Abstract
T lymphocytes (T cells) orchestrate adaptive immune responses that clear pathogens from infected hosts. T cells recognize short peptides (p) derived from antigenic proteins bound to protein products of the MHC genes. Recognition occurs when T cell receptor (TCR) proteins expressed on T cells bind sufficiently strongly to antigen-derived pMHC complexes on the surface of antigen-presenting cells. A diverse repertoire of self-pMHC-tolerant TCR sequences is shaped during development of T cells in the thymus by processes called positive and negative selection. Combining computational models and analysis of experimental data, we parsed the contributions of positive and negative selection to the design of TCR sequences that recognize antigenic peptides with specificity, yet also exhibit cross-reactivity. A dominant role for negative selection in mediating antigen specificity of mature T cells and a molecular mechanism for TCR recognition of antigen are described.
Keywords: statistical mechanics, T cell antigen specificity, thymic selection
Because T cell receptor (TCR) genes undergo stochastic somatic rearrangement, most T cells express a distinct TCR, thereby enabling the T cell population to recognize many different antigenic short peptide (p)MHC complexes. TCR recognition of pMHC is both specific and degenerate. It is specific, because if a TCR recognizes a particular pMHC complex, most mutations to the peptide amino acids abrogate recognition (1, 2). It is degenerate because a given TCR can interact productively with several antigenic peptides (3). pMHC complexes where the peptide is derived from the cell's own proteins are also displayed on antigen-presenting cell (APC) surfaces. TCRs are self-tolerant because they bind weakly to these “self”-pMHC complexes, thereby avoiding frequent autoimmune responses.
The diverse, specific/degenerate, and self-tolerant T cell repertoire is designed during T cell development in the thymus (4–8). Immature T cells (thymocytes) interact with a variety of self-pMHC molecules expressed on the surface of thymic epithelial cells as well as hematopoietically derived macrophages and dendritic cells. Thymocytes expressing a TCR that binds with high affinity to any self-pMHC molecule are deleted in the thymus (a process called negative selection). However, a thymocyte's TCR must also bind sufficiently strongly to at least one type of self pMHC complex to receive survival signals and emigrate from the thymus (a process called positive selection).
Signaling events, gene transcription programs, and cell migration during T cell development in the thymus have been studied extensively (4–14). Despite important advances, how interactions with self-pMHC complexes in the thymus shape the peptide-binding properties of selected TCR amino acid sequences such that mature T cells exhibit their special properties is poorly understood.
Recent experiments carried out by Huseby et al. (1, 2) provided important clues in this regard. These experiments determined differences in how T cells interact with foreign (antigenic) pMHC depending on whether they developed in conventional mice that display a diverse array of self-pMHC complexes in the thymus or if they develop in mice that were engineered to express only one type of peptide in the thymus. For T cells that develop in conventional mice, T cell recognition of antigenic pMHC was found to be sensitive to most mutations of the antigenic peptide's amino acids. In contrast, T cells selected in mice with only one type of peptide in the thymus were much more peptide-degenerate, with some T cells being tolerant to most mutations of antigenic peptide amino acids.
We reasoned that a detailed understanding of the origin of these experimental results may shed light on the broader question of how the thymus designs diverse self-tolerant TCR sequences that mediate specific/degenerate antigen recognition. Toward this end, we studied a computational model of thymic selection. Our main conclusions can be summarized as follows. Avoiding negative selection against diverse peptides in the thymus imposes strong constraints on the amino acid composition of the peptide contact residues of selected TCRs. Specifically, TCR peptide contact residues are greatly enriched in amino acids that bind weakly to all other amino acids, a result consistent with our analysis of available crystal structures of TCR–pMHC complexes. We show that such TCRs recognize antigenic peptides via multiple modest interactions, each of which contributes a significant fraction of the binding affinity required for recognition. Therefore, mutations to most peptide amino acids abrogate recognition, thus conferring specificity. Positive selection is important for many properties, such as MHC restriction, but not antigen specificity. Our results, and a model for TCR recognition of antigen that emerges from it, illuminate how thymic selection meets the apparently conflicting demands of antigen specificity, cross-reactivity, and self-tolerance.
Model Development
To describe the interactions between TCRs and pMHC complexes, we represent them as strings of sites (Fig. 1A). Each site on a TCR can interact with the corresponding site on a pMHC molecule. Such “string models” for studying TCR–pMHC interactions have been used to study various issues, including thymic selection (12, 14, 15), and employed simplified representations of amino acids (e.g., a string of numbers, bits, etc.). From the standpoint of our work, the most pertinent result revealed by these past studies are calculations showing that negative selection reduces TCR cross-reactivity. The mechanistic reasons underlying this numerical result or how it relates to amino acid sequences of selected TCRs were not described. Our goal was to elucidate how the diversity of endogenous peptides bound to host MHC proteins encountered in the thymus determines the amino acid sequences of peptide contact residues on selected TCRs and how such TCRs are antigen specific while also being cross-reactive and self-tolerant.
