Abstract
The cell envelope (CE) is a specialized structure that is important for barrier function in terminally differentiated stratified squamous epithelia. The CE is formed inside the plasma membrane and becomes insoluble as a result of cross-linking of constituent proteins by isopeptide bonds formed by transglutaminases. To investigate the earliest stages of assembly of the CE, we have studied human epidermal keratinocytes induced to terminally differentiate in submerged liquid culture as a model system for epithelia in general. CEs were harvested from 2-, 3-, 5-, or 7-d cultured cells and examined by 1) immunogold electron microscopy using antibodies to known CE or other junctional proteins and 2) amino acid sequencing of cross-linked peptides derived by proteolysis of CEs. Our data document that CE assembly is initiated along the plasma membrane between desmosomes by head-to-tail and head-to-head cross-linking of involucrin to itself and to envoplakin and perhaps periplakin. Essentially only one lysine and two glutamine residues of involucrin and two glutamines of envoplakin were used initially. In CEs of 3-d cultured cells, involucrin, envoplakin, and small proline-rich proteins were physically located at desmosomes and had become cross-linked to desmoplakin, and in 5-d CEs, these three proteins had formed a continuous layer extending uniformly along the cell periphery. By this time >15 residues of involucrin were used for cross-linking. The CEs of 7-d cells contain significant amounts of the protein loricrin, typically expressed at a later stage of CE assembly. Together, these data stress the importance of juxtaposition of membranes, transglutaminases, and involucrin and envoplakin in the initiation of CE assembly of stratified squamous epithelia.
INTRODUCTION
One important function of stratified squamous epithelia is to provide a physical barrier against the environment and protection for the tissues internal to them. Much of this barrier function is provided by a cell envelope (CE), which is a specialized structure formed just inside the plasma membrane as the cells terminally differentiate (Reichert et al., 1993; Simon, 1994; Nemes and Steinert, 1999). In all stratified squamous epithelia, the CE commonly consists of an ∼10-nm-thick (Jarnik et al., 1998) macromolecular assembly of highly insoluble proteins built from some or many of the following components (Steinert and Marekov, 1995, 1997; Steinert et al., 1998a; Robinson et al., 1997): annexin I, cystatin α, elafin, filaggrin, involucrin, loricrin, type II keratin intermediate filament proteins, pancornulins, small proline-rich proteins (or cornifins), trichohyalin, and various cell junctional proteins, including desmoplakin, envoplakin, and periplakin. These proteins become insoluble by cross-linking together by disulfide bonds and Nε-(γ-glutamyl)lysine isopeptide bonds formed by the action of transglutaminases (TGases) (Greenberg et al., 1991; Reichert et al., 1993; Simon, 1994; Melino et al., 1999; Nemes and Steinert, 1999). Note that in orthokeratinizing epithelia such as the epidermis, hair cuticle, or rodent forestomach, the CE is often termed the cornified cell envelope. In these cases, water barrier function is an additional function of the epithelium, which is contributed in part by an ∼5-nm-thick layer of lipids attached on the exterior of the CE (Swartzendruber et al., 1987; Wertz et al., 1989; Elias and Menon, 1991; Downing et al., 1993; Jackson and Elias, 1993; Zahn and Gattner, 1997; Marekov and Steinert, 1998; Nemes et al., 1999a).
From a practical experimental perspective, the CE is operationally defined as that which remains insoluble after exhaustive extraction with denaturants and reducing agents (Steinert, 1995; Steinert and Marekov, 1995, 1997). However, because the density of isolated CEs is less than might be expected (∼0.7 g/cm3; Jarnik et al., 1998), it is conceivable that additional components including calcium-binding proteins or other cell peripheral proteins might be associated with CEs in vivo and that may be lost by these isolation procedures (Robinson et al., 1997). Although most studies to date have been performed on CEs made by the epidermis, it has become clear that there are differences in the protein contents of the CEs formed by various epithelia and isolated this way. For example, CEs of the epidermis are enriched in glycine and serine, which we now know is due to the ∼80% content of loricrin (Steven and Steinert, 1994; Steinert, 1995; Steinert et al., 1998a; Jarnik et al., 1996, 1998). However, in a variety of hyperproliferative skin diseases, the glycine and serine content is much lower because of the diminished content of loricrin (Reichert et al., 1993). CEs of the hair cuticle are enriched in cysteine because of the presence of as yet uncharacterized sulfur-rich proteins (Zahn and Gattner, 1997). Also, it has been reported that CEs of internal epithelia or epidermal keratinocytes grown in culture are enriched with prolines because of a high content of various members of the small proline-rich family of CE proteins (Steven and Steinert, 1994; Jarnik et al., 1996). Thus all of these data suggest that the spectrum and amounts of proteins used to construct the CE vary in accordance with the particular barrier function requirements of the epithelium (Steinert et al., 1998a,b).
However, a large body of data suggests that virtually all stratified squamous epithelia express the protein involucrin. Indeed, several pieces of evidence suggest that involucrin is used early in the formation of CE structures. First, mathematical modeling of amino acid compositions of CEs recovered from a variety of sources has revealed that involucrin is commonly present in CEs (Steven and Steinert, 1994; Steinert, 1995; Jarnik et al., 1996). Second, several expression and labeling studies have revealed that involucrin is deposited at the cell periphery before other proteins such as loricrin (Rice and Green, 1977, 1979; Simon and Green, 1984, 1985, 1988; Yaffe et al., 1992, 1993; Crish et al., 1993; Hohl et al., 1995; Ishida-Yamamoto et al., 1996, 1997). Third, timed proteolysis experiments of foreskin epidermal CEs revealed that involucrin epitopes could be exposed, and involucrin-containing peptides were released, only after extended times of digestion after removal of the dense cytoplasmic overlayer of loricrin (Steinert, 1995). However, when bound ceramides on the exterior side were first removed by a saponification reaction, involucrin was exposed and could be readily released early during the digestion, leaving a loricrin polymer that was more resistant to proteolysis (Steinert and Marekov, 1997). Similarly, involucrin peptides were readily recoverable from the CEs from “immature” keratinocytes, which contained only limited amounts of loricrin (Steinert and Marekov, 1997). In addition, we have shown that involucrin is a major substrate for the covalent attachment of ceramide lipids (Marekov and Steinert, 1998). Together, these data suggest that involucrin must have been deposited in the intimate vicinity of the membrane during early stages of CE assembly and where it would be readily accessible for ceramide attachment. Finally, three of the major protein partners to which involucrin and/or lipids were attached include desmoplakin, the major cytoplasmic constituent of desmosomes, and two novel cell peripheral proteins, envoplakin and periplakin (Steinert and Marekov, 1997; Steinert et al., 1998a; Ruhrburg et al., 1996, 1997; Ruhrberg and Watt, 1997).
Nevertheless, important temporal and physical questions concerning how and where CE assembly is initiated remain to be addressed. In this study, we have generated CEs in cultured human epidermal keratinocytes as a model system for stratified squamous epithelia in general. We have used both immunogold labeling and sequencing of peptides to explore the earliest stages of CE assembly and then followed the progression and fates of accumulated cross-linked proteins as these cells terminally differentiate. Our new data document that an appropriate alignment of cellular membranes, TGase enzyme(s), and the structural proteins involucrin, envoplakin, and perhaps periplakin are involved in the initiation of CE assembly.
