Abstract
Rapid rebound of plasma viremia in patients after interruption of long-term combination antiretroviral therapy (cART) suggests persistence of low-level replicating cells or rapid reactivation of latently infected cells. To further characterize rebounding virus, we performed extensive longitudinal clonal evolutionary studies of HIV env C2-V3-C3 regions and exploited the temporal relationships of rebounding plasma viruses with regard to pretreatment sequences in 20 chronically HIV-1-infected patients having undergone multiple 2-week structured treatment interruptions (STI). Rebounding virus during the short STI was homogeneous, suggesting mono- or oligoclonal origin during reactivation. No evidence for a temporal structure of rebounding virus in regard to pretreatment sequences was found. Furthermore, expansion of distinct lineages at different STI cycles emerged. Together, these findings imply stochastic reactivation of different clones from long-lived latently infected cells rather than expansion of viral populations replicating at low levels. After treatment was stopped, diversity increased steadily, but pretreatment diversity was, on average, achieved only >2.5 years after the start of STI when marked divergence from preexisting quasispecies also emerged. In summary, our results argue against persistence of ongoing low-level replication in patients on suppressive cART. Furthermore, a prolonged delay in restoration of pretreatment viral diversity after treatment interruption demonstrates a surprisingly sustained evolutionary bottleneck induced by punctuated antiretroviral therapy.
Keywords: HIV-1, latent reservoir, structured treatment interruption, viral diversity, coreceptor usage
It is unclear to what extent HIV-1 replication under current combination antiretroviral therapy (cART) persists in vivo. In patients with long-term suppression of viremia, plasma HIV RNA rebounds within days or weeks when cART is interrupted. This suggests persistence of low-level replication and/or rapid reactivation of latently infected cells. Gradual and continuous evolution would be expected in the former and stochastic reappearance of phylogenetically distinct viruses in the latter case.
Despite the success of current cART (1–3), eradication of HIV is not possible because of the persistence of a reservoir of latently infected cells with a very long half-life (4–8). Furthermore, low-level replication in some patients may lead to replenishment of the latent reservoir, thus raising the bar for eradication even higher (7, 9–12). The complexity of anatomical and cellular compartments harboring replication-competent virus may further add to the difficulty of eradicating HIV (13, 14). If cART—even after prolonged duration—is stopped, HIV viremia rebounds within days to weeks, often reaching very high peaks, before stabilizing at close to pretreatment HIV-RNA levels (15–18). Whether these viral set points can be lowered by initiating cART early during primary HIV infection remains to be determined (19–23).
The characteristics of rebounding virus have not been fully elucidated as yet. In particular, it is not known whether the rebounding virus originates from reactivated latently infected cells or arises from cells involved in the process of low-level replication (24–27). To address this question, we took advantage of the Swiss Spanish Intermittent Treatment Trial (15, 16) and performed extensive clonal studies of plasma HIV RNA before cART, during strictly defined structured treatment interruptions (STI) and during the subsequent prolonged treatment interruption. To our knowledge, a longitudinal study of this type entailing multiple planned STIs has not been achieved up to now. We specifically aimed to ascertain the origin of rebounding virus based on phylogeny: (i) If rebounding virus stems from low-level replication, one expects gradual and continuous evolution, similar to what is observed in viremic patients over time. (ii) If rebounding virus originates from reactivated, latently infected cells, one expects stochastic reappearance of phylogenetically distinct viruses.
Results
Phylogenetic Relationship of Rebounding Plasma Virus.
To study characteristics of replicating HIV plasma virus, a total of 1,753 clonal sequences of the gp120 C2-V3-C3 region were generated from 20 HIV-1-infected patients over a median of 6.5 years (range 3.3–8.6) by using an established RT-PCR and cloning strategy (28). Cross-contamination between subjects and contamination with laboratory strains could be excluded by highly specific clustering of individual patient sequences in a phylogenetic reconstruction including all sequences from our study subjects (29). Analyses using the GARD genetic algorithm failed to show any evidence of intrapatient recombination. All patients had enrolled in the Swiss HIV Cohort Study while untreated. Subsequently they received cART for a median of 2.7 years (range 1.5–3.4), before participating in a treatment interruption trial (Swiss Spanish Treatment Interruption Trial) (15, 16). Neighbor-joining phylogenetic trees of pretreatment sequences and of each set of STI-derived clones within each patient are shown in supporting information (SI) Fig. S1. Phylogenetic trees were also generated for each patient by using the maximum-likelihood method. Individual trees including clonal sequences from the pretreatment and from the latest time point after the prolonged treatment interruption are depicted in Fig. S2. Clearly distinct clades were observed in most patients. However, in a few subjects (e.g., 111, 112, and 127), the virus populations isolated at the different time points appeared to be overlapping. Cladistic evaluation of interpopulation gene flow (by Slatkin–Maddison test) showed statistically significant genetic segregation between pretherapy and postinterruption viral populations in all patients except 118. Each sequence can theoretically be tracked to a most recent common ancestral sequence (MRCA), which is represented by the most distal node in a phylogenetic tree, common to all sequences from an individual patient (30–32). Pretreatment sequences were closest to the MRCA in 15 of the 20 patients, and sequences obtained years later after the last treatment interruption exhibited the highest distance to the MRCA in 13 patients (Fig. 1Left). In contrast, a totally different picture was found if maximum likelihood trees were generated for nine patients also including the clonal sequences obtained during the structured treatment interruptions and frequently thereafter (Fig. 1 Right and Fig. S3). In 8 of these 9 patients, sequences obtained during short treatment interruptions (n = 2) or obtained during the prolonged treatment interruption (n = 6) were genetically closest to the MRCA. Thus, the continuous relationship between time of sampling and structure of the phylogenetic tree was lost, suggesting that rebounding virus was phylogenetically older than virus studied at the pretreatment time point. This finding argues against the possibility that rebounding virus during STI originates from viral populations undergoing low-level persistent replication during suppressive therapy.