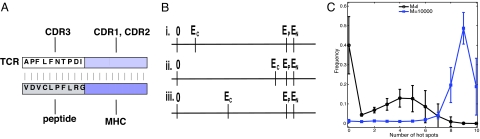
Fig. 1.
A simple model recapitulates differences in specificities of T cells selected in mice with one or many types of peptides in the thymus (1, 2). (A). Schematic description of the model. The interactions between CDR1 and CDR2 regions of the TCR and conserved residues on the MHC are described by a TCR-dependent energy equal to Ec. Amino acids on the peptide (and variant MHC residues) as well as the corresponding contact residues on the CDR3 loops of the TCR are treated explicitly, and their interactions are described in Model Development (Eq. 1). (B) Cartoon representation of the three regimes of values of TCR-MHC interactions (Ec). In these regimes the TCR–MHC interactions are (i) weak, (ii) strong, and (iii) moderate compared with the threshold for negative selection, EN. (C) Selection against many peptides in the thymus results in a larger number of hot spots characterizing antigen recognition. The frequencies of occurrence of one, two, three, etc., hot spots (defined in Results) on MHC-bound antigenic peptide moieties recognized by selected TCRs. For TCRs that develop in a thymus with many types of self-peptides (blue curve, M = 10,000 peptides) many sites on the antigenic peptide moiety are hot spots. For TCRs that develop in a thymus with only one type of self-pMHC complex (black curve, M = 1 peptide) there are far fewer hot spots, indicating less specific (more degenerate or cross-reactive) recognition.
The specific features of our model were chosen to address these issues and to relate our results closely to known experimental data such as that of Huseby et al. (1, 2). Because Huseby et al. used transgenic mice that expressed a single type of MHC, we divided the string of sites on the pMHC molecule into a conserved part representing the MHC and a variable part representing the peptides. One could also view the variable sites more generally as representative of the peptides and the variable residues of the MHC. The CDR1 and CDR2 loops of the TCR mostly contact MHC residues, whereas the CDR3 loop primarily contacts the peptide residues. We partitioned the TCR interaction sites in to two parts: a region representing the CDR1 and CDR2 loops and a part that mimics the CDR3 loop. Because the CDR3 loops are hypervariable, the amino acids of the peptide contact residues of the CDR3 region are explicitly considered, whereas those of the less variable CDR1 and CDR2 regions are not (Fig. 1A). For ease of reference, the CDR3 sites are called, “variable.” These variable sites represent only those CDR3 amino acids that contact peptide amino acids (or variable MHC residues). Thus, we do not explicitly treat the conformation of the CDR3 loop, which would be necessary if the entire sequence of CDR3 amino acids was considered. Similarly, because peptides bound to MHC are short, peptide conformation is not an important variable. Although we vary the peptide length (data not shown), most results we present are for peptides that are 10 aa long.
We generate panels of TCR and self pMHC molecules on the computer by picking amino acids for the peptides and peptide contact residues on the CDR3 loops of the TCR according to the probabilities with which amino acids appear in the human (or mouse) proteome (16) (Table S1). Antigenic peptides are generated using the frequency of occurrence of amino acids in Listeria monocytogenes, a common bacterial pathogen (17). To assess the effects of thymic selection as well as antigen recognition, we evaluate the energy of interaction between TCR-pMHC pairs. The interaction energy between the CDR1 and CDR2 regions of TCRs and the MHC is given a value equal to Ec (and it is varied to describe different TCRs). The total interaction energy equals the sum of Ec and the value obtained by aligning the TCR and pMHC amino acids that are treated explicitly and adding the pairwise interactions between corresponding amino acids. For a given TCR–pMHC pair, the total interaction energy is
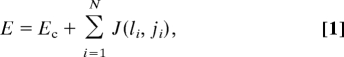 |
where Ec is defined above, and J (li, ji) is the interaction energy between the ith amino acids on the variable part of the TCR (li) and the peptide (ji), respectively, and N is the length of the variable regions. The matrix J encodes the values of interaction energies between specific types of amino acids. For most results presented, J was taken to be the parameterized potential due to Miyazawa and Jernigan (MJ matrix) which has been used fruitfully to study proteins (18, 19). However, we also used other potentials (vide infra), including ones where the interaction between a pair of juxtaposed amino acids depends on the neighboring residues, to show that our qualitative results and mechanistic insights are independent of this choice [supporting information (SI) Fig. S1]. We express energy values in units of the thermal energy, kBT, where kB is Boltzmann's constant, and T is absolute temperature. At 37°C, the thermal energy equals 0.6 Kcal/mole. We emphasize that the purpose of our study is not to compute specific values of energies but to use them to obtain qualitative mechanistic insights.