MATERIALS AND METHODS
Culturing of Normal Human Epidermal Keratinocytes (NHEK Cells) and Isolation of CE Fragments
Cryopreserved NHEK cells (Clonetics, San Diego, CA) were plated at a density of 5 × 103 cells/cm2 on calf skin collagen (Sigma, St. Louis, MO)-coated dishes in serum-free keratinocyte growth medium (Clonetics), supplemented with 60 μg/ml bovine pituitary extract, and in low Ca2+ (0.05 mM). After ∼3 d when they had reached >90% confluency, the cells were transferred to this medium containing high Ca2+ (0.6 mM) and 25 μg/ml calcium ionophore A23187 (Calbiochem, La Jolla, CA) to facilitate terminal differentiation (Kim et al., 1995; Candi et al., 1998a). Attached cells were recovered after 2, 3, 5, or 7 d. Also collected were detached cells sloughed into the medium between 6 and 7 d and pooled with the 7-d attached cells. To isolate CEs, these four batches of cells in two to four separate experiments were separately pelleted and washed in PBS and then extracted in boiling SDS buffer containing Tris-HCl, pH 8.8, 1 mM EDTA, and 10 mM dithiolthreitol. Samples were examined by Normarski optics to assess gross morphologies. Then the preparations were exhaustively extracted by sonication by repeated boiling in this buffer as described previously (Steven and Steinert, 1994; Steinert, 1995; Steinert and Marekov, 1995, 1997). The CE fragments were pelleted through 20% Ficoll in this buffer to remove adherent (that is, non–cross-linked) solubilized proteins and finally washed three times in PBS to remove most SDS. Typical yields of CEs per 100-mm dish of cultured keratinocytes were 2 d cells, 0.01 mg; 3 d cells, 0.15 mg; 5 d cells, 0.6 mg; and 7 d cells, 1.5 mg.
Immunogold Electron Microscopy
Pellets of CE fragments (0.1–1 mg) were treated for preimbedding, reacted with the several antibodies listed below, and then labeled with protein A-gold of diameter of 5, 10, or 15 nm (Mehrel et al., 1990; Steinert, 1995; Steinert and Marekov, 1997). Affinity-purified antibodies used were rabbit polyclonal anti-human keratin 1 (1:200 dilution) (Steinert and Marekov, 1997), rabbit polyclonal anti-human keratin 6 (Roop et al., 1984), rabbit polyclonal anti-human keratin 10 (1:200 dilution) (Roop et al., 1984), goat polyclonal anti-human loricrin (1:100) (Hohl et al., 1991), rabbit polyclonal anti-mouse small proline-rich protein 1/3 (SPR1/3; 1:500) (which cross-reacts with human proteins; Kartasova et al., 1996), mouse monoclonal anti-human involucrin (1:50) (Biomedical Technologies, Stoughton, MA), rabbit polyclonal anti-human desmoplakin (1:100) (a gift from Dr. R.D. Goldman, Northwestern University Medical School, Chicago, IL); rabbit polyclonal anti-rat plectin (1:50) (Sigma), mouse monoclonal AE-13 (a gift from Dr. T.-T. Sun, New York University, New York, NY), and mouse monoclonal anti-human annexin I and mouse monoclonal anti-human integrin β4 (1:100) (both from Life Technologies, Gaithersburg, MD). We also prepared rabbit polyclonal anti-human envoplakin and periplakin antibodies using as immunogens synthetic peptides of sequences Cys-Tyr-Arg-Ser-Ala-Ser-ProThr-Val-ProArg-Ser-Leu-Arg and Met-Ser-Ile-Gln-Glu-Leu-Ala-Val-Leu-Val-Ser-Gly-Gln-Lys, corresponding to their terminal residues (Ruhrburg et al., 1996) and (Ruhrburg et al., 1997), respectively. After affinity purification, both were used at 1:200 dilutions. To remove some apparent keratin epitopes, the anti-envoplakin antibody was further purified on a column coupled with the mixed keratins expressed in preconfluent epidermal keratinocytes grown in low-Ca2+ medium. Both antibodies recognized only single bands corresponding to their known sizes on Western blots of keratinocyte extracts. The numbers of gold particles per unit length were counted for ≥50 μm of both desmosomal or interdesmosomal sites in at least two different preparations of CEs of each period. In CEs from 7-d cells, the locations of the desmosomal remnants were often no longer discernible, but except for desmoplakin, most epitopes when present were uniformly distributed. In all cases, preimmune sera were used as antibody controls, and the numbers of gold particles were uniformly <1/μm.
Protein Chemistry Procedures
Protein and peptide amounts were quantitated by amino acid analysis after acid hydrolysis (110°C, in vacuo for 22 h). Mathematical modeling estimates of the amounts of various proteins present in CE preparations based on amino acid compositions were calculated exactly as before (Steven and Steinert, 1994; Steinert, 1995). In the case of 2-d CEs, to rationalize the calculated values, it was necessary to assign a substantial fraction (14%) to a GenBank average amino acid composition. Because of their generally very similar compositions in the quantitatively major amino acids, we were unable to reliably separately estimate the amounts for desmoplakin and envoplakin. Accordingly, we used a combined average composition termed “plakins.” Isodipeptide amounts were quantitated by amino acid analysis after total proteolytic digestion (Hohl et al., 1991). Aliquots of CEs from 3- and 6- to 7-d cultured cells were resuspended in 0.1 M N-ethylmorpholine-acetate, pH 8.3, and digested at 37°C with 1% (wt) trypsin (Sigma, sequencing grade) for 2–6 h. The released solubilized peptides were freed of undigested material by centrifugation at 14,000 × g, dried, and redissolved in water containing 10% acetonitrile and 0.09% trifluoroacetic acid. They were then resolved by HPLC using a 10–90% acetonitrile gradient over a 120-min period exactly as before (Steinert and Marekov, 1995, 1997). Empirically, we found that <5% of the isopeptide cross-link of the tryptic solubilized material before fractionation was present in peptides that eluted with <25% acetonitrile before 45 min; >95% could be accounted for in discrete peptide peaks that eluted later in the gradient. Thus all those peptide peaks that eluted after 45 min were collected, dried, and subjected to microsequencing as before after covalent attachment onto a solid support. Most such peptides contained two or more sequences adjoined by one or more cross-links. Because most sequences were derived from readily recognizable known CE proteins, assignment of sequence and cross-link information was straightforward (Steinert and Marekov, 1995, 1997; Steinert et al., 1998). In addition, some of the more prominent peaks that eluted before 45 min were sequenced even though they possessed a single sequence and no cross-link.
RESULTS
A number of earlier studies have reported the expression characteristics of some individual or pairs of precursor proteins of the CE barrier structure of terminally differentiating keratinocytes, but to date these studies have provided few data on how, when, or where the assembly of this structure is initiated. In this study, we have used a combination of immunogold electron microscopy and protein chemistry using CEs recovered from NHEK cells from four different times during differentiation in submerged liquid culture. Together, these studies have afforded new information on both the sites and temporal order of the assembly of >10 proteins used to form the CE.
Modeling Data Reveal That Involucrin, Envoplakin, Desmoplakin, and SPRs Are Major Components of the CEs Used in This Study
Before exhaustive extraction by sonication and passage through a Ficoll gradient (to remove adherent noncrosslinked proteins), we examined the CE preparations by Normarski optics to assess their morphologies. We found that CEs from all four periods were flexible and fragile and rarely were recovered as intact cell body structures (our unpublished results). Based on earlier amino acid composition analyses, these presumably represent immature CEs (reviewed by Reichert et al., 1993) and thus are appropriate for studies on the earliest stages of CE assembly.
Table 1 lists more quantitative information on the properties of the CE preparations used in this study. The amounts recovered and the isopeptide cross-link contents presumably reflect important initiation and developmental aspects of CE formation. The minimal yield of 11–12 nmol of cross-link/mg of protein corresponds to approximately one cross-link per 600 residues and is <15% of that present in epidermal stratum corneum CEs. The amino acid compositions were used to calculate likely protein contents by application of established mathematical modeling algorithms (Steven and Steinert, 1994; Steinert, 1995). In the case of the 2-d CEs, it was necessary to assign a substantial fraction to the GenBank average composition to obtain positive (and thus biologically relevant) values for known CE proteins. The overall robustness of these calculations was evident from the fact that in all cases the root mean square residuals were low (our unpublished data), and the values totaled near 100% without additional constraints. The high GenBank content of 2-d CEs presumably reflects the presence of large numbers of other proteins each in individually minor amounts, which had become insoluble as a result of cross-linking. Some of these may represent desmosomal plaque and other plasma membrane proteins, because the purified CE remnants retained a double cell membrane structure apparently held together at desmosomes (see Figures 1, 3, and 4). Notably however, almost 50% of the protein mass of 2-d CEs was involucrin, and the cross-link yield reflects almost exactly one cross-link per mole of involucrin, which would be sufficient to form a macromolecular structure. In the later CEs, desmoplakin, envoplakin, SPRs, and involucrin together formed 70–80% of the total, and there were some variations in the relative amounts. The 6- to 7-d CEs contained significant but minor amounts of loricrin, suggestive of a trend toward CE maturation typical of intact epidermis (Table 1). Together, these data illustrate that desmoplakin, envoplakin, involucrin, and SPRs are the quantitatively major protein species involved in the earliest stages of CE assembly. Thus each of these proteins was investigated in more detail.
