, comparative literature-curation analysis alone identifies many genetic interactions conserved between S. pombe and S. cerevisiae.
Table 1.
Summary of overlapping literature-curated genetic interactions in S. pombe and S. cerevisiae
| Genetic interaction type | Interaction source |
Overlapping interactions |
|||
|---|---|---|---|---|---|
| Detectable S. cerevisiae | Detectable S. pombe | Observed no. (%) | Average no. expected ± SD (%) | P | |
| Synthetic lethal + synthetic sick | 10,737 | 273 | 62 (23) | 1.3 ± 1.3 (0.5) | <0.0001 |
| Synthetic lethal | 7,249 | 185 | 33 (18) | 0.7 ± 0.9 (0.4) | <0.0001 |
| Synthetic sick | 4,152 | 61 | 10 (16) | 0.2 ± 0.4 (0.3) | <0.0001 |
| Phenotypic enhancement | 1,347 | 170 | 18 (11) | 0.4 ± 0.6 (0.2) | <0.0001 |
| Phenotypic suppression | 559 | 123 | 9 (7) | 0.1 ± 0.4 (0.1) | <0.0001 |
| All interactions | 12,643 | 566 | 89 (16) | 3.5 ± 2.3 (0.9) | <0.0001 |
Only detectable genetic interactions involving two genes with an ortholog in both species are considered; thus, the S. cerevisiae dataset shown here is smaller than the full ScBioGRID set, and the S. pombe dataset is smaller than the full SpGI-Overlap dataset (see Table S1). The percent observed overlapping values are always calculated with respect to the number of detectable S. pombe interactions.
S. pombe Synthetic Genetic Array (SpSGA) Analysis.
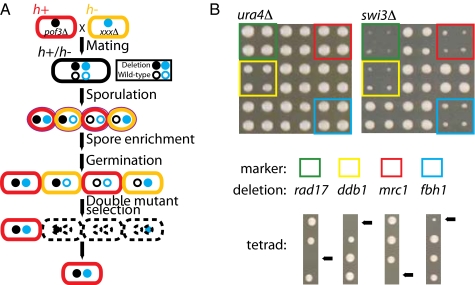
Literature-curated datasets may be biased in a number of ways, and therefore may not enable us to estimate the true degree of genetic network overlap. To enable us to rapidly identify large numbers of genetic interactions in S. pombe experimentally, we developed a method called SpSGA (Fig. 1A and SI Methods). Using SpSGA, we reliably detect known synthetic lethal interactions (Fig. S1). SpSGA does not require specialized screening strains or chemical selections to isolate recombinant double mutants (Fig. S3), making it comparable but relatively simpler to implement than the recently described S. pombe epistasis mapper method (24). One important limitation of the SpSGA approach, however, is that it makes use of heat treatment (3 days at 42°C) to select against unmated haploid cells. The SpSGA method may therefore not be appropriate for use with some temperature-sensitive strains if the conditional allele is required for spore viability.
Fig. 1.
(A) Outline of the SpSGA method. Cells of opposite mating type (h+, h−) are mated on minimal SPA media and allowed to sporulate for 3 days at 26 °C. Then, to enrich for spores, mating plates are transferred to 42° for 3 days—a treatment that kills unmated haploid cells. Following spore enrichment, cells are transferred to rich medium to allow for germination, then transferred again to double-drug medium to select for recombinant double-mutant progeny. S. pombe haploids do not mate on rich medium; therefore, selection for a specific haploid mating type is not required. (B) A portion of the final 1536-formatted miniarray screening plate (YES+Nat+G418) showing a comparison of two queries, a representative control, ura4::kanMX4 (ura4Δ), and a representative query gene, swi3::kanMX4 (swi3Δ). Novel genetic interactions are identified between swi3Δ and mrc1Δ, rad17Δ, ddb1Δ, and fbh1Δ and confirmed by tetrad dissection. Arrows indicate the position of the double deletion mutant, which in one of every four cases is inviable or slow growing.