Recent experiments show that negative selection occurs when the TCR–pMHC interaction affinity exceeds a sharply defined threshold (9). Because affinity correlates directly with the free energy (or energy) gained upon binding, in our model, if the interaction energy between a TCR and self-pMHC is more attractive than (exceeds) a threshold value, EN, this TCR is negatively selected. It is possible that the off-rate characterizing TCR–pMHC binding, rather than affinity, determines ligand potency, and, indeed, ligands that induce positive and negative selection are separated by a sharp boundary in off-rate as well. Off-rate correlates with the free-energy barrier associated with dissociation of the TCR–pMHC complex. For a related set of reactions, this barrier and the binding energy scale similarly (20) (Linear Free-Energy Relationships in SI Text) and so use of the interaction energy should correlate with trends in off-rate as well. The ability of a pMHC ligand to stimulate positive selection does not go to zero abruptly (9). In our model, if the interaction energy between a particular TCR–pMHC pair exceeds a threshold value, Ep, the TCR is positively selected. Replacing the soft threshold associated with positive selection with a sharp boundary does not affect qualitative results (Fig. S2) because we find that the characteristics of peptide binding residues on selected T cells are largely shaped by negative selection. The effects of varying Ep and EN over wide ranges are described in the context of our results.
Results
Selection Against Many Endogenous pMHC Molecules Is Required for Antigen-Specific TCR Sequences.
We first tested whether our computational model could recapitulate the experimental observation (1, 2) that T cell recognition of an antigenic peptide is sensitive to mutations at many peptide sites for T cells selected against many endogenous thymic peptides, whereas very few sites on the antigenic peptide are important for recognition for T cells selected in mice that express one type of peptide in the thymus.
For a specific choice of the interaction energy between the CDR1/CDR2 region of the TCR and the MHC (Ec), a panel of one million sequences of TCR peptide contact residues was generated by choosing different amino acids for the variable region according to the frequency with which they appear in the human proteome (results for mouse are in Fig. S3). In the case where there was only one type of self pMHC complex in the thymus, the interaction energy between each TCR and a MHC-bound peptide moiety representative of the human proteome was computed by using the MJ interaction energy matrix and Eq. 1. Only those TCRs that have interaction energies lying between the positive- and negative-selection thresholds (Ep and EN) were selected. The selected T cells were then challenged with many antigenic peptides characteristic of L. monocytogenes (17). A TCR was considered to recognize an antigenic pMHC if the interaction energy exceeded the negative-selection threshold, EN. In this way, panels of selected T cells that recognize different antigens were generated. Each amino acid on the antigenic peptides was then mutated to the 19 other possibilities, and recognition by the reactive TCRs was again assessed. If more than half the mutations at a particular amino acid site led to abrogation of recognition for an originally reactive T cell, the site was labeled a “hot spot.” This procedure was repeated 1,000 times with a different panel of preselection TCRs, and choices for the peptide in the thymus and antigenic peptides to obtain statistics on the number of hot spots characterizing interactions between a typical antigenic peptide and selected TCRs.
For many types of peptides in the thymus, we generated a panel of 10,000 self-peptides using amino acid frequencies characteristic of the human proteome (16). The results we obtain are qualitatively robust if at least 100 types of pMHC complexes are in the thymus (Fig. S4). Pathologically large numbers of peptides in the thymus result in deletion of all thymocytes. Interaction energies of the panel of TCRs with self-pMHCs were calculated. A TCR was positively selected if it interacted with at least one such pMHC with an energy that exceeded the positive-selection threshold (Ep). To avoid negative selection, a TCR must not interact with any self-pMHC with an energy that exceeds the negative selection threshold (EN). Hot spots characterizing antigen recognition were determined in the manner described above.
Although the interaction energy between the CDR1 and CDR2 regions of the TCR and MHC (Ec) varies continuously as residues on the CDR1 and CDR2 regions change, TCRs can be grouped into three classes based on the relative values of Ec and the negative-selection threshold, EN (Fig. 1B): (i) TCR–MHC interactions are very weak (Ec and EN are separated by a large value); (ii) TCR–MHC interactions are very strong (Ec and EN are separated by a small value); (iii) TCR–MHC interactions are moderate in scale (Ec and EN are separated by a moderate value). Based on recent experimental data (9), for results reported, the difference between Ep and EN is taken to be relatively small (5 kBT). For completeness, we consider cases where this gap is large, and the qualitative results are unchanged (see Results for Cases Where the Gap Between the Positive and Negative Selection Thresholds Is Large, and TCR-MHC Interactions Are Weak in SI Text).