Table 1.
Amino acid compositions and calculated protein contents of CEs
| CEs from | 2 d | 3 d | 5 d | 6–7 d | Epidermisa |
| Yield (mg per dish) | 0.01 | 0.15 | 0.55 | 1.75 | |
| Amount of cross-link in CEs (nmol/mg protein) | 12.1 | 10.7 | 29.3 | 31.2 | 89.2 |
|---|---|---|---|---|---|
| Amino acid contentb | |||||
| Asp + Asn | 4.60 | 4.35 | 4.35 | 4.60 | |
| Thr | 2.80 | 2.95 | 3.80 | 3.35 | |
| Ser | 4.20 | 4.30 | 4.55 | 6.85 | |
| Glu + Gln | 32.80 | 28.95 | 26.95 | 24.35 | |
| Pro | 8.75 | 12.85 | 12.25 | 11.75 | |
| Gly | 6.80 | 7.95 | 8.45 | 10.50 | |
| Ala | 6.45 | 3.55 | 3.90 | 3.90 | |
| Val | 5.60 | 5.85 | 6.00 | 5.85 | |
| Cys | 2.55 | 3.85 | 3.90 | 4.00 | |
| Ile | 1.85 | 1.35 | 1.65 | 2.00 | |
| Leu | 10.80 | 7.65 | 7.55 | 7.25 | |
| Tyr | 0.95 | 1.85 | 1.95 | 2.00 | |
| Phe | 1.05 | 1.90 | 2.05 | 1.95 | |
| Lys | 7.45 | 8.90 | 8.45 | 8.00 | |
| Arg | 3.55 | 3.50 | 3.35 | 3.50 | |
| Modeled % amounts of major CE proteinsb | |||||
| Loricrin | 0 | 5 | 8 | 19 | 78 |
| SPRs | 10 | 27 | 26 | 21 | 5 |
| Involucrin | 48 | 28 | 28 | 24 | 5 |
| Keratins | 9 | 6 | 5 | 6 | 2 |
| Plakinsc | 12 | 24 | 21 | 19 | 2 |
| Filaggrin | 0 | 0 | 0 | 2 | 1 |
| Cystatin α | 4 | 2 | 2 | 2 | 2 |
| Elafin | 0 | 2 | 2 | 2 | 2 |
| GenBank averaged | 14 | 8 | 5 | 4 | 2 |
Data are from Steinert and Marekov (1997).
Numbers do not round to 100, because some minor residues or proteins were omitted from the calculations.
These include average compositions of desmoplakin and envoplakin.
This is included to rationalize the modeled estimates (Steven and Steinert, 1994): in principle, a high content suggests the presence of a large range of other proteins each in very minor amounts.
Figure 1.
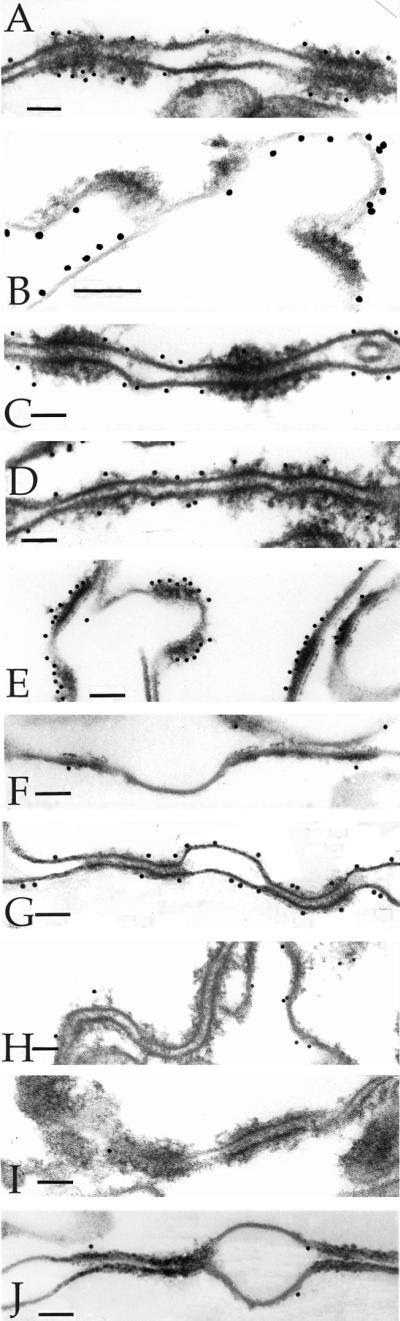
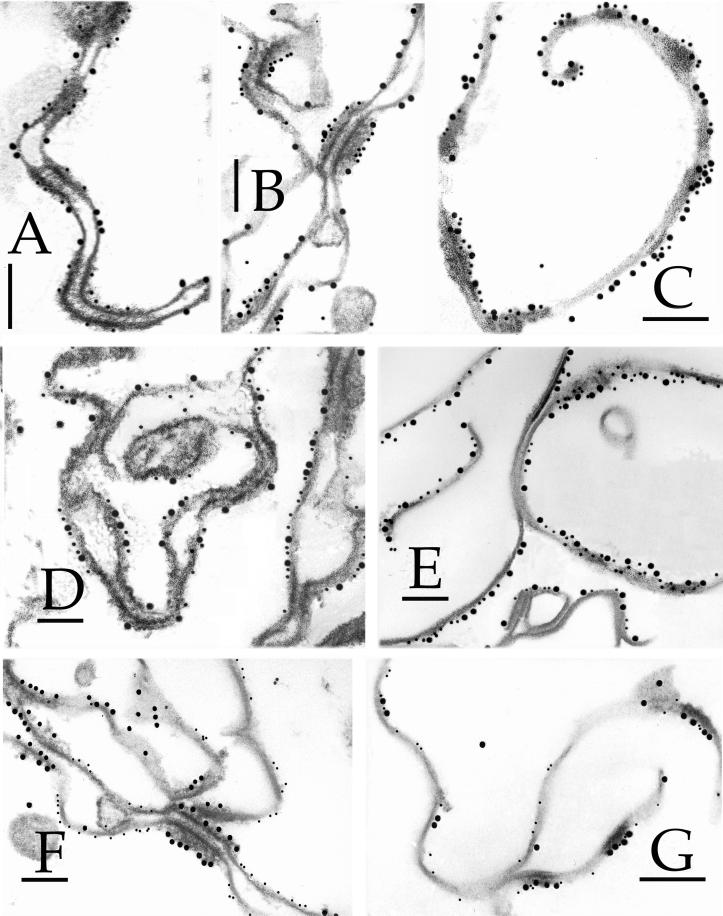
Detection of epitopes of 3-d CEs by immunogold electron microscopy. The epitopes were tested with 10-nm gold particles, and were (A) desmoplakin, (B) envoplakin, (C) periplakin, (D) involucrin, (E) keratin 1, (F) keratin 10, (G) SPR1, (H) loricrin, (I) plectin, and (J) annexin I. Bars, 100 nm.
Figure 3.
Use of double-labeling immunogold electron microscopy to monitor distributions of epitopes in 2-d (A), 3-d (B, D, and F), 5-d (C), or 6- to 7-d (E and G) CEs. Combinations used were (A–C) desmoplakin (5-nm particles) and involucrin (15 nm), (D and E) envoplakin (5 nm) and involucrin (15 nm), and (F and G) envoplakin (5 nm) and desmoplakin (15 nm). Bars, 100 nm.
Figure 4.
Use of double-labeling immunogold electron microscopy to monitor distribution of epitopes for SPRs (15-nm particles) and envoplakin (5 nm). (A) 2-d CEs, (B) 3-d, (C) 5-d, and (D) 7-d. Bars, 100 nm.