Fig. 1.
Time relationship of rebound sequences to the MRCA. (Left) Results from phylogenetic trees (Fig. S2) containing the clonal sequences from pretreatment (pre) and from late time point after treatment stop (post). For each patient, the MRCA was inferred, and sampling times of the clones with the highest and lowest MRCA distance are indicated. (Right) Analysis of trees obtained from nine patients who were sampled intensively (Fig. S3) including also the clonal sequences obtained during the structured treatment interruptions (STI) and frequently thereafter. (Two identical clones from different time points had the highest distance to the MRCA in patient 106.)
Changes of Viral Diversity Over Time.
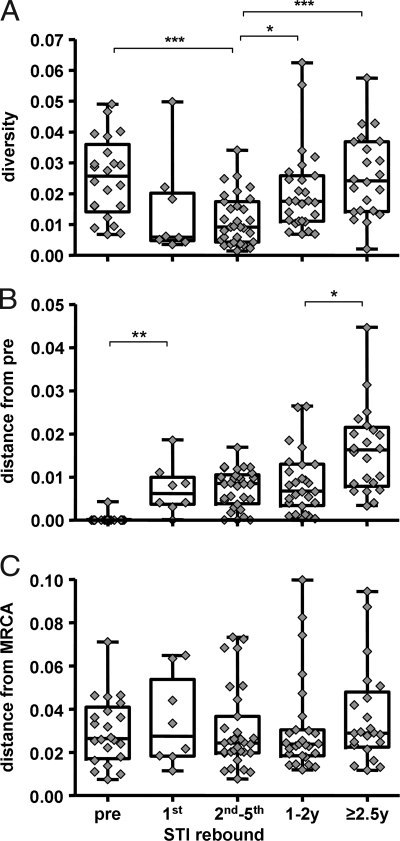
To date, it is not known to what extent viral diversity may change during treatment interruptions when compared with pretreatment viral diversity. For this reason, we calculated viral diversity of clonal sequences of the C2-V3-C3 region longitudinally, at pretreatment, during the four STIs and sequentially thereafter (Fig. 2A). There was a sharp drop in diversity within the first STI (week 2; mean ± SD 1.44 ± 1.59%). When comparing diversities of the second to the fifth viral rebound, diversity remained significantly lower than pretreatment levels (1.15 ± 0.81% versus 2.51 ± 1.27%; P < 0.001). Diversity increased again after 1–2 years to a mean of 2.05 ± 1.35% (P = 0.024) and only reached pretreatment diversity levels with a delay of 2.5 years or later (2.57 ± 1.34%). In general, during the short 2-week STIs, relatively homogenous virus populations emerged, suggesting mono-, or oligoclonal origin. Interestingly, however, rebounding virus often was phylogenetically distinct from one interruption to the next in a given patient (e.g., patients 120, 124, and 109 first rebound; individual trees are shown in Fig. S3). These observations reinforce the hypothesis that reactivation of latent cells is the origin of viral rebound rather than persistent low-level replication.
Fig. 2.
Changes over time in viral diversity of clonal C2-V3-C3 sequences (A), average genetic distances (net divergence) from the pretreatment time point (B), and of average branch lengths to the MRCA (C) from pretreatment to the first, second to fifth interruption, 1–2 year time points, and to time points later than 2.5 years estimated by Tamura-Nei six-parameter model. Asterisks indicate the level of statistical significance (*, P < 0.05; **, < 0.01; ***, < 0.001; Mann–Whitney test with Bonferroni correction for multiple comparisons).
Evolution of Genetic Pairwise Distances.