Very few preselection TCRs with CDR1 and CDR2 loops that interact very weakly with conserved MHC (case i) are positively selected if Ep and EN are relatively close (Table S2). In effect, they are not MHC restricted. These TCRs are irrelevant for our studies of how thymic selection shapes antigen-specific peripheral T cells. TCRs with CDR1 and CDR2 loops that interact very strongly with MHC (Ec close to or greater than EN, case ii) are negatively selected with very high probability (Table S2) and so are not relevant for our studies of understanding the origin of how thymic selection results in antigen specificity in the periphery. Not surprisingly then, our studies focus on TCRs with values of Ec that correspond to moderate interactions between the CDR1/CDR2 loops and MHC (case iii). These TCRs are positively selected with high probability and must avoid negative selection to emerge into the periphery (Table S2).
Fig. 1C shows the frequency of hot spots resulting from our calculations when the conserved TCR–MHC interactions are moderate in scale (EN − Ec taken to be 40 kBT for the results). For TCR selected against many types of peptides, a large fraction of the antigenic peptide's amino acids are hot spots. In contrast, when TCR are selected against one type of peptide in the thymus, very few antigenic peptide amino acids are hot spots. This mirrors previous experimental observations (2). We also find that for moderate TCR–MHC interactions, the ability of T cells to mature when only one type of peptide is present in the thymus is limited by positive selection, whereas T cell survival is limited by negative selection when there are many types of peptides in the thymus (Fig. S5). Because our computational model recapitulates known experimental data (Fig. 2 and refs. 1 and 2), we used the model to obtain insights into the mechanistic origins of antigen specificity.
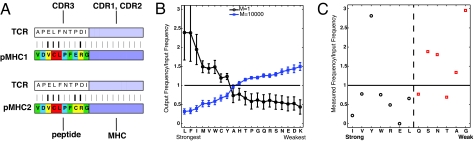
Fig. 2.
Consequences of avoiding negative selection on the composition of peptide contact residues of selected TCRs. (A) Schematic description of frustration due to negative selection. The thickness of the bars (or color of peptide amino acids: strong, red; moderate, yellow; weak, blue; very weak, green) is proportional to the interaction energy between TCR and pMHC residues. When developing in a thymus with only one type of endogenous peptide, a TCR that results in a few strong interactions and several weak or moderate interactions with this peptide can survive selection. This is because the total interaction energy falls between the positive- and negative-selection thresholds. The sequence of TCR peptide contact residues shown, that survives selection against one type of peptide in the thymus, would likely be negatively selected when there are many types of peptides in the thymus. For example, a peptide that differs by one amino acid from the first one (shown as a change from E to C) may lead to an additional moderate interaction energy that is sufficient to increase the total interaction energy past the negative selection threshold. (B and C) Selection against many types of peptides in the thymus results in selected TCRs with peptide contact residues with an enhanced frequency of amino acids that interact weakly with all other amino acids. The ordinate is the ratio of the frequencies of occurrence of an amino acid in the peptide contact residues of selected TCRs to preselection TCRs. (B) For the computational results, the abscissa is a list of amino acids ordered according to the maximum energy (as per the MJ interaction potential) with which it interacts with all other amino acids. The qualitative results are robust to changes in potential (Fig. S1 B, D, and F). (C) The ordinate was obtained by analyzing the 18 available crystal structures of TCR-pMHC (I) complexes as described in the text. Amino acids were classified as strongly interacting (IVYWREL) or weakly interacting (QSNTAG) following ref. 23.
Frustration During Negative Selection Strongly Constraints Selected TCR Sequences.
For a TCR to emerge from the thymus when only one type of pMHC complex is present therein, the binding energy of the TCR for this pMHC must lie in the interval between EN and Ep. Because the interaction energy between the TCR's peptide contact residues and the peptide's amino acids is a sum over individual contact energies (Eq. 1), many sequences of peptide contact residues on the TCR can satisfy this criterion. A type of selected sequence that occurs with high probability is one where a small number of TCR residues make strong contacts with the corresponding peptide amino acids, and all of the others make irrelevant (i.e., weak) contacts (Fig. 2A). A TCR with such a sequence of peptide contact residues on the TCR would almost certainly be negatively selected when many types of peptides are present in the thymus. This is because it will likely encounter another peptide in the thymus that can differ by only a single amino acid, leading to an additional significant interaction and a total energy that exceeds EN.
Thus, surviving negative selection presents a frustrating situation because a TCR that avoids negative selection with one peptide in the thymus could be negatively selected by another peptide. Positive selection does not present this problem because, once a TCR receives survival signals by binding a single peptide more strongly than Ep, interactions with other peptides are only relevant for negative selection. The frustration associated with subsequently avoiding negative selection by all these diverse pMHCs is the dominant constraint determining peripheral TCR sequences.
To explore how this frustration influences the character of the peptide contact residues of selected TCRs, we developed an analytical approximation (Methods) that suggested that the peptide contact residues on selected TCRs are greatly enriched in amino acids that bind weakly to other amino acids.