Immunogold Electron Microscopy Reveals Different Temporal Fates of CE Protein Precursors and Other Junctional Proteins
Initially, we used immunogold electron microscopy with single antibodies to decorate CE fragments from 3-d (Figure 1) and 6- to 7-d (Figure 2) cultured NHEK cells. Based on these data, we next performed a series of double-labeling experiments with several combinations of pairs of antibodies on 2-, 3-, 5-, and 6- to 7-d CEs to obtain more specific information about location and temporal aspects (Figures 3–5). In parallel, we counted the numbers of gold particles per micrometer of CE fragment to obtain semiquantitative information. These data are summarized in Table 2 and in the following comments for each antigen.
Figure 2.
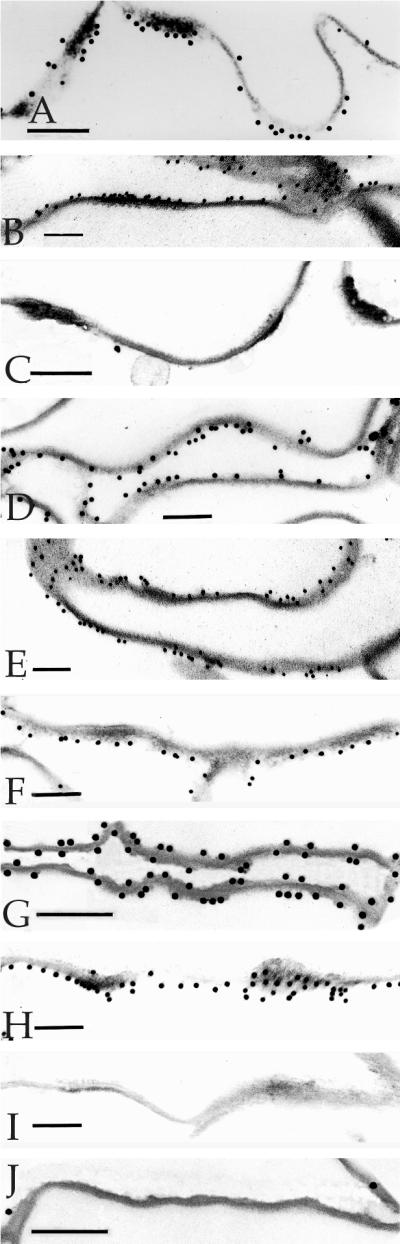
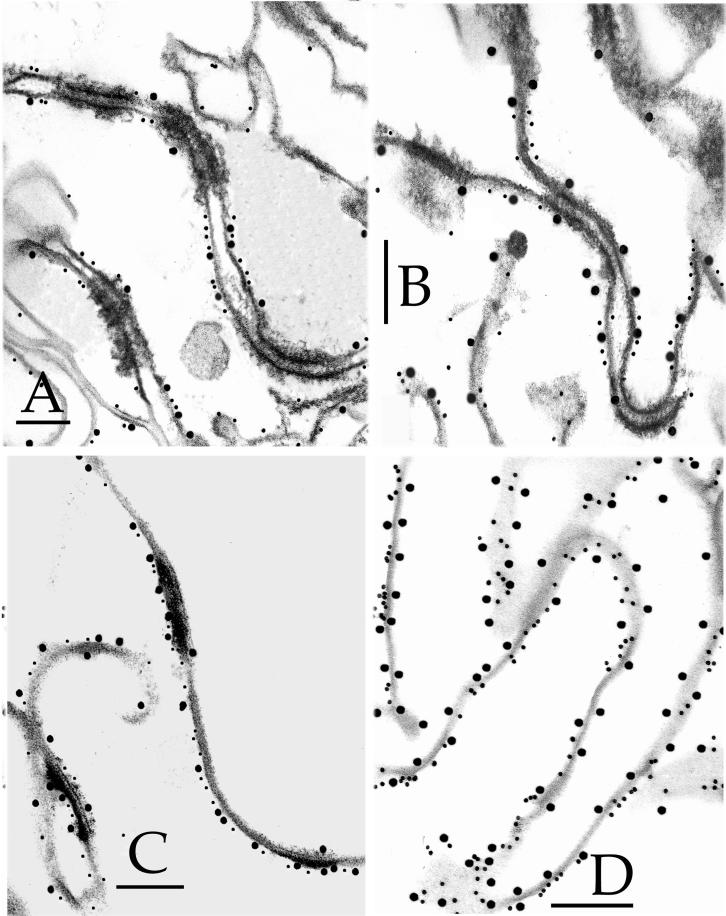
Detection of epitopes on 6- to 7-d CEs by immunogold electron microscopy. The epitopes were tested with 10-nm gold particles and were (A) desmoplakin, (B) envoplakin, (C) periplakin, (D) involucrin, (E) keratin 1, (F) keratin 10, (G) SPR1, (H) loricrin, (I) plectin, and (J) annexin I. Bars, 100 nm.
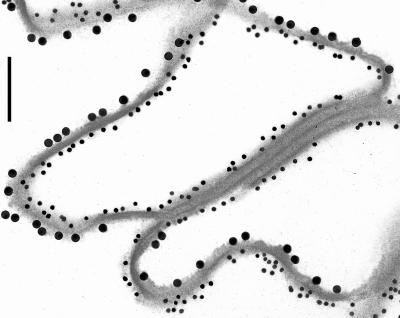
Figure 5.
Use of double-labeling immunogold electron microscopy to monitor distribution of epitopes for envoplakin (15-nm particles) and loricrin (5 nm) in 6- to 7-d CEs. Note that the two labels occur of different sides. Bar, 100 nm.
Table 2.
Summary of fates of major CE precursor proteins
| Protein | Days in culture
|
|||
|---|---|---|---|---|
| 2 | 3 | 5 | 6–7 | |
| Involucrin | Interdesmosomal | Mostly interdesmosomal | Equally distributed | Equally distributed |
| 19 (1161) | 17 (1034) | 14 (744) | 12 (658) | |
| 1 (68) | 5 (304) | 13 (692) | 13 (709) | |
| Envoplakin | Interdesmosomal | Interdesmosomal | Equally distributed | |
| 26 (1537) | 35 (2073) | 19 (1079) | ||
| <1 (23) | 1 (77) | 17 (971) | ||
| Periplakin | Interdesmosomal | None detectable | ||
| 9 (511) | <1 (3) | |||
| <1 (12) | <1 (8) | |||
| Desmoplakin | At desmosomes | At desmosomes | At desmosomes | |
| 19 (1005) | 27 (1898) | 41 (2157) | ||
| <1 (19) | 2 (106) | 2 (103) | ||
| SPR 1 | Interdesmosomal, only one side | Mostly interdesmosomal, only one side | Equally distributed, mostly one side | Equally distributed, equally both sides |
| 5 (272) | 21 (1291) | 19 (1245) | 22 (1433) | |
| <1 (23) | 8 (492) | 18 (1191) | 20 (1307) | |
| Loricrin | Not significant | Equally distributed, only one side | ||
| <1 (27) | 29 (1914) | |||
| <1 (20) | 25 (1655) | |||
| Keratin 1 | At desmosomes | Equally distributed | ||
| 15 (1150) | 14 (618) | |||
| 1 (76) | 18 (805) | |||
| Keratin 5 | At desmosomes | At desmosomes | ||
| 31 (1779) | 16 (844) | |||
| 1 (67) | <1 (38) | |||
| Keratin 10 | Not significant | Equally distributed | ||
| <1 (34) | 10 (665) | |||
| 1 (50) | 8 (540) | |||
| Plectin | Not significant | Not significant | ||
| <1 (10) | <1 (5) | |||
| <1 (5) | <1 (1) | |||
| Annexin 1 | Not significant | Not significant | ||
| <1 (13) | <1 (17) | |||
| <1 (9) | <1 (11) | |||
In each case, the first and second rows of numbers refer to the gold particles per micrometer at interdesmosomal and desmosomal regions, respectively. The total numbers of particles counted are shown in parentheses.
Desmoplakin.
This protein is the major cytoplasmic constituent of desmosomes. Epitopes of the antibody used reside along the rod domain (Green et al., 1990). Gold particles were closely associated only with desmosomes and desmosomal remnants of all CE samples (Figures 1A, 2A, and 3, A–C, F, and G), indicating that at least portions of desmoplakin survive terminal differentiation maturation events and become permanently cross-linked to the CE, and that these epitopes are not subsequently dispersed along the entire CE fragment.