To further quantify potential viral evolution in a time-dependent manner, we generated pairwise distances from the pretreatment time point to the first, second, third, fourth, and fifth interruption and furthermore to the 1- to 2-year time points and to time points later than 2.5 years. There was a significant increase of pairwise distance at the first interruption (mean distance 0.72 ± 0.58%; P = 0.006; Fig. 2B). Thereafter, the mean distance remained relatively stable over the remaining short-term STIs (0.73 ± 0.44%) and up to the year 1–2 time points (0.85 ± 0.71%) and only thereafter increased to 1.61 ± 0.98% after 2.5 years or later (P = 0.028). The inferred net divergence of intraindividual sequences from the first (pretreatment) and last time points sampled of 1.6% (range 0.1–4.5%) together with antiretroviral drug-free intervals of 3.2 years (range: 0.3–4.2) corresponds to an average increase in viral population divergence of 0.8% per year during the period without antiretroviral therapy. This is in agreement with previously reported divergence rates of 1% per year (33). Evolutionary rates were, on average, 1% per year, which is also in line with earlier reports (33). To further characterize whether significantly increased pairwise genetic distances observed during the short treatment interruptions reflect forward evolution or just mirror rebounding viruses of different phylogeny, we further performed an analysis calculating genetic distances from clones obtained at the different time points with regard to their relationship to the MRCA.
Evolution of Genetic Distances to the MRCA.
According to the coalescence theory (30–32), the branch length to the MRCA is proportional to the number of elapsed generations. Thus, if significant evolution has occurred between time points, one would expect a significant difference in their respective branch lengths to the MRCA. As shown in Fig. 2C, mean distances to the MRCA to any studied time point remained remarkably stable at 0.032 ± 0.020 (average values 0.030, 0.034, 0.030, 0.031, and 0.036) over the whole time period observed. Thus, with regard to the MRCA, no significant evolution could be detected between any investigated time points. This is in agreement with the hypothesis that the majority of viral variants rebounding during STI and of circulating plasma viruses sampled after treatment was stopped were already present before the initiation of cART. However, it must be noted that the variability of the observed branch lengths to the MRCA was relatively high in comparison with the average distance itself. Moreover, it is not completely certain that the MRCA based on coalescent methodology always captures the correct root. Although the external sequences included in the phylogenies as outgroup evolved very early in the global HIV epidemic, a possible influence on the MRCA position relative to the more recently evolved patient sequences cannot be excluded.
Evolution of Coreceptor Usage Phenotype.
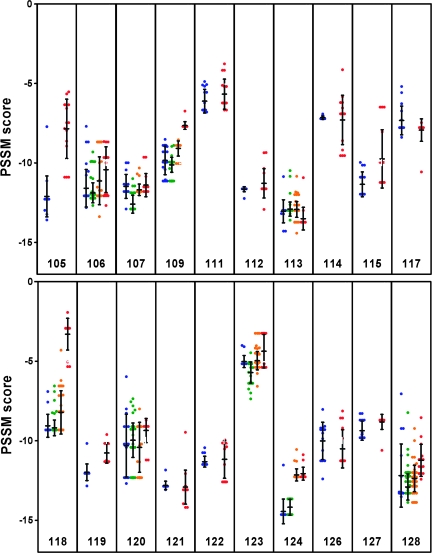
We have previously shown that no switch from R5 to X4 viruses during STIs was detectable by using a phenotypic coculture-based assay (34). Here, we generated position-specific scoring matrix (PSSM) scores (35) longitudinally for all clonal sequences available of our 20 patients (Fig. 3). These analyses confirmed our previous findings that no R5-to-X4 switch took place during STIs. However, patient 118 showed an increase of the average score from −9.1 (range −9.3 to −7.2) at 18 months after entering SSITT to −3.3 (range −5.3 to −1.9) 4 years after SSITT. This patient concurrently showed a remarkable increase of plasma viremia from 5,864 to 102,000 copies/ml (1.5 and 4.2 years after first STI, respectively) and had to initiate treatment again 4.2 years after SSITT because his CD4 count fell to <300 cells per microliter. When longitudinal PSSM scores from 20 patients were studied, we found a significant (P < 0.01) average increase of at least one point in seven patients. In addition, we found a parallel overall increase in plasma viral load, suggesting that increasing affinity for the CXCR4 coreceptor may reflect subtle clinical progression of the disease. Furthermore, these findings highlight that the switch from R5 to X4 is a gradual process that may occur during prolonged treatment interruption rather than a rapid “on/off” phenomenon.
Fig. 3.
Predictions of coreceptor use in the aligned V3 loop amino acid sequences by position-specific scoring matrix (PSSM, subtype B SI/NSI matrix). Scores of individual clones are shown longitudinally from pretreatment (blue dots), during the short STI (green) to the last time point (red). Scores above approximately −2 are predictive of X4 coreceptor phenotype. Bars denote mean and SD.