Negative Selection Against Many Peptides Results in TCR Sequences with Peptide Contact Residues Enriched in Weakly Interacting Amino Acids.
To test this suggestion, we first examined the amino acid compositions of the peptide contact residues of the selected TCRs obtained from our computer simulations. When there are many types of peptides in the thymus, peptide contact residues of selected TCRs are enriched in amino acids that interact weakly with other amino acids, whereas strongly interacting amino acids are attenuated (Fig. 2B). The opposite is true when T cell selection is mediated by a single peptide species in the thymus, with preferential selection of TCR that contain strongly interacting amino acids. In Fig. 2B, amino acids were ordered according to the maximum value of the strength with which each amino acid interacts with all others. The nature of the MJ interaction potential is such that this order also reflects the ordering obtained by considering the average value of the interaction energy of an amino acid with all others. The qualitative results shown in Fig. 2B are robust to changes in the interaction potential (Fig. S1). Using different potentials only changes the identities of the amino acids that interact weakly or strongly or the criterion used to define interaction strength. For example, if a potential is such that the order of amino acids obtained by using the average interaction energies with other amino acids is quite different from that obtained by considering the largest interaction energies, the qualitative results in Fig. 2B are obtained if we use the latter quantities to order amino acids.
Do experimental data support our conclusion that frustration due to negative selection skews the mature T cell repertoire to TCRs composed of peptide contact residues enriched in amino acids that bind weakly to other amino acids? We analyzed the 18 available crystal structures of TCR bound to class I pMHC complexes to obtain the frequency with which different amino acids are represented at residues of the TCR that contact the peptide (21). All TCR moieties that contact peptide amino acids were considered, and two methods were used to identify these contact residues. One was to define a contact as a position where a water molecule does not fit in the gap between a TCR residue and a peptide amino acid. In the other method, residues in contact have their Cα atoms within 6.5 Å of each other. The qualitative results are the same for both methods (Fig. S6), and in Fig. 2C, we show results using the second criterion.
Whereas the qualitative computational results (Fig. 2B) are independent of interaction potential, to compare the experimental data with this prediction, we need to know whether a particular amino acid is “weak” or “strong” in reality. We have used two different prescriptions to order the amino acids according to the strength of their interactions with other amino acids. One is to use the MJ matrix, but the order thus obtained has been criticized because it overemphasizes hydrophobic interactions and considers interactions between charged amino acids to be weak (22). Data obtained by examining the stability of thermophiles are proposed to be better suited for analyzing the strength of interactions between amino acids (23), and posit that the strongly interacting amino acids are IVYWREL, and the weakly interacting ones are QNSTAG (23).
Fig. 2C shows results where amino acids are divided into two classes (weak and strong) according to this prescription. The data obtained from crystal structures are in qualitative agreement with the theoretical prediction in that weakly interacting amino acids are enriched on peptide contact residues of the TCR, and strongly interacting amino acids are attenuated. Using the MJ matrix leads to similar results (Fig. S6), except that charged amino acids (R, E, K), which are “weak” according to the MJ matrix, are additional outliers. Tyrosine is considered to be a strongly interacting amino acid by either approach, but is well represented in the TCR–peptide contact residues. This may be because a germ-line-encoded tyrosine interacts with a conserved MHC residue that is close to the peptide amino acids (24, 25), and so it may interact ubiquitously with peptide amino acids.
Our results suggest that negative selection against many types of thymic peptides results in mature TCRs with peptide contact residues that interact weakly with other amino acids. How does this influence their antigen specificity?
Antigen Specificity Is the Result of TCR Residues Binding Peptides via Multiple Moderate Interactions.
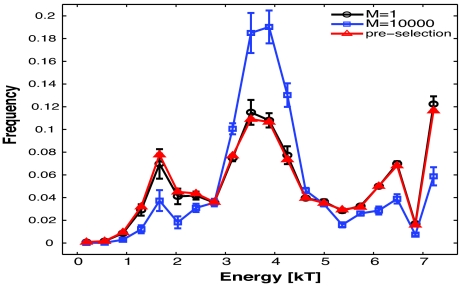
In our model, the interaction energy between an antigenic peptide and residues of a TCR that recognizes it is the sum of 10 numbers, with each number being the interaction energy between an amino acid on the peptide and the corresponding TCR contact site (Fig. 1A). We computed the values of these site–site interaction energies using all our TCR–antigenic peptide pairs. In Fig. 3, we compare the frequency with which each value of these interaction energies occurs for three cases: preselection TCRs, TCRs that developed in a thymus with many types of pMHC, and TCRs that developed in a thymus with one type of pMHC.
Fig. 3.