Envoplakin.
Envoplakin is a recently discovered cell peripheral keratinocyte protein and is a member of the plakin family, because it shares major structural and organizational similarities with desmoplakin in particular (Ruhrburg et al., 1996; Ruhrberg and Watt, 1997). It is typically expressed in the epidermis after commitment to terminal differentiation in immediate suprabasal cells, well before the first appearance of involucrin in the upper spinous cell layers (Ruhrburg et al., 1996). In addition, previous sequencing analyses from this laboratory have documented many cross-links between envoplakin and various other CE proteins, including involucrin and type II keratins (Steinert and Marekov, 1997; Candi et al., 1998b), and furthermore, it serves as a template for ester-linked ceramides in the epidermis (Marekov and Steinert, 1998). We made an antibody using a synthetic peptide corresponding to the carboxyl-terminal 14 amino acid residues of human envoplakin, which after affinity purification to remove minor amounts of keratin epitopes, recognized only a 220-kDa band on SDS extracts of foreskin epidermal keratinocytes (our unpublished data). This epitope was localized along the plasma membrane at interdesmosomal sites of 2-d (Figure 4A) and 3-d (Figures 1B, 3, D and F, and 4B) CEs. In 5-d (Figure 4C) or 6- to 7-d (Figures 2B, 3, E and G, and 4D) CEs, the epitopes had become essentially equally distributed along one side of the cell periphery.
Periplakin.
Periplakin is also a new member of the plakin family, is similarly located along the cell periphery of terminally differentiating keratinocytes, and is predicted to interact directly to form a heterodimer and perhaps multimeric complexes with envoplakin (Ruhrburg et al., 1997; Ruhrberg and Watt, 1997). Although we have not recorded any cross-links of periplakin with itself or other CE proteins, it may serve as a substrate for ester-linked ceramides (Marekov and Steinert, 1998). We made a new affinity-purified antibody to the carboxyl-terminal 14 residues of human periplakin, which recognized only a single band of 190 kDa (our unpublished data). This epitope decorated the plasma membrane at interdesmosomal sites of 3-d CEs (Figures 1C) but could no longer be detected in significant amounts in older CEs (e.g., Figure 2C for 6- to 7-d CEs). This may be due to loss or masking of this epitope.
Involucrin.
Consistent with earlier immunogold observations on whole tissue or cultured cell samples (Ishida-Yamamoto et al., 1996, 1997), in three separate preparations of 2-d CEs, we found that most gold particles localized to interdesmosomal sites (Figure 3A and Table 1). In 3-d CEs, labeling density was consistent, but a significant minority of gold particles were also located at desmosomal remnants (Figures 1D and 3, B and D, and Table 1). However, in 5-d (Figure 3C) and 6- to 7-d (Figures 2D and 3E) CEs, involucrin had become equally distributed along one side of the cell periphery.
Keratins 1, 6, and 10.
The epitopes for the type II keratins 1 (Figure 1E) and 6 (our unpublished data) were initially seen at desmosomal remnants in 3-d CEs. In 6- to 7-d CEs, the epitopes for keratin 1 had become uniformly distributed (Figure 2E), but those for keratin 6 remained predominantly at desmosomal remnants (our unpublished data). In contrast, keratin 10 epitopes were evident only in the 6- to 7-d CEs (compare Figures 1F and 2F).
Loricrin.
This CE protein was not significantly detectable in 3-d CE preparations (Figure 1H), but in 6- to 7-d CEs, it uniformly decorated one side of the CE fragments, which was opposite to that of envoplakin (Figures 2H and 5). It has been shown previously (Mehrel et al., 1990; Steven et al., 1990) that loricrin is located on the cytoplasmic side of CE fragments, and that it is expressed abundantly only in advanced, differentiating cultured cells (Steven and Steinert,1994; Jarnik et al., 1996, 1998). Accordingly, we interpret the present data to mean that in 7-d CEs, loricrin had formed a substantial layer on the cytoplasmic side, which had occluded the epitopes for envoplakin and periplakin. This may explain why envoplakin epitopes were detectable only on the opposite side.
SPR1.
The anti-mouse antibody used in these studies also recognized the human SPR1a/b and 3 proteins (Kartasova et al., 1996). The labeling mirrored that of involucrin in that, in 2-d CEs, the observed modest degree of labeling occurred only at interdesmosomal sites (Figure 4A); in 3-d CEs, significantly more decoration occurred, and some was observed at desmosomal sites also (Figures 2G and 4B); labeling was uniformly distributed in 5-d CEs (Figure 4C) mostly on one side; but in 7-d CEs, labeling occurred uniformly on both sides (Figures 2G and 4D). We have shown previously that SPRs form a cross-linked amalgam with loricrin in vivo (Steinert et al., 1998a,b). We interpret the latter observation the same way as for loricrin in that SPR epitopes were present both within the cytoplasmic, loricrin-rich layer with as well as on the side closer to the membrane.
Several Other Potential CE or Junctional Proteins Were Negative.
Plectin is an additional member of the plakin family, is an important structural component of hemidesmosomes, and is retained in desmosomes of differentiating cells (Rezniczek et al., 1998). However, the epitopes of the antibody we used were not present on isolated CE fragments (Figures 1I and 2I). Annexins constitute a family of important peripheral proteins in many cell types, including annexin 1 in epidermal keratinocytes. One report has suggested that it may be a cross-linked component of cultured keratinocyte CEs (Robinson et al., 1997; Robinson and Eckert, 1998). However, we were unable to detect it in any of the CEs used in this study (Figures 1J and 2J). We also used commercially available antibodies to β4-integrin, plakophilin, plakoglobin, desmoglein 3, and desmocollin 3a. However, epitopes for none of these antigens were recognized on the CEs used in this study (our unpublished results). In each case, we cannot exclude the possibility that the epitopes for the antibodies were lost and/or had become masked.
Sequencing of Cross-linked Peptides Reveals Temporal Differences in the Incorporation of Protein Precursors into the CE
Recovery and Protein Origins of CE Proteins.
CEs from two sources were recovered in 5- to 10-mg amounts for protein sequencing procedures: those of attached cells after 3 d in culture and those pooled from 7-d attached and 6- to 7-d sloughed cells. These were subjected to trypsin digestion, whereupon >97 and ∼93%, respectively, of the CE protein mass was solubilized. Likewise, >93% of the isopeptide cross-link was solubilized in each case. The tryptic peptides were then resolved by HPLC as before (Steinert and Marekov, 1995, 1997; Steinert et al., 1998a) yielding ∼200 individual major and minor peptide peaks (our unpublished results). We found empirically that those peaks eluted by <25% acetonitrile contained only trace substoichiometric amounts of cross-link, whereas the 170–180 peaks eluted by >25% acetonitrile accounted for >87% of the cross-link (Table 3). Each of the latter was sequenced to completion. In the case of those from 3-d CEs, 151 contained multiple sequences because of the presence of one (125), two (17), three (7), or four (2) cross-links, yielding 337 separate peptide “branches.” In the 6- to 7-d CEs, the 157 peptides contained one (84), two (38), three (21), or four (13) cross-links and 434 peptide branches. Whereas the proteins of origin of almost all branches were identifiable (Table 3), and the glutamine and/or lysine residues used for cross-linking in each case could be placed in the known protein sequences, only 27 branches possessed sequences that could not be reconciled in searches of existing databases. The remaining 40 peptide peaks did not contain a cross-link but contained 30- to 50-residue-long stretches of known CE proteins. As estimated by amino acid analysis, the ∼7% of tryptic insoluble protein of 6- to 7-d CEs consisted almost entirely of loricrin.
Table 3.