Discussion
Our results reveal four major findings: (i) We show that HIV rebounding after STI in patients with prolonged suppression to <50 copies per milliliter was often phylogenetically closer to the MRCA than pretreatment viruses. (ii) Despite total observational periods of 4–8 years, we found no temporal structure of rebounding virus in regard to pretreatment sequences in the assembled phylogenies. Clonal sequences obtained during different STIs often clustered with different pretreatment sequences. This implies that the rebounding virus populations emerged by expansion of distinct subclades preexisiting in the pretreatment samples. These observations suggest that the rebounding virus originates from reactivation of latently infected cells rather than from viral lineages continuously replicating at low levels. (iii) We demonstrate that viral diversity during STI was very low compared with pretreatment diversity and remained so for a prolonged period, reflecting a sustained evolutionary bottleneck introduced by suppressive cART. (iv) STIs did not provoke a switch in coreceptor usage; however, overall predicted CXCR4 usage gradually increased in one-third of the patients, suggesting that evolution of coreceptor phenotype is continuous rather than an on/off phenomenon. Overall, these observations suggest strong and surprisingly lasting effects of cART and STI on HIV population genetics.
Over the last decade, a vibrant debate has focused on whether low-level replication continues in patients with viral loads <50 copies per milliliter or not. Some have argued that persistent spliced and unspliced cellular HIV RNA, persistent episomal HIV DNA, and persistent or intermittently detectable HIV plasma RNA are signs of ongoing low-level HIV replication (36–38). However, it has been shown by several groups now that neither episomal HIV DNA (39–42) nor persistence of cellular transcripts (43–46) or low-level plasma viremia (11, 12, 47, 48) are reliable markers for the detection of ongoing low-level replication. Another argument against continuous ongoing low-level replication comes from a lack of viral evolution in the latent reservoir and in lymphoid organs, when patients with <50 copies per milliliter are studied (4, 7, 8, 49). Furthermore, on a population level, the lack of emergence of drug-resistance mutations in patients with <50 copies per milliliter in the long term also strongly argues against ongoing replication (von Wyl V, et al. 15th Conference on Retroviruses and Opportunistic Infections, February 3–6, 2008, Boston, Abstract 896). Taken together, these studies underline the difficulty in proving or disproving the existence of low-level replication.
Here, we took advantage of a well controlled structured treatment-interruption trial in patients who had sustained suppression of plasma viremia to <50 copies per milliliter before treatment interruption and performed longitudinal clonal sequencing of the envelope C2-V3-C3 domain to look for evidence that low-level replication during suppressive cART is the source of rebounding virus upon treatment interruption and to examine the overall effects of punctuated cART on intrapatient HIV population genetics.
Several earlier studies had already attempted to characterize rebounding plasma virus after discontinuation of highly active antiretroviral therapy. Chun et al. (24) analyzed the patterns of HIV env heteroduplexes, and Zhang et al. (25) and Martinez Picado et al. (26) examined the length polymorphisms of the env variable loops V1-V2 and V4-V5 to detect differences between rebounding plasma virus and those isolated from tissue reservoirs. Phylogenetic analyses of clonal sequences isolated from virion RNA as well as cell-associated HIV RNA and DNA were also performed in three patients each by Imamichi et al. (50) and Martinez Picado et al. (26). In aggregate, these investigators found evidence for both reactivation of virus from the latent reservoir and ongoing low-level replication of HIV in tissue compartments.
Genetic evolution and increasing diversity are characteristics of HIV replication (51–53). Thus, if low-level viral replication occurred continuously, one would expect that rebounding virus reflected genetic shifts resulting from the numerous replication cycles that had occurred despite treatment (30). During successful cART, viral replication may be at such low levels that immune responses lack adequate stimulation to maintain good response (54, 55) and so would exert less selective pressure. Minimal viral replication in the absence of significant immune selection pressure might therefore be associated with limited numbers of adaptive mutations. Nonetheless, one might still expect temporal structure in the data because of continual accumulation of synonymous substitutions. In the absence of low-level replication, one would expect to see rebounding virus with genetic composition similar to pretreatment viruses or, potentially, to ancestral viruses because of reactivation of older, latently infected cells. Interestingly, in eight of nine patients in whom we performed extensive longitudinal env cloning studies, we found that viruses closest to the MRCA did not originate from pretreatment clones but from clones derived from rebounding viruses. The striking lack of a temporal relationship between rebounding virus and pretreatment viruses strongly suggests that rebounding virus originates from reactivated, latently infected cells rather than from a cellular pool or compartment engaged in low-level viral replication. Further support for this interpretation comes from the fact that viruses varied between different treatment stops. From one interruption to the next, distinct viral populations could emerge, again suggesting that distinct clones of latently infected cells were reactivated upon stopping cART. Another argument favoring the reactivation hypothesis is the lack of evolution from pretreatment viruses to rebounding viruses. If low-level replication occurred despite viral suppression, one would expect a higher genetic distance to the MRCA from rebounding viruses when compared with pretreatment viruses. This was not observed in our patients, nor were the genetic distances of rebounding virus to the individual MRCA associated with the duration of treatment before STI. Sequences from pretreatment and from the latest time points >2.5 years after start of STI were often situated on very separate branches of the phylogenetic trees. This may account for a large distance between the two but a similar distance to the MRCA. The late samples at >2.5 years exhibited increasing divergence from pretreatment variants (Fig. 2B) but remained equidistant from the MRCA (Fig. 2C). This seemingly paradoxical observation also suggests that stochastic reactivation of minor virus populations instead of slow within-host replication had influenced the structural relationship of the observed phylogeny.