Distribution of amino acid–amino acid contact energies (in units of kT, described in text) characterizing interactions between selected reactive TCRs and antigenic peptides suggest the basis for specificity. The distribution of interaction energies between individual amino acids on peptide contact residues on the TCR and antigenic peptides are shown. The distribution for TCRs that develop in a thymus with many endogenous peptides (blue curve) is very different from that for preselection TCRs (red curve). The distribution of contact energies is not significantly altered for TCRs that develop in a thymus with only one type of peptide (black curve) compared with preselection TCRs.
Our results indicate that, compared with the preselection TCRs, antigen recognition by TCRs selected against many types of pMHC complexes is mediated by fewer strong and weak amino acid–amino acid interactions, resulting in a pronounced enhancement of moderate interactions. This result is consistent with experimental observations of Savage and Davis (26). This focusing on moderate interactions is because negative selection constrains mature TCR peptide contact residues to be composed of weakly interacting amino acids (Fig. 2). The weakly interacting amino acids on the TCR bind to strongly interacting amino acids on antigenic peptides (Fig. S7) resulting in multiple moderate scale interactions that add up to a total binding energy that is large enough for recognition. Because antigen recognition is mediated by multiple interactions of moderate value, each contact makes a significant contribution to the total interaction energy necessary for recognition. Therefore, disrupting most interactions by mutating peptide amino acids results in abrogation of recognition. This is the origin of antigen specificity. This prediction is consistent with measurements reported for the B3K 506 TCR, which was selected against many types of pMHC complexes in the thymus and recognizes the 3K–IAb pMHC (1). Many mutations of the antigenic peptide correspond to moderate ΔΔG values, and each contributes significantly to recognition.
When there is one type of pMHC complex in the thymus, the peptide-binding residues of selected TCRs are not subject to the important constraints of avoiding negative selection against many types of peptides, and moderate amino acid–amino acid interactions do not dominate (Fig. 3). Strongly interacting amino acids are represented more than in the preselection repertoire (Fig. 2B), resulting in a small enhancement of strong interactions between amino acids (Fig. 3). These strong interactions make dominant contributions to the total interaction energy required for antigen recognition (see also Fig. 2A). Thus, mutating the antigenic peptide amino acids that contact strongly interacting amino acid residues on the TCR should abrogate recognition, but mutations at most other sites should have little impact. This is reflected in the experimental data reported by Huseby et al. (1). For one example, consider the YAe62.8 TCR, which is selected against a single type of peptide in the thymus and recognizes variants of the 3K-IAb antigenic peptide. Most mutations to the antigenic peptide result in small changes in ΔΔG, but one mutation results in a large change. This one major peptide contact dominates the interaction energy with the others being irrelevant, and this is the origin of enhanced cross-reactivity.
TCRs that survive negative selection against many types of peptides are quite diverse because many sequences are consistent with the constraint that peptide contact residues are predominantly composed of amino acids that interact weakly with all others.
Discussion
Although important clues were provided by the experimental data reported by Huseby et al. (1, 2), a mechanistic understanding of how thymic selection designs TCR sequences that are simultaneously antigen specific, cross-reactive, diverse, and self-tolerant remained unclear. Our computational studies shed light on these issues.
If a TCR receives survival signals from a self-pMHC complex, it is positively selected. Interactions with the other peptides expressed in the thymus are then only relevant for negative selection. Positive selection ensures MHC restriction, enables weak binding of TCRs to self pMHC, and influences the fraction of T cells that survive thymic selection. Thus, it mediates important properties. However, antigen specificity appears to be determined by the requirement that positively selected T cells must survive negative selection.
TCR sequences must simultaneously avoid being negatively selected by many endogenous MHC-bound peptides, and this imposes strong constraints on the nature of the peptide contact residues of selected TCRs. We find that this is why, in mature T cells, these residues are enriched in amino acids that interact weakly with other amino acids (referred to as “weak” amino acids). For a selected TCR to recognize an antigenic peptide in the periphery, it must bind to it with an affinity that exceeds a threshold. This can occur only if the peptide is composed of amino acids that are among the strongest binders of the corresponding weak amino acids of the TCR's peptide contact residues (Fig. S7), resulting in a number of moderate scale interactions that sum up to exceed the threshold affinity required for recognition. Because each moderate interaction contributes a substantial fraction of the overall affinity, disrupting most of them (via mutations) abrogates recognition. Thus, antigen specificity emerges because TCR residues that contact the peptide are enriched in amino acids that interact weakly with other amino acids. It is worth remarking that weakly binding amino acids are not always the mediators of recognition; TCR selected against one type of peptide do not exhibit this behavior (1, 2), and the EGFR receptors-binding sites are cysteine rich (27).
Because the amino acids treated explicitly in our model include variable MHC residues, our results are also consistent with data showing that TCR selected against many peptides are also MHC specific. We note in passing that we have also studied the alloreactivity of selected TCRs (data not shown). Our findings suggest that the relative importance of the peptide (compared with the MHC) in mediating alloreactive responses depends on how different the allo- and endogenous MHCs are vis-à-vis their interaction energies with the CDR1 and CDR2 loops of a particular TCR (Ec in our model); the greater this difference, the less important the peptide.