Occurrences of sequences of proteins in CEs from cultured NHEK cells
| CE experiment
|
3 d
|
6–7 d
|
||
|---|---|---|---|---|
| Protein | Number | Yield | Number | Yield |
| Cystatin α | 6 | 150 (1.7) | 11 | 870 (3.2) |
| Desmoplakin | 31 | 1110 (12.3) | 30 | 3050 (11.2) |
| Elafin | 5 | 60 (0.7) | 11 | 650 (2.4) |
| Envoplakin | 60 | 1040 (11.5) | 66 | 2830 (10.4) |
| Involucrin | 86 | 2650 (31.2) | 110 | 6550 (23.4) |
| Keratin 1 | 7 | 90 (0.9) | 19 | 480 (1.8) |
| Keratin 2e | 1 | 10 (0.1) | 2 | 30 (0.1) |
| Keratin 5/6 | 19 | 300 (3.2) | 36 | 1510 (5.6) |
| Keratin 10 | 3 | 60 (0.2) | ||
| Loricrin | 5 | 260 (2.7) | 42 | 3390 (14.5) |
| SPR 1 or 3 | 70 | 1930 (21.0) | 56 | 3360 (12.4) |
| SPR 2 | 40 | 930 (10.6) | 29 | 2340 (8.6) |
| Unidentified | 9 | 430 (4.4) | 18 | 1520 (5.6) |
| No. of peptides | 151 | 157 | ||
| No. of peptide branches | 339 | 434 | ||
| Cross-link recovery | 9.0 of 10.7 (84%) | 27.2 of 31.2 (87%) | ||
Yield is in picomoles. Percentages of molar total are shown in parentheses.
The identified proteins listed in Table 3 document 12 known CE species of which desmoplakin, envoplakin, involucrin, and SPRs were the most abundant. The calculated molar yields of each are generally very similar to those estimated by mathematical modeling of amino acid compositions (Table 1). The glutamines and/or lysines used in the proteins desmoplakin, elafin, envoplakin, keratins, loricrin, and SPRs were the same as those identified in previous sequencing experiments on CEs recovered from in vivo foreskin epidermis (Steinert and Marekov, 1995, 1997; Steinert et al., 1998a,b; Candi et al., 1998b). We have reported elsewhere for these cultured NHEK cells the detailed analyses of the glutamines and lysines used for cross-linking to the proteins loricrin (Candi et al., 1995, 1999), SPRs (Steinert et al., 1998b) and keratins (Candi et al., 1998b).
Analyses of Sequences from Recovered Peptides Reveal Temporal Changes in Protein Partners and Glutamine and Lysine Residues Used for Cross-Links.
Table 4 summarizes the protein partners cross-linked together in the 3- and 6- to 7-d CEs. Notably, there were marked shifts in frequency in several cases. In 3-d CEs, envoplakin, involucrin, SPR1, and SPR2 were largely cross-linked to themselves; there were many cross-links between envoplakin and involucrin as well as involucrin and SPRs but very few cross-links between desmoplakin and involucrin or desmoplakin and envoplakin. In 6- to 7-d CEs, there were many more cross-links between desmoplakin and involucrin, desmoplakin and envoplakin, or involucrin and type II keratins. Likewise, there were still many cross-links between envoplakin and involucrin. And as expected, almost all loricrin cross-link partners appeared in the 6- to 7-d CE sample.
Table 4.
Frequency of cross-linking between protein partners in cultured NHEK CEs
| No. of peptides | Cα | DP | EF | EP | IV | K1 | K2e | K5 | K10 | LO | R1 | R2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cystatin α (Cα) | 17 | 0 | |||||||||||
| Desmoplakin (DP) | 61 | 1, 0 | 0 | ||||||||||
| Elafin (EF) | 16 | 0 | 0, 1 | 0 | |||||||||
| Envoplakin (EP) | 126 | 1, 1 | 1, 11 | 0, 1 | 16, 7 | ||||||||
| Involucrin (IV) | 196 | 0, 3 | 2, 17 | 0, 2 | 14, 11 | 21, 12 | |||||||
| Keratin 1 (K1) | 26 | 0, 1 | 0 | 0, 1 | 1, 2 | 4, 9 | 0 | ||||||
| Keratin 2e (K2e) | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Keratin 5 (K5) | 55 | 1, 2 | 0 | 1, 3 | 4, 11 | 4, 23 | 0 | 0 | 0 | ||||
| Keratin 10 (K10) | 3 | 0 | 0 | 0, 1 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Loricrin (LO) | 47 | 0, 3 | 0, 2 | 0, 1 | 0, 1 | 0, 3 | 1, 4 | 1, 2 | 1, 9 | 0, 2 | 0, 3 | ||
| SPR1 (R1) | 128 | 2, 2 | 8, 6 | 3, 1 | 17, 14 | 23, 12 | 1, 2 | 0 | 2, 5 | 0 | 1, 6 | 11, 8 | |
| SPR2 (R2) | 69 | 0 | 6, 3 | 0, 1 | 6, 4 | 18, 12 | 0 | 0 | 1, 1 | 0 | 1, 6 | 1, 0 | 7, 2 |
In each case, the numbers refer to occurrences in 3- or 6- to 7-d CE preparations.
Furthermore, analysis of the involucrin data revealed use of Lys-62 with Gln-133 or Gln-496, two novel pairs involved in cross-link formation not seen in previous analyses of CEs of intact epidermis (Steinert and Marekov, 1997; Table 5). Specifically, the data reveal a marked shift in residues used: whereas these three residues were used 26 times in 3-d CEs, they were seen only three times in 6- to 7-d CEs. Notably, Gln-496 was subsequently observed in 6- to 7-d CEs and those from intact epidermis to be cross-linked almost exclusively with desmoplakin.
Table 5.
Progressive cross-linking of involucrin using different Gln and Lys residues
| Cross-links | CEs from: | Cultured cells
|
Epidermisa | |
|---|---|---|---|---|
| 3 d | 6–7 d | |||
| Involucrin–involucrin | Gln-133-Lys-62 | 325 | 100 | |
| Gln-465-Lys-468 | 40 | 250 | 850 | |
| Gln-465-Lys-485 | 25 | 245 | 565 | |
| Gln-465-Lys-508 | 40 | 120 | ||
| Gln-489-Lys-468 | 195 | 415 | ||
| Gln-489-Lys-485 | 285 | 420 | ||
| Gln-489-Lys-508 | 90 | |||
| Gln-496-Lys-62 | 445 | 165 | ||
| Involucrin–envoplakin | Lys-378 | 35 | 120 | |
| Lys-468 | 150 | |||
| Lys-485 | 245 | 640 | ||
| Lys-508 | 85 | 35 | ||
| Involucrin–desmoplakin | Gln-456 | 140 | ||
| Gln-495 | 170 | |||
| Gln-496 | 20 | 1145 | 985 | |
| Involucrin–SPRs | Gln-308 | 30 | ||
| Gln-309 | 115 | |||
| Gln-368 | 50 | 65 | ||
| Gln-369 | 280 | 445 | ||
| Gln-426 | 85 | 85 | ||
| Gln-427 | 245 | 515 | ||
| Gln-455 | 65 | 70 | ||
| Gln-456 | 465 | 590 | 265 | |
| Involucrin–loricrin | Gln-308 | 35 | ||
| Gln-309 | 76 | |||
| Gln-369 | 165 | |||
| Gln-425 | 70 | |||
| Gln-426 | 115 | 260 | ||
| Gln-455 | 25 | |||
| Gln-456 | 57 | 80 | ||
Yield is in picomoles per milligram of CE protein.
Data are from saponified foreskin CEs of Steinert and Marekov (1997).
Together, these data document progressive changes of both protein partners and residue use that presumably reflect initiation and then maturation of CE formation.
DISCUSSION
An Elaborated Model for the Initiation of CE Formation in Epithelia
An early event in the terminal differentiation program of stratified squamous epithelia is the expression of a variety of unique proteins whose likely ultimate fate is assembly into the CE barrier structure. Historically, this process was typically monitored by the expression of the marker protein involucrin (Watt and Green, 1981; Pillai and Bickle, 1991), and indeed, involucrin was found to decorate the cell peripheries of differentiating keratinocytes by immunogold electron microscopy (Ishida-Yamamoto et al., 1996, 1997). Based on these observations, existing dogma has suggested that CE assembly is initiated by cross-linking of involucrin. More recent work has shown that the expression of two novel members of the plakin family, envoplakin (Ruhrberg et al., 1996) and periplakin (Ruhrberg et al., 1997), are also valuable early markers for terminal differentiation and CE formation in some stratified squamous epithelia, and in fact, their expression clearly precedes that of involucrin by several cell layers in the epidermis, for example. Using a combination of high-resolution immunogold labeling and protein sequencing of peptides generated from CEs formed in early differentiating NHEK cells, we have provided robust support for the hypothesis that involucrin, envoplakin, and perhaps periplakin are critical components of the earliest stages of CE formation, and we provide for the first time molecular details on how this process might occur.