We focused our first experimental analysis of genetic interactions in S. pombe on a set of 222 genes involved in conserved cellular processes, such as DNA damage checkpoint activation and repair, chromatin remodeling, intracellular transport, and other functions (Table S3). We specifically interrogated this set of functions because they have been examined extensively within the current, but largely incomplete, S. cerevisiae genetic interaction network (6, 7, 25). We isolated double mutants from an orthogonal array of 222 (Kan-marked) × 222 (Nat-marked) gene deletion strains. Double-mutant colony size was scored using the SGA-score algorithm, which quantitates genetic interactions based on colony size, assigning negative scores to SS/SL interactions and positive scores to epistatic genetic interactions (see SI Methods).
Following the removal of a few strains whose deletion allele appeared to be in the incorrect chromosomal location as determined by linkage analysis (Table S3), we were ultimately able to obtain data for 16,860 unique double mutants. We focused on SL/SS interactions and highlighted the 8% most extreme interactions (669 total) for further analysis (Table S4, and see Fig. S2a and Methods for a rationalization of this cutoff). We call this our SpSGA-derived high-confidence (SpSGA HC) dataset. We confirmed 353/397 (89%) interactions in the SpSGA HC dataset by random spore analysis (RSA) (Table S4). Thus our dataset has an 89% positive predictive value (see SI Methods). In the SpSGA HC dataset we identify 10/11 (91%) detectable literature-curated synthetic lethal genetic interactions (Table 2 and Table S4), suggesting we have captured the vast majority of true positive interactions. However, 18/150 (12%) randomly selected double mutants falling between the 9% and 100% data cutoff showed a synthetic sick interaction by random spore analysis (data not shown). Therefore, the number of true positive genetic interactions within the complete SpSGA dataset is potentially greater than the 669 SpSGA HC interactions. Overall, in comparison with genome-wide screens with S. cerevisiae (6), we observe a high level of genetic interactions between the genes examined. This is likely attributable to the fact that many of the genes selected for analysis are functionally related, and genetic interactions tend to occur amongst functionally related genes (6).
Table 2.
Comparison of the SpSGA HC dataset with experimentally derived and literature-curated data for S. pombe and S. cerevisiae
| Experimentally derived dataset | No. of interactions | Comparison dataset | No. of interactions | No. of detectable overlapping interactions | Observed no. of overlapping interactions (%) | Expected no. of overlapping interactions ± SD (%) | P |
|---|---|---|---|---|---|---|---|
| SpSGA HC | 669 | SpGI-Overlap | 341 | 11 | 10 (91) | 0.3 ± 0.6 (3) | <0.0001 |
| ScBioGRID | 14,566 | 559 | 72 (14) | 4.8 ± 3 (0.9) | <0.0001 | ||
| ScSGA HC | 742 | 240 | 54 (23) | 6 ± 3.2 (2.5) | <0.0001 |
The SpSGA HC dataset, containing 669 interactions, is compared to SpGI-Overlap, ScBioGRID, and ScSGA HC-derived datasets. For SpGI-Overlap and ScBioGRID, the number of interactions reported here (341 and 14,566, respectively) are the nonredundant set of SL and SS interactions that are common to SpIOB and SpBIOGRID, with no differentiation between SL and SS interactions. The number of detectable overlapping interactions is the total number of known genetic interactions in these datasets where both genes were also examined in SpSGA. The small number of detectable overlapping interactions (11) relative to the full set of SL/SS interactions in the SpGI-Overlap dataset reflects the fact that most interactions examined here by SpSGA have not previously been studied. SD = standard deviation.
In total, 99% (659/669) of the SpSGA HC interactions are not found in SpGI-Overlap. For example, we detect novel interactions between swi3Δ and both mrc1Δ and rad17Δ (Fig. 1B). The S. cerevisiae ortholog of swi3, CSM3, had previously been shown to have synthetic lethal interactions with MRC1/mrc1 and RAD24/rad17 (6), showing that we are able to detect conserved interactions using SpSGA. In total, overlapping the SpSGA HC dataset against the literature-curated ScBioGRID dataset identified 64 conserved interactions (of 398 potentially detectable interactions; 16%), which is significantly more than expected by chance (10,000 randomized networks, P < 0.0001; Table 2 and Table S2).