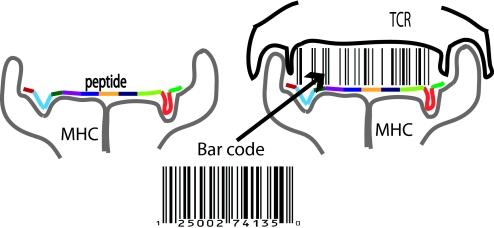
Our results suggest a model for specificity of TCR–antigenic pMHC recognition that is different from Fisher's “lock and key” metaphor for the specificity with which an enzyme binds its substrate. It also appears to be different from that applicable to specificity of antibody–antigen interactions where shape complementarity and multiple weak interactions are inextricably coupled (28). Shape complementarity is important for TCR recognition of antigen in two ways (Fig. 4). First, it plays a key role in peptide binding to the MHC groove, and hence influences antigen presentation. Secondly, shape complementarity is possibly important in mediating interactions of the TCR with MHC moieties, which results in orienting the TCR in a way that juxtaposes its peptide contact residues with the peptide. Indeed, it has been suggested that if the peptide has a conformation that is not relatively flat, it disrupts TCR–MHC interactions, thereby preventing positive selection (29). But, these TCR–MHC interactions required for positive selection and binding of peripheral TCRs to MHC in the proper orientation do not confer peptide specificity.
Fig. 4.
A bar code scanning model for specificity of TCR recognition of antigenic peptides. The thickness of the lines in the cartoon is proportional to the strength of TCR–peptide interactions.
Once properly oriented, a TCR scans the relatively flat conformation of the short peptide, and recognizes the epitope if a number of peptide amino acids correspond to strong binders for the weak peptide contact residues of this TCR. For reasons described above, recognition is specific because each resulting interaction is moderate. Shape complementarity seems to be decoupled from the origin of specificity. TCR recognition of antigen is analogous to scanning a flat “bar code” for the appropriate number of moderately thick lines. In this metaphor, the moderately thick lines represent moderate interactions mediated by peptide amino acids that are strong binders for the weak amino acids that comprise the TCR's peptide contact residues. This bar-code model also makes vivid why specificity and cross-reactivity can coexist. For example, consider a situation where any three of four contacts with the peptide amino acids need to be of moderate scale for recognition; i.e., three of the four lines need to be moderately thick. If a particular peptide satisfies this criterion (say, lines 1, 3, and 4 are moderately thick), mutations at any one of these sites will abrogate recognition (specificity). But another peptide that leads to lines 1, 2, and 3 being moderately thick will also be recognized by this TCR (cross-reactivity). One might say that TCRs scan a bar code and recognize statistical patterns—ones that have a sufficient number of moderately thick lines.
We hope that the results we have reported will motivate experimental and computational studies that will ultimately elucidate how one of nature's intriguing designers (the thymus) works and how its aberrant regulation can contribute to autoimmune disease. An important question unresolved by our studies is how variability in expression levels of different types of endogenous peptides in the thymus influences the T cell repertoire.
Methods
How Negative Selection Against Many Peptides Constrains Selected TCR Sequences.
The probability (P) that a TCR characterized by a sequence of peptide contact residues, l⃗ = {l1, l2, l3,…}, is not negatively selected can be written as:
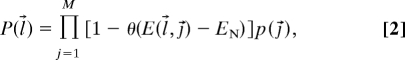 |
where M is the number of peptides in the thymus, E(l⃗, j⃗) is the absolute value of the interaction energy between the TCR and a peptide composed of a sequence of amino acids, j⃗ = {j1, j2, j3, …}, which occurs with probability p(j⃗). The step function, θ, represents the negative selection threshold. Approximations (described in Probability that a TCR Will Escape Negative Selection in SI Text) allowed us to rewrite Eq. 2 as:
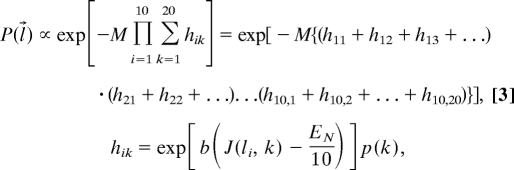 |
where b is a positive constant.
Eq. 3 suggests that if any of the quantities, hik, becomes large, the probability of survival of that TCR becomes small, and that hik becomes large if the TCR's peptide contact residues interact strongly with its corresponding peptide amino acid. Thus, TCR with a high probability of survival must be composed of peptide contact residues that bind weakly to other amino acids.
Supplementary Material
Acknowledgments.