Role of Involucrin in CE Initiation
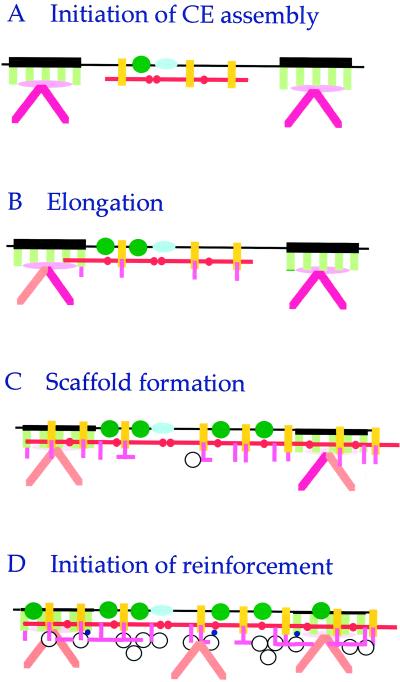
Mammalian involucrins are thought to have evolved by tandem duplications of glutamine- and glutamic acid-rich sequences spanning between the amino-terminal (“head”) and carboxyl-terminal (“tail”) domains (Green and Djian, 1992). These head and tail domain sequences have been highly conserved during mammalian evolution, supporting the notion that these two domains, or more specifically at least certain glutamine and lysine residues within them, have been retained because they are functionally important. On the other hand, most of the estimated 46-nm length of human involucrin is contributed by the central peptide repeat domain. Our analyses of in vivo sequencing data involving involucrin, which documented cross-linking through many widely separated glutamine and lysine residues (Steinert and Marekov, 1997), support the notion that the expansion of the numbers of central domain repeats was driven by the evolutionary benefit of increasing the number of possible TGase substrate sites for several other CE proteins. Thus involucrin functions by cross-bridging widely separated CE proteins and in this way is ideally suited to serve as a scaffold for CE assembly (Yaffe et al., 1992).
Our new data document how involucrin is first deployed to form this scaffold. The sequencing data of 3-d CEs revealed (Table 5) that primarily only three residues of involucrin are initially used for cross-linking: Gln-133 with Lys-62 on the head domain, which would give rise to head-to-head oligomerization; and Gln-496 with Lys-62, which would give rise to head-to-tail oligomerization. Notably, Gln-496 has been identified in in vitro assays as the most reactive Gln residue of involucrin (Simon and Green, 1988; Nemes et al., 1999b). Together with the immunogold labeling data, which showed initial interdesmosomal labeling (Figures 1D and 3, A, B, and E), we conclude that simple oligomerization of involucrin through these head and tail domain sequences represents a very early step of CE formation (Figure 6). Subsequently, Gln-496 was cross-linked primarily to desmoplakin (Table 4; Steinert and Marekov, 1997; Steinert et al., 1998a), which is consistent with the immunogold labeling data showing that with time involucrin spreads from interdesmosomal to desmosomal sites (Figure 3, B–E). The reason for the reduced numbers or absence of cross-links involving these three residues of involucrin in older CEs and those of intact epidermis (Table 5) is most likely technical in nature: quantitatively minor peptide peaks eluted by HPLC are difficult to identify in a background of much larger peaks involving different cross-links (see peptide yield data of Table 3 compared with the 10-fold higher yield of CEs).
Figure 6.
New model for the early stages of CE assembly in stratified squamous epithelia. (A) Our immunogold and sequencing data indicate that a very early step in CE assembly involves the head-to-head and head-to-tail oligomerization of involucrin (red bars) at interdesmosomal regions of the plasma membrane, in close juxtaposition with the TGase 1 enzyme (green circles), envoplakin (yellow rods), and perhaps periplakin (blue ovoids). (B) subsequently in 3-d CEs, involucrin deposition elongates to the desmoplakin (green rods) component of desmosomes, and SPR (pink rods) proteins become attached. (C) By 6-d CEs, an involucrin–envoplakin–SPR continuum or scaffold has formed along the entire cell periphery. (D) In 7-d or more mature sloughed cell CEs, this scaffold serves as a platform for the subsequent addition of other reinforcement proteins, including loricrin (white circles) and other proteins (small blue circles). Also, the keratin chain composition of the keratin intermediate filaments at desmosomes changes from mostly keratins 5, 6, and 14 (dark pink rods) to keratins 1, 2e, and 10 (orange rods) located along the entire periphery. In all models, the lower side is cytoplasmic. Note that to recover recognizable CE fragments with associated desmosomal remnants after 2-d culture, many other cell peripheral proteins presumably have become cross-linked in minor amounts, but we were unable to identify these (e.g., plectin and integrins; pink ovoids) by sequencing or immunogold electron microscopy.
Possible Coordinate Roles of Envoplakin and Periplakin in CE Initiation
The recovery of even small amounts of insoluble CEs in 2-d cultured cells implies that the fragments already contain sufficient cross-links to form a recognizable macromolecular structure (Figures 1, 3, and 4). This would require at least one cross-link per protein chain, and indeed the recovered yield of cross-link (approximately one per 600 residues) is consistent with this (Table 1). Nevertheless, of all of the protein species that constitute the isolated 2-d CEs, mathematical modeling of the amino acid analysis data suggested that involucrin accounts for ∼50% of the total protein. This further implies that involucrin might have become associated or cross-linked to something else at the membrane surface in addition to itself. We can think of two possibilities for this. First, involucrin might have been initially transported to and retained at the cell membrane by cross-linking to various S100 calcium-binding proteins and/or the ubiquitous annexin system (Robinson et al., 1997; Robinson and Eckert, 1998). Recently, we have proposed a second, more passive alternative method. Using an experimental in vitro synthetic lipid vesicle system that mimics the composition of eukaryotic plasma membranes, we found that involucrin spontaneously binds to membranes at the submicromolar concentrations of Ca2+ expected in early differentiating keratinocytes (Nemes et al., 1999b). Furthermore, we found that the TGase 1 enzyme is likely to initiate cross-linking of involucrin, because of several TGase enzymes known to be present in keratinocytes, only TGase 1 can bind to membranes through its acyl lipid anchors. Moreover, it specifically activates Gln-496, Gln-133, and minor amounts of three other head domain glutamine residues of juxtaposed involucrin molecules. Thus our new data raise the possibility that involucrin may anchor to the cell periphery by becoming cross-linked to another major interdesmosomal protein.
The data of this paper suggest that envoplakin is a quantitatively plausible candidate at early stages of CE assembly (Tables 1–4 and Figure 6). Envoplakin in fact is expressed demonstrably earlier than involucrin in the epidermis: it first appears in the immediate suprabasal cell layer as opposed to the higher spinous layers for involucrin. Whereas a simple Ca2+-dependent mechanism for the association of involucrin with plasma membranes was proposed recently (Nemes et al., 1999b), it is not yet clear how envoplakin may associate with plasma membranes (Ruhrberg et al., 1996, 1997). Our new data indicate that envoplakin and involucrin are colocalized at interdesmosomal sites in CEs from 2-d cells (Figures 3A and 4A); envoplakin becomes extensively cross-linked to itself in 3-d CEs (Table 4); envoplakin–involucrin interchain cross-linking has frequently occurred in 3-d CEs (Table 5); in CEs of 5-d cultured cells (Figure 4C), envoplakin, like involucrin, had formed an inter- and intradesmosomal continuum; and by 6- to 7-d CEs, both envoplakin and involucrin had become extensively cross-linked with desmoplakin. In this way envoplakin and involucrin may consolidate scaffold formation for subsequent stages of CE assembly (Figure 6). These considerations of our new data thus afford robust support for the initial suggestion on the possible role of envoplakin in CE scaffold formation (Ruhrberg et al., 1997; Ruhrberg and Watt, 1997). Furthermore, because our in vitro data have suggested that involucrin first becomes cross-linked by the membrane-anchored TGase 1 enzyme (Nemes et al., 1999b), it is tempting to speculate that envoplakin is also cross-linked to itself and involucrin by TGase 1. In this way, the coordinated juxtaposition on membranes of TGase 1 (linked through its acyl lipid adducts), involucrin (linked through Ca2+), envoplakin, and perhaps other proteins seems essential for the initiation of CE assembly (Figure 6). Accordingly, further work will be necessary to test these hypotheses to identify the molecular mechanisms by which envoplakin become associated with membranes and to explore how envoplakin and involucrin become cross-linked together.