Finally, the SpSGA method enables detection of unique genetic interactions between genes that are not found in S. cerevisiae. For example, in the SpSGA HC dataset we detect synthetic lethal interactions between swi3Δ and both ddb1Δ and fbh1Δ (Fig. 1B), two genes involved in the DNA damage response that have sequence orthologs in H. sapiens but not S. cerevisiae (26, 27) (Table S3). Thus, SpSGA can provide insight into the function of conserved genes that cannot be studied using S. cerevisiae SGA methods.
Experimental Comparison of Genetic Interaction Networks.
Examining the degree of genetic network conservation between species using literature-curated data alone, which is not systematic, may be prone to bias. To begin to address this question in a more unbiased way, we used the established S. cerevisiae SGA method (5) to generate a genetic interaction dataset for an orthogonal matrix of 227 S. cerevisiae genes orthologous to those examined in S. pombe (Table S5). The resultant dataset, containing 17,455 unique double mutants, was scored using the identical SGA-score algorithm applied to the SpSGA dataset and, similarly, the extreme 8% SL/SS genetic interactions (742 total) were extracted for comparative analysis (Table S6). We call this our ScSGA-derived high-confidence (ScSGA HC) dataset.
With comparable high-quality, side-by-side SpSGA HC and ScSGA HC datasets for orthologous genes, we were in a position to attempt to identify both conserved and nonconserved interactions, thereby providing a direct, experimental measure of the true extent of conservation of genetic interactions. Restricting our analysis to only those genes with a single predicted ortholog in both species, we observe 23% overlap (54/240 unique detectable interactions) between the SpSGA HC dataset and the ScSGA HC dataset, significantly more than would be expected by chance (versus 10,000 random networks, P < 0.0001; Table 2 and Table S2). Varying the percentage of the most extreme SL/SS interactions included in the comparison between 1% and 40% has little effect; within this range the percentage overlap of genetic interactions never falls below 19% (Fig. S2b). Given the observed positive and negative predictive values for the SpSGA HC dataset, we estimate the true conservation of SL/SS genetic interactions within our dataset to be ∼29% (see SI Methods). Additional sampling of genetic interactions will be required to confirm whether the observed degree of overlap is representative of the overall extent of genetic network conservation on a genome-wide level.
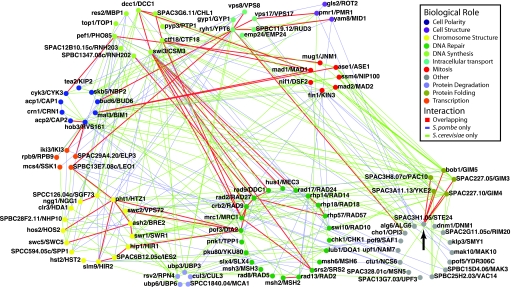
Most genetic interactions in the SpSGA HC and ScSGA HC datasets are species specific, suggesting extensive functional rewiring (Fig. 2). For example, we reconfirm our previous observations from S. cerevisiae (5) that genes encoding members of the prefoldin protein-folding complex, such as PAC10, GIM5, GIM3, GIM4, and YKE2, buffer mitotic spindle formation and cell polarity, consistent with the role of this complex in tubulin and actin folding (ref. 28; Fig. 2). However, similar interactions were not observed in S. pombe (Fig. 2), suggesting that in this organism, actin and tubulin folding via the prefoldin complex could be redundant or that other complexes fulfill these functions altogether. Given this difference, the conservation of genetic interactions between prefoldin complex genes and SPAC3H1.05/STE24, which encodes a CAAX protease that processes prenylated proteins (29, 30), is notable. The consequences of the conservation of the interactions between SPAC3H1.05/STE24 and the prefoldin complex in both yeasts are unclear, but may suggest an important role for this complex in the activity of this conserved protease or one of its substrates.
Fig. 2.
Overlap of the SpSGA HC and ScSGA HC networks. Genes were assigned to single “biological role” categories manually, as described in SI Methods. Eighty-seven SpSGA unique interactions (edges) are in blue, 143 ScSGA unique interactions are in brown, and 54 overlapping interactions are in red. The number of ScSGA unique interactions is larger than the number of SpSGA unique interactions because the ScSGA HC dataset is larger than the corresponding SpSGA HC dataset (see Table 2). A black arrow indicates the position of SPAC3H1.05/STE24, a gene discussed in the text.