Some additional unexpected results merit mentioning. When compared with pretreatment plasma virus, viral diversity sharply dropped to very low levels in most patients during the first treatment interruption. This is evidence that viral rebound after suppression to <50 copies per milliliter is mono- or oligoclonal. Even more striking: It took >2 years after STI to reach the pretreatment levels of viral diversity. This viral-rebound pattern in chronic infection resembles in many ways the situation found in acute HIV infection, where the recipient is also infected by mono-or oligoclonal HIV, and viral diversity then increases on average by 1% per year (33). Similar situations in chronically infected patients upon stopping cART were unexpected and demonstrate that a genetic bottleneck, introduced by full viral suppression can result in prolonged effects on viral-population structure. It is possible that persisting neutralizing antibody responses and HIV-specific CTL responses raised earlier against pretreatment viruses led to purifying selection of viruses, and these immune pressures were no longer present at the time of rebound. This phenomenon could also explain the emergence of viruses during STIs that, in many cases, were closer to the MRCA than the pretreatment viruses, suggesting that they went into latency at an earlier stage. At the time of rebound, immune responses against these early viruses may already have decreased more than those against the pretreatment viruses that were present at high levels before treatment.
In summary, the finding of homogeneous HIV populations during short structured treatment interruptions implies mono- or oligoclonal origin of the rebounding virus. Expansion of distinct lineages at different STIs suggests stochastic reactivation of different clones of long-lived latently infected cells rather than expansion of populations of low-level replicating virus. A prolonged delay in restoration of pretreatment viral diversity after treatment interruption demonstrates a surprisingly sustained evolutionary bottleneck induced by punctuated antiretroviral therapy. These data suggest that cART introduced significant effects on further evolution of the viral population.
Materials and Methods
Study Subjects.
We included all patients who were enrolled into the Swiss-Spanish Intermittent Treatment Trial (SSITT) at the University Hospital Zürich, provided that a pretreatment plasma sample was available and PCR amplification was successful. Two patients were excluded because they did not complete the SSITT trial. Patients had undetectable viral loads (<50 copies per milliliter) for at least 6 months and then underwent four consecutive STI cycles (2 weeks off and 8 weeks on treatment), followed by a longer fifth treatment interruption of indefinite duration. Nine patients were sampled intensively. The detailed patient characteristics with identical patient identification numbers have been reported previously (17, 28, 29, 34, 40, 44, 56). Written informed consent was obtained from all patients according to the guidelines of the Ethics Committee of the University Hospital Zurich.
Sequencing and Phylogenetic Analyses.
RNA extraction from plasma and subsequent PCR amplification, molecular cloning, and bidirectional sequencing of the C2–C3 region of HIV-1 env was performed as described (28). To test the representativity of the isolated virus populations, we compared the results of our cloning strategy with those of limiting dilution analysis in two patient samples. A total of 67 clonal env C2-V3-C3 sequences was examined. All viral quasispecies identified by maximum-likelihood analysis after terminal-dilution cloning were also detected by our molecular cloning method (Table S1). Conversely, four minority species were not found by limiting dilution. This indicates that our method was not excessively affected by resampling bias. Approximately 16 individual clones were obtained at each time point. Lasergene software version 5.08 (DNASTAR) was used for editing and alignment. Intrapatient recombination was tested by GARD (Genetic Algorithm for Recombination Detection), using the single breakpoint-detection method (57). These analyses failed to show evidence of recombination in sequences from any of the 20 individuals in this study. No putative breakpoints were identified, and a single phylogeny rather than multiple segment-specific phylogenies best described the data in all cases (Akaike information criterion scores were not improved with multiple trees). Genetic distances were estimated by the Tamura–Nei six-parameter model using MEGA version 4 (58). Tajima's D tests performed in MEGA4 and DnaSP (59) revealed that sequences from nearly all individuals in our study were evolving neutrally (evidence for purifying selection without appreciable impact on coalescence times was found in subjects 112 and 117). Neighbor-joining phylogenetic trees were inferred by MEGA 4. Maximum-likelihood phylogenetic trees were inferred by DNAML run on a Unix system of the University of Zurich by using randomized input order, global rearrangements, and multiple jumble options (PHYLIP Phylogeny Inference Package version 3.6 distributed by J. Felsenstein, Department of Genetics, University of Washington, Seattle). The Slatkin–Maddison test (60), a cladistic measure of interpopulation gene flow, was performed to evaluate degree of viral population shift between time points. The following outgroup sequences were included in the analysis: HIV-1 subtype B isolates HXB2 (GenBank accession no. K03455), ADA (M60472), YU2 (M93258), BAL (M68893), JRFL (U63632), NL4–3 (U26942), subtype A isolate U455 (M62320), subtype C isolate ETH2220 (U46016), subtype D isolate ELI (K03454), subtype F isolate BR020 (AF005494), and the HIV group M envelope fragment ZR59a-2 (AF030527) isolated from a historical plasma sample collected in 1959 (61).