Likewise, although we have not yet found cross-linked peptides involving periplakin, we have identified possible periplakin–ceramide adducts (Marekov and Steinert, 1998), which requires that it too is present in very close proximity to the plasma membrane surface. Periplakin was found to colocalize with envoplakin (Ruhrberg et al., 1997), and based on common patterns in the distributions of charged residues on their rod domains, it is possible that envoplakin and periplakin may copolymerize or even form heterodimers. These ideas allow the possibility that periplakin is also involved in the earliest stages of CE assembly. Again, it is possible that minor periplakin-containing peptides might have been obscured in the HPLC profile by the quantitatively larger peptide peaks derived from the more abundant involucrin and SPR proteins. Thus further work will be necessary to explore the role of periplakin. Finally, the apparent redundant coparticipation of involucrin, envoplakin, and perhaps periplakin in forming an initial scaffold suggests that loss of one protein by mutation may not seriously interrupt CE assembly in stratified squamous epithelia.
Roles of SPRs in CE Formation
In 2-d CEs and beyond, we found colocalization of SPRs, envoplakin, and involucrin (Figures 3, A–E, and 4). Notably, the labeling density of SPRs was initially low but increased fourfold in older CEs (Table 2). Because sequencing revealed that SPRs were cross-linked to involucrin through multiple Gln residues and not Gln-496 (Table 5), these data may mean that cross-linking of SPRs occurs subsequent to oligomerization of involucrin and envoplakin. Alternatively, their cross-linking may have occurred independently. Because the sites used for SPR cross-linking to involucrin were also found to be used by TGases in in vitro solution assays (Nemes et al., 1999b), it is possible that SPR cross-linking occurs through other cytosolic TGases, including perhaps soluble forms of TGase 1. Indeed, we have suggested that cytosolic TGases likely cross-link SPR1 proteins first into small oligomers that are subsequently attached to a growing CE structure by further cross-linking by other TGases, including the membrane-anchored TGase 1 enzyme (Candi et al., 1999).
We have provided evidence that SPRs serve as cross-bridging proteins in CE structures and in this way might modulate the biomechanical properties of the CEs and thus the entire epithelium in which they are expressed (Steinert et al., 1998a,b). Likewise, we suggest that abundant SPR occurrence in early CEs formed by cultured cells may serve to strengthen the structure during the initial stages of assembly.
Summary: A Common Assembly Mechanism of CEs in Stratified Squamous Epithelia?
The present studies with CEs recovered from cultured NHEK cells apparently mimic many aspects of CE assembly observed in vivo in intact foreskin epidermis. We are particularly struck by the coincidence of the large amounts of proteins such as involucrin, envoplakin, and SPRs in the cultured cell CEs as was also found in immature and saponified foreskin epidermal CEs, as well as the specific glutamine and lysine residues used in each protein for cross-linking. Such data therefore afford assurance that cultured NHEK cells are indeed a valid system for extrapolation to the earliest times of CE formation in intact skin.
This said, our data also support the notion that the NHEK system is likely to be of value as well in elucidating the early events in the formation of CE barrier structures during the terminal differential programs in many other types of stratified squamous epithelia. A variety of proteins such as involucrin, envoplakin, and SPRs are known to be coexpressed at least during the earliest stages of differentiation in other epithelia. One cogent extant example is the case of the periderm present early during the second trimester of human development, which precedes epidermal formation by many weeks. The single-cell periderm layer forms a limited CE barrier structure that is composed largely of involucrin and SPRs (Watt et al., 1989; Holbrook and Wolff, 1993; Akiyama et al., 1999; Lee et al., 1999). Expression of envoplakin and periplakin in human periderm has not yet been reported. Another example consists of CEs from human gingiva tissue, which on the basis of mathematical modeling and protein sequencing data consist of >60% SPRs and ∼10% each of envoplakin, involucrin, and loricrin (our unpublished results). Similarly, cultured esophageal epithelial cells or other airway epithelial cell types (An et al., 1993), vaginal and uterine epithelia (Jetten et al., 1996), and several other types of internal epithelia (Fujimoto et al., 1997) abundantly coexpress involucrin and SPRs. Accordingly, our new data raise the possibility that the initial events involved in CE formation may be common for a wide range of stratified squamous epithelia, for which the cultured epidermal keratinocyte system serves as a convenient model.
ACKNOWLEDGMENTS
We thank Drs. Eleonora Candi, Ulrike Lichti, and Edit Tarcsa for useful discussions during the course of this work.
Abbreviations used:
- CE
cell envelope
- NHEK
normal human epidermal keratinocyte
- SPR
small proline-rich protein
- TGase
transglutaminase
REFERENCES
- Akiyama M, Smith LT, Yoneda K, Holbrook KA, Hohl D, Shimizu H. Periderm cells form cornified cell envelope in their regression process during human epidermal development. J Invest Dermatol. 1999;112:903–909. doi: 10.1046/j.1523-1747.1999.00592.x. [DOI] [PubMed] [Google Scholar]
- An G, Tesfaigzi J, Chuu Y-J, Wu R. Isolation and characterization of the human spr1 gene and its regulation of expression by phorbol ester and cyclic AMP. J Biol Chem. 1993;268:10977–10982. [PubMed] [Google Scholar]
- Candi E, Lahm A, Ceci R, Rossi A, Kim I-G, Ciani B, Melino G, Steinert PM. Transglutaminase 1 mutations in lamellar ichthyosis: loss of activity due to failure of activation by proteolytic processing. J Biol Chem. 1998a;273:13693–13702. doi: 10.1074/jbc.273.22.13693. [DOI] [PubMed] [Google Scholar]
- Candi E, Melino G, Pei G, Tarcsa E, Marekov LN, Steinert PM. Bacterially-expressed human loricrin: biochemical, structural and transglutaminase substrate properties of the major epidermal cornified cell envelope structural protein. J Biol Chem. 1995;270:26382–26390. doi: 10.1074/jbc.270.44.26382. [DOI] [PubMed] [Google Scholar]
- Candi E, Tarcsa E, DiGiovanna JJ, Compton JG, Elias PM, Marekov LN, Steinert PM. A highly conserved lysine residue on the head domain of type II keratins is essential for the attachment of keratin intermediate filaments to the cornified cell envelope through isopeptide crosslinking by transglutaminases. Proc Natl Acad Sci USA. 1998b;95:2067–2072. doi: 10.1073/pnas.95.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Tarcsa E, Idler WW, Marekov LN, Steinert PM. Transglutaminase crosslinking properties of the small proline rich 1 family of cornified cell envelope proteins. J Biol Chem. 1999;274:7226–7237. doi: 10.1074/jbc.274.11.7226. [DOI] [PubMed] [Google Scholar]
- Crish JF, Zaim TM, Eckert RL. Tissue-specific and differentiation-appropriate expression of the human involucrin gene in transgenic mice: an abnormal epidermal phenotype. Differentiation. 1993;53:191–200. doi: 10.1111/j.1432-0436.1993.tb00708.x. [DOI] [PubMed] [Google Scholar]
- Downing DT, Stewart ME, Wertz PW, Strauss JS. Lipids of the epidermis and the sebaceous glands. In: Fitzpatrick TB, Eisen AZ, Wolff K, Freedberg IM, Austen KF, editors. Dermatology in General Medicine. 4th ed. New York: McGraw-Hill; 1993. pp. 210–221. [Google Scholar]
- Elias PM, Menon GK. Stratum corneum lipids. Adv Lipid Res. 1991;24:1–26. doi: 10.1016/b978-0-12-024924-4.50006-7. [DOI] [PubMed] [Google Scholar]
- Fujimoto