Our analysis of divergent genetic interactions reveals interesting gene-specific differences. At a higher level of analysis, an important question is whether groups of genes, functioning together in the same biological process, exhibit large-scale changes in genetic interaction patterns, such that two processes are more or less likely to be associated with each other by genetic interactions in one species or another. Though our data hint at this possibility, additional efforts to increase the number of validated interactions in both species will be required to address this question with adequate statistical power.
Toward a Core Eukaryotic Genetic Interaction Network.
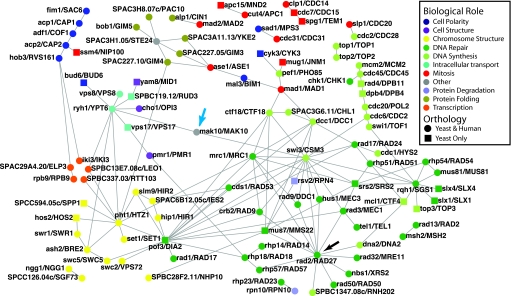
Previous work provided conflicting evidence about whether genetic interactions are conserved between species (8, 10, 31). By integrating data from similar literature-curated and experimentally derived sources, we find strong evidence that many SL/SS interactions are conserved between two distantly related yeasts, S. pombe and S. cerevisiae. We propose that these data, further supplemented with additional data from diverse organisms, may make it possible to define a core eukaryotic genetic interaction network. As a first step toward this goal, we have assembled all conserved yeast interactions identified in this study into a single network (the conserved yeast network, or CYN) containing 144 unique interactions between 106 genes (Fig. 3). In this network, all interactions involve genes with 1:1 orthologous relationships in S. pombe vs. S. cerevisiae (see Table S2 for details). Examination of the CYN reveals conserved interactions spanning numerous processes, including DNA repair, cytokinesis, chromatin remodeling, and intracellular trafficking. The conserved edges of genetic interaction networks highlight important functional connections that might otherwise be overlooked if found in only one species. One such example is the interaction between mak10/MAK10 (Fig. 3, blue arrow), which encodes a component of the N-terminal acetyltransferase C (NatC) complex (32), and swi3/CSM3, a component of the replication fork protection machinery required for efficient DNA replication. A prediction suggested by the conserved network is that N-terminal acetylation of one or more proteins might be critical for efficient DNA replication in eukaryotes.
Fig. 3.
The conserved yeast network (CYN). SL/SS interactions common to both S. pombe or S. cerevisiae were identified by overlapping literature-curated and experimentally derived datasets, as described (see SI Methods and Table S2). Genes were assigned to a single biological role category as described in SI Methods. The blue arrow indicates the position of mak10/MAK10, and the black arrow indicates the position of rad2/RAD27, both of which are discussed in the text.
Many of the genes in the CYN have predicted human orthologs (Fig. 3, round node shape, and Table S3), and it is tempting to speculate that conserved genetic interactions are more likely to involve genes that are also conserved throughout eukaryotic evolution. However, the absence of sequence conservation in higher organisms does not necessarily preclude yeast-specific genes (Fig. 3, square node shape) from playing important roles in conserved networks. For example, several yeast-only genes, such as pof3/DIA2, mus7/MMS22, and srs2/SRS2, have many conserved interactions and appear to play central roles in the CYN.
Some of the genes in the CYN have human orthologs implicated in cancer. It is possible to selectively kill certain tumor cells by exploiting synthetic lethal interactions (4, 33), but the identification of useful mutant combinations in mammals is laborious. We suspect that this process could be facilitated using the CYN to predict SL/SS interactions likely to be also conserved in humans. For example, in humans and mice, loss-of-function mutations of the S. pombe rad2 ortholog, FEN1, are associated with lung cancers (34). In the conserved yeast network we observe interactions between the FEN1 ortholog, rad2/RAD27, and potentially drug-sensitive interactors, including the RecA family ATPase rhp57/RAD57 and the kinase rad3/MEC1 (Fig. 3, black arrow). A prediction of the conserved yeast network is that inhibition of one of these conserved synthetic lethal interactors may be sufficient to kill cells harboring FEN1 mutations. Testing of this prediction, along with future efforts to generate reagents and techniques allowing genetic interactions to be studied in additional species, should provide a better understanding of the core eukaryotic genetic “wiring diagram” and help guide efforts to rationally select targets for therapeutic intervention.