We thank Profs. Herman Eisen and Eugene Shakhnovich for fruitful discussions and comments. This work was supported by National Institutes of Health (NIH) Grant 1-PO1-AI071195-01 and a NIH Director's Pioneer award (to A.K.C.). E.S.H. was supported in part by Beckman Young Investigator and Searle Scholar awards.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808081105/DCSupplemental.
References
- 1.Huseby ES, et al. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- 2.Huseby ES, et al. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Unanue ER. Antigen-presenting function of the macrophage. Annu Rev Immunol. 1984;2:395–428. doi: 10.1146/annurev.iy.02.040184.002143. [DOI] [PubMed] [Google Scholar]
- 4.von Boehmer H, et al. Thymic selection revisited: How essential is it? Immunol Rev. 2003;191:62–78. doi: 10.1034/j.1600-065x.2003.00010.x. [DOI] [PubMed] [Google Scholar]
- 5.Werlen G, Hausmann B, Naeher D, Palmer E. Signaling life and death in the thymus: Timing is everything. Science. 2003;299:1859–1863. doi: 10.1126/science.1067833. [DOI] [PubMed] [Google Scholar]
- 6.Siggs OM, Makaroff LE, Liston A. The why and how of thymocyte negative selection. Curr Opin Immunol. 2006;18:175–183. doi: 10.1016/j.coi.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: Learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 8.Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 9.Daniels MA, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 10.Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte–stromal cell interactions visualized by two-photon microscopy. Science. 2002;296:1876–1880. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- 11.Borghans JAM, Noest AJ, De Boer RJ. Thymic selection does not limit the individual MHC diversity. Eur J Immunol. 2003;33:3353–3358. doi: 10.1002/eji.200324365. [DOI] [PubMed] [Google Scholar]
- 12.Detours V, Mehr R, Perelson AS. A quantitative theory of affinity-driven T cell repertoire selection. J Theor Biol. 1999;200:389–403. doi: 10.1006/jtbi.1999.1003. [DOI] [PubMed] [Google Scholar]
- 13.Scherer A, Noest A, de Boer RJ. Activation-threshold tuning in an affinity model for the T-cell repertoire. Proc R Soc London Ser B. 2004;271:609–616. doi: 10.1098/rspb.2003.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Detours V, Perelson AS. Explaining high alloreactivity as a quantitative consequence of affinity-driven thymocyte selection. Proc Natl Acad Sci USA. 1999;96:5153–5158. doi: 10.1073/pnas.96.9.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao DL, Davenport MP, Forrest S, Perelson AS. The effects of thymic selection on the range of T cell cross-reactivity. Eur J Immunol. 2005;35:3452–3459. doi: 10.1002/eji.200535098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flicek P, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moszer I, Glaser P, Danchin A. Subtilist—A relational database for the Bacillus subtilis Genome. Microbiol UK. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Tang C, Wingreen NS. Nature of driving force for protein folding: A result from analyzing the statistical potential. Phys Rev Lett. 1997;79:765–768. [Google Scholar]
- 19.Miyazawa S, Jernigan RL. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 20.Edwards JO. Correlation of relative rates and equilibria with a double basicity scale. J Am Chem Soc. 1954;76:1540–1547. [Google Scholar]
- 21.Kaas Q, Ruiz M, Lefranc MP. IMGT/3Dstructure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acids Res. 2004;32:D208–D210. doi: 10.1093/nar/gkh042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorenson JM, Head-Gordon T. The importance of hydration for the kinetics and thermodynamics of protein folding: Simplified lattice models. Biophys J. 1999;76:A109–A109. doi: 10.1016/S1359-0278(98)00068-6. [DOI] [PubMed] [Google Scholar]
- 23.Zeldovich KB, Berezovsky IN, Shakhnovich EI. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 2007;3:62–72. doi: 10.1371/journal.pcbi.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng D, et al. Structural evidence for a germline-encoded T cell receptor—Major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 25.Dai S, et al. Crossreactive T cells spotlight the germline rules for alpha beta T cell–receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savage PA, Davis MM. A kinetic window constricts the T cell receptor repertoire in the thymus. Immunity. 2001;14:243–252. doi: 10.1016/s1074-7613(01)00106-6. [DOI] [PubMed] [Google Scholar]
- 27.Abe Y, et al. Disulfide bond structure of human epidermal growth factor receptor. J Biol Chem. 1998;273:11150–11157. doi: 10.1074/jbc.273.18.11150. [DOI] [PubMed] [Google Scholar]
- 28.Perelson AS, Oster GF. Theoretical studies of clonal selection—Minimal antibody repertoire size and reliability of self-non-self discrimination. J Theor Biol. 1979;81:645–670. doi: 10.1016/0022-5193(79)90275-3. [DOI] [PubMed] [Google Scholar]
- 29.Schumacher TNM, Ploegh HL. Are MHC-bound peptides a nuisance for positive selection. Immunity. 1994;1:721–723. doi: 10.1016/s1074-7613(94)80013-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.