Coreceptor Usage Prediction.
Predictions of coreceptor use were made by scoring the V3 amino acid sequences according to position-specific scoring matrices trained on the syncytium-inducing phenotype (PSSM SI/NSI) and on the X4 or R5 coreceptor phenotype (PSSM X4/R5) (35) (software available at http://ubik.microbiol.washington.edu). This method derives a score by using precalculated matrices for the 35 aa forming the V3 loop. Two different alignments were generated for V3 sequences consisting of only 34 aa; in these cases, the resulting slightly different PSSM score values were averaged. In addition, we used a support vector machine-based method (WetCat algorithm) to classify the aligned V3 loop sequences according to coreceptor phenotype (62). In general, this algorithm and the subtype B matrices X4R5 and SINSI yielded comparable results. CXCR4-using virus was sporadically detected with each method, but no clear switch in coreceptor usage was found in the combined analysis. Our longitudinal analysis of coreceptor usage relied on the SINSI scores. One sequence was excluded because it contained a larger gap in the V3 loop.
Statistical Analyses.
Standard statistical analyses were performed by using GraphPad Prism version 5. The two-sided Mann–Whitney test was used to compare average distances. Bonferroni correction for multiple comparisons was carried out to detect type I error. Two-sided Fisher's exact test was used for 2 × 2 contingency tables.
Supplementary Material
Acknowledgments.
We thank the participating patients for their commitment; Christine Schneider, Christina Grube, and Roland Hafner for excellent patient care; Esther Beerli, Friederike Burgener, Christine Leemann, Tuyet Trinh Lu, Barbara Niederöst, and Bärbel Sauer for laboratory support; and Ingrid Nievergelt and Christine Vögtli for administrative assistance. This work was supported by Swiss National Science Foundation (SNF) Grants 3345-65168 and 3100AO-103748/1 (to H.F.G. and A.T.), PP00B-102647 (to A.T.), and 320000-116035 (to H.F.G.); the Swiss HIV Cohort Study (SHCS) (SNF Grant 33CSC0-10878), SHCS Project No. 290 (to H.F.G.); a grant from the EMDO Foundation (to H.F.G.); and a research grant from the Kanton of Zürich. A.T. is the Elizabeth Glaser Scientist supported by the Elizabeth Glaser Pediatric AIDS Foundation. J.K.W. was supported by National Institutes of Health Grant R01 NS51132.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF424791–EF426096, DQ002058–DQ002345, AY656534–AY656549, AY656440–AY656471, AY656249–AY656312, AY375663–AY375678, AY375616–AY375630, and AY375568–AY375583).
eUniversity Hospital, CH-4031 Basel, Switzerland;
fRegional Hospital, CH-6903 Lugano, Switzerland;
gNational Center for Retroviruses, CH-8028 Zurich, Switzerland;
hUniversity Hospital, CH-1011 Lausanne, Switzerland;
iUniversity Hospital and
jDepartment of Sociology, University of Geneva, CH-1211 Geneva, Switzerland;
kUniversity Hospital, CH-8091 Zurich, Switzerland;
lDepartment of Social and Preventive Medicine,
mUniversity Hospital, and
nInstitute of Infectious Diseases, University of Bern, CH-3012 Bern, Switzerland;
oInstitute of Medical Microbiology, University of Basel, CH-4051 Basel, Switzerland;
pCantonal Hospital, CH-9007 St. Gallen, Switzerland;
qChildren's Hospital, CH-9007 St. Gallen, Switzerland;
rCantonal Institute of Microbiology, CH-6500 Bellinzona, Switzerland;
sUniversity Children's Hospital, CH-8032 Zurich, Switzerland;
tInstitute of Social and Preventive Medicine, University of Lausanne, CH-1010 Lausanne, Switzerland;
uUniversity Children's Hospital, CH-4005 Basel, Switzerland; and
vInstitute for Clinical Microbiology and Immunology, CH-9000 St. Gallen, Switzerland.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804192105/DCSupplemental.