Methods
Literature-Curated Datasets.
S. pombe genetic interactions were curated manually (the SpIOB dataset) or obtained from BioGRID 2.0.39 (the SpBioGRID dataset). Literature curation for the SpIOB dataset was performed as follows. Searching of the PubMed database (www.ncbi.nlm.nih.gov/sites/entrez) identified 7699 articles of interest containing the keywords pombe, fission yeast, or schizosaccharomyces. The title of each article was read manually, resulting in the selection of 1974 articles likely to contain genetic interaction information. Each of the 1974 articles was read, and genetic interactions were extracted manually and recorded in a custom database. All candidate interactions stored in the database were reviewed and approved by a final expert curator before being added to the SpIOB dataset. For S. cerevisiae, datasets involving interactions not confirmed by random spore analysis or tetrad dissection were not considered.
Bioneer S. pombe Gene Deletion Library.
A set of 2663 single-gene deletion strains (genotype: geneX::kanMX4 h+ ade6-M210 ura4-D18 leu1–32), where geneX indicates any one of the genes in the collection, were generated using standard gene replacement methods. Details concerning the construction and verification of the deletion collection are available at http://pombe.bioneer.co.kr/technic_infomation/construction.jsp.
Yeast Strains, Strain Construction, and Strain Manipulations.
All S. pombe and S. cerevisiae strains were generated and grown using standard protocols and manipulated using a Singer RoToR plate handling robot (Singer Instruments).
S. pombe SGA (SpSGA).
The SpSGA method is described in the legend to Fig. 1 and in SI Methods.
S. pombe Miniarray Screening and Confirmation.
We selected a set of 222 genes for analysis by SpSGA (Table S3). A total of 215 gene deletions were from the Bioneer library. To construct the miniarray, G418-resitant starting strains (genotype: h+ geneX::kanMX4) were switched to Nat-resistant miniarray strains (genotype: h- geneX::natMX4) using standard PCR, transformation, and selection techniques. We independently isolated kanMX4 and natMX4 deletions for seven genes of interest (Table S3). All 222 Nat-marked strains were arrayed in 384 format and screened against the collection of 222 Kan-marked strains in batches of 10–20 queries at a time. Up to three batches were processed per week. Genetic interactions were subsequently confirmed using random spore analysis or tetrad dissection as described in SI Methods.
S. cerevisiae Strains, Culturing, and S. cerevisiae SGA (ScSGA).
S. cerevisiae strains were cultured and SGA data collected for 227 strains exactly as described (6), except the resultant interaction data were filtered for potential linkage involving genes lying within 100 kbp on the same chromosome, rather than 50 kbp. All potential interactions were processed using the identical SGA-score algorithm applied to the SpSGA data.
Computational Analysis and Genetic Network Representation.
Overlapping genetic interactions were identified using custom PERL and MATLAB scripts that are available upon request. The statistical significance of overlap was determined in comparison to 10,000 computationally generated networks that maintained the same global network topology as the observed network, but randomly shuffled links between genes. P values were computed empirically and represent the likelihood of seeing the observed degree of overlap between networks in the 10,000 randomly shuffled networks.
Additional Methods.
Additional methods used in this paper are described in SI Methods.
Supplementary Material
Acknowledgments.
We thank Bryan-Joseph San Luis, Alan Jiao, Renée Brost, Yiqun Chen, and Hongwei Zhu for help with screening and confirmation; Harry Singer and Carl Singer for help with robotic protocols; and Michael Costanzo, Huiming Ding, Bilal Sheikh, and Quaid Morris for help with data analysis. This work was supported by a postdoctoral fellowship from the National Cancer Institute of Canada (to S.J.D.), National Institutes of Health Roadmap Grant U54RR020839, “Technology Center for Networks and Pathways” (to A.P.), a Canadian Cancer Society grant (to D.D.), and Canadian Institutes of Health Research Grant GSP-41567 (to B.A. and C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806261105/DCSupplemental.
References
- 1.Maxwell CA, et al. Genetic interactions: The missing links for a better understanding of cancer susceptibility, progression and treatment. Mol Cancer. 2008;7:4. doi: 10.1186/1476-4598-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartman JLt, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291(5506):1001–1004. doi: 10.1126/science.1056072. [DOI] [PubMed] [Google Scholar]
- 3.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8(6):437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 4.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278(5340):1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 5.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 6.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303(5659):808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 7.Pan X, et al. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16(3):487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 8.Byrne AB, et al. A global analysis of genetic interactions in Caenorhabditis elegans. J Biol. 2007;6(3):8. doi: 10.1186/jbiol58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38(8):896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 10.Tischler J, Lehner B, Fraser AG. Evolutionary plasticity of genetic interaction networks. Nat Genet. 2008;40(4):390–391. doi: 10.1038/ng.114. [DOI] [PubMed] [Google Scholar]
- 11.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 12.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 13.Simmer F, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1(1):E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani R, St Onge RP, Hartman JLt, Giaever G, Roth FP. Defining genetic interaction. Proc Natl Acad Sci USA. 2008;105(9):3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3(11):838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 16.Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci USA. 2000;97(21):11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunnerhagen P. Prospects for functional genomics in Schizosaccharomyces pombe. Curr Genet. 2002;42(2):73–84. doi: 10.1007/s00294-002-0335-6. [DOI] [PubMed] [Google Scholar]
- 18.Clarke L. Centromeres of budding and fission yeasts. Trends Genet. 1990;6(5):150–154. doi: 10.1016/0168-9525(90)90149-z. [DOI] [PubMed] [Google Scholar]
- 19.Rustici G, et al. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36(8):809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 20.Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24(7):841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt MW, Houseman A, Ivanov AR, Wolf DA. Comparative proteomic and transcriptomic profiling of the fission yeast Schizosaccharomyces pombe. Mol Syst Biol. 2007;3:79. doi: 10.1038/msb4100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decottignies A, Sanchez-Perez I, Nurse P. Schizosaccharomyces pombe essential genes: A pilot study. Genome Res. 2003;13(3):399–406. doi: 10.1101/gr.636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4(10):861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- 25.Reguly T, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5(4):11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zolezzi F, Fuss J, Uzawa S, Linn S. Characterization of a Schizosaccharomyces pombe strain deleted for a sequence homologue of the human damaged DNA binding 1 (DDB1) gene. J Biol Chem. 2002;277(43):41183–41191. doi: 10.1074/jbc.M207890200. [DOI] [PubMed] [Google Scholar]
- 27.Sakaguchi C, Morishita T, Shinagawa H, Hishida T. Essential and distinct roles of the F-box and helicase domains of Fbh1 in DNA damage repair. BMC Mol Biol. 2008;9:27. doi: 10.1186/1471-2199-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Fanarraga M, Avila J, Guasch A, Coll M, Zabala JC. Review: Postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J Struct Biol. 2001;135(2):219–229. doi: 10.1006/jsbi.2001.4386. [DOI] [PubMed] [Google Scholar]
- 29.Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275(5307):1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 30.Tam A, Schmidt WK, Michaelis S. The multispanning membrane protein Ste24p catalyzes CAAX proteolysis and NH2-terminal processing of the yeast a-factor precursor. J Biol Chem. 2001;276(50):46798–46806. doi: 10.1074/jbc.M106150200. [DOI] [PubMed] [Google Scholar]
- 31.Tarailo M, Tarailo S, Rose AM. Synthetic lethal interactions identify phenotypic “interologs” of the spindle assembly checkpoint components. Genetics. 2007;177(4):2525–2530. doi: 10.1534/genetics.107.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polevoda B, Sherman F. NatC Nalpha-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. J Biol Chem. 2001;276(23):20154–20159. doi: 10.1074/jbc.M011440200. [DOI] [PubMed] [Google Scholar]
- 33.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5(9):689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 34.Zheng L, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13(7):812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.