References
- 1.Egger M, et al. Impact of new antiretroviral combination therapies in HIV infected patients in Switzerland: prospective multicentre study. Swiss HIV Cohort Study. Br Med J. 1997;315:1194–1199. doi: 10.1136/bmj.315.7117.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palella FJ, Jr, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 3.Walmsley S, et al. Lopinavir-ritonavir versus nelfinavir for the initial treatment of HIV infection. N Engl J Med. 2002;346:2039–2046. doi: 10.1056/NEJMoa012354. [DOI] [PubMed] [Google Scholar]
- 4.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 5.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Chun TW, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 7.Strain MC, et al. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: Intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci USA. 2003;100:4819–4824. doi: 10.1073/pnas.0736332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunthard HF, et al. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J Virol. 1999;73:9404–9412. doi: 10.1128/jvi.73.11.9404-9412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havlir DV, et al. Productive infection maintains a dynamic steady state of residual viremia in human immunodeficiency virus type 1-infected persons treated with suppressive antiretroviral therapy for five years. J Virol. 2003;77:11212–11219. doi: 10.1128/JVI.77.20.11212-11219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strain MC, et al. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J Infect Dis. 2005;191:1410–1418. doi: 10.1086/428777. [DOI] [PubMed] [Google Scholar]
- 11.Havlir DV, et al. Prevalence and predictive value of intermittent viremia with combination HIV therapy. J Am Med Assoc. 2001;286:171–179. doi: 10.1001/jama.286.2.171. [DOI] [PubMed] [Google Scholar]
- 12.Greub G, et al. Intermittent and sustained low-level HIV viral rebound in patients receiving potent antiretroviral therapy. AIDS. 2002;16:1967–1969. doi: 10.1097/00002030-200209270-00017. [DOI] [PubMed] [Google Scholar]
- 13.Wong JK, et al. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DM, et al. Long-term persistence of transmitted HIV drug resistance in male genital tract secretions: implications for secondary transmission. J Infect Dis. 2007;196:356–360. doi: 10.1086/519164. [DOI] [PubMed] [Google Scholar]
- 15.Fagard C, et al. A prospective trial of structured treatment interruptions in human immunodeficiency virus infection. Arch Intern Med. 2003;163:1220–1226. doi: 10.1001/archinte.163.10.1220. [DOI] [PubMed] [Google Scholar]
- 16.Oxenius A, et al. Stimulation of HIV-specific cellular immunity by structured treatment interruption fails to enhance viral control in chronic HIV infection. Proc Natl Acad Sci USA. 2002;99:13747–13752. doi: 10.1073/pnas.202372199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer M, et al. HIV RNA in plasma rebounds within days during structured treatment interruptions. AIDS. 2003;17:195–199. doi: 10.1097/00002030-200301240-00009. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz L, et al. HIV dynamics and T-cell immunity after three structured treatment interruptions in chronic HIV-1 infection. AIDS. 2001;15:F19–27. doi: 10.1097/00002030-200106150-00001. [DOI] [PubMed] [Google Scholar]
- 19.Goujard C, et al. Interruption of antiretroviral therapy initiated during primary HIV-1 infection: Impact of a therapeutic vaccination strategy combined with interleukin (IL)-2 compared with IL-2 alone in the ANRS 095 Randomized Study. AIDS Res Hum Retroviruses. 2007;23:1105–1113. doi: 10.1089/aid.2007.0047. [DOI] [PubMed] [Google Scholar]
- 20.Hecht FM, et al. A multicenter observational study of the potential benefits of initiating combination antiretroviral therapy during acute HIV infection. J Infect Dis. 2006;194:725–733. doi: 10.1086/506616. [DOI] [PubMed] [Google Scholar]
- 21.Streeck H, et al. Immunological and virological impact of highly active antiretroviral therapy initiated during acute HIV-1 infection. J Infect Dis. 2006;194:734–739. doi: 10.1086/503811. [DOI] [PubMed] [Google Scholar]
- 22.Smith DE, Walker BD, Cooper DA, Rosenberg ES, Kaldor JM. Is antiretroviral treatment of primary HIV infection clinically justified on the basis of current evidence? AIDS. 2004;18:709–718. doi: 10.1097/00002030-200403260-00001. [DOI] [PubMed] [Google Scholar]
- 23.Kaufmann DE, et al. Limited durability of viral control following treated acute HIV infection. PLoS Med. 2004;1:e36. doi: 10.1371/journal.pmed.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chun TW, et al. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med. 2000;6:757–761. doi: 10.1038/77481. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, et al. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J Clin Invest. 2000;106:839–845. doi: 10.1172/JCI10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Picado J, et al. Viral evolution during structured treatment interruptions in chronically human immunodeficiency virus-infected individuals. J Virol. 2002;76:12344–12348. doi: 10.1128/JVI.76.23.12344-12348.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey JR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joos B, et al. Low human immunodeficiency virus envelope diversity correlates with low in vitro replication capacity and predicts spontaneous control of plasma viremia after treatment interruptions. J Virol. 2005;79:9026–9037. doi: 10.1128/JVI.79.14.9026-9037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joos B, et al. Positive in vivo selection of the HIV-1 envelope protein gp120 occurs at surface-exposed regions. J Infect Dis. 2007;196:313–320. doi: 10.1086/518935. [DOI] [PubMed] [Google Scholar]
- 30.Kingman JFC. On the Genealogy of Large Populations. J Appl Prob. 1982;19:27–43. [Google Scholar]
- 31.Hudson RR. Properties of a neutral allele model with intragenic recombination. Theor Popul Biol. 1983;23:183–201. doi: 10.1016/0040-5809(83)90013-8. [DOI] [PubMed] [Google Scholar]
- 32.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shankarappa R, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trkola A, et al. Human immunodeficiency virus type 1 fitness is a determining factor in viral rebound and set point in chronic infection. J Virol. 2003;77:13146–13155. doi: 10.1128/JVI.77.24.13146-13155.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen MA, et al. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, et al. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 37.Sharkey M, Triques K, Kuritzkes DR, Stevenson M. In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J Virol. 2005;79:5203–5210. doi: 10.1128/JVI.79.8.5203-5210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pomerantz RJ. Residual HIV-1 infection during antiretroviral therapy: The challenge of viral persistence. AIDS. 2001;15:1201–1211. doi: 10.1097/00002030-200107060-00002. [DOI] [PubMed] [Google Scholar]
- 39.Bushman F. Measuring covert HIV replication during HAART: The abundance of 2-LTR circles is not a reliable marker. AIDS. 2003;17:749–750. doi: 10.1097/01.aids.0000050868.71999.09. [DOI] [PubMed] [Google Scholar]
- 40.Fischer M, et al. Shifts in cell-associated HIV-1 RNA but not in episomal HIV-1 DNA correlate with new cycles of HIV-1 infection in vivo. Antiviral Ther. 2003;8:97–104. [PubMed] [Google Scholar]
- 41.Brussel A, et al. Longitudinal monitoring of 2-long terminal repeat circles in peripheral blood mononuclear cells from patients with chronic HIV-1 infection. AIDS. 2003;17:645–652. doi: 10.1097/00002030-200303280-00001. [DOI] [PubMed] [Google Scholar]
- 42.Pierson TC, et al. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J Virol. 2002;76:4138–4144. doi: 10.1128/JVI.76.8.4138-4144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer M, et al. Attenuated and nonproductive viral transcription in the lymphatic tissue of HIV-1-infected patients receiving potent antiretroviral therapy. J Infect Dis. 2004;189:273–285. doi: 10.1086/380797. [DOI] [PubMed] [Google Scholar]
- 44.Fischer M, et al. Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J Infect Dis. 2004;190:1979–1988. doi: 10.1086/425983. [DOI] [PubMed] [Google Scholar]
- 45.Kaiser P, et al. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol. 2007;81:9693–9706. doi: 10.1128/JVI.00492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frenkel LM, et al. Multiple viral genetic analyses detect low-level human immunodeficiency virus type 1 replication during effective highly active antiretroviral therapy. J Virol. 2003;77:5721–5730. doi: 10.1128/JVI.77.10.5721-5730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palmer S, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci USA. 2008;105:3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunthard HF, et al. Residual human immunodeficiency virus (HIV) Type 1 RNA and DNA in lymph nodes and HIV RNA in genital secretions and in cerebrospinal fluid after suppression of viremia for 2 years. J Infect Dis. 2001;183:1318–1327. doi: 10.1086/319864. [DOI] [PubMed] [Google Scholar]
- 50.Imamichi H, et al. Human immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J Infect Dis. 2001;183:36–50. doi: 10.1086/317641. [DOI] [PubMed] [Google Scholar]
- 51.Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 52.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 53.Ho DD, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 54.Gray CM, et al. Frequency of class I HLA-restricted anti-HIV CD8+ T cells in individuals receiving highly active antiretroviral therapy (HAART) J Immunol. 1999;162:1780–1788. [PubMed] [Google Scholar]
- 55.Frahm N, et al. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004;78:2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metzner KJ, et al. Emergence of minor populations of human immunodeficiency virus type 1 carrying the M184V and L90M mutations in subjects undergoing structured treatment interruptions. J Infect Dis. 2003;188:1433–1443. doi: 10.1086/379215. [DOI] [PubMed] [Google Scholar]
- 57.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: A genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 58.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 59.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 60.Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu T, et al. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 62.Pillai S, Good B, Richman D, Corbeil J. A new perspective on V3 phenotype prediction. AIDS Res Hum Retroviruses. 2003;19:145–149. doi: 10.1089/088922203762688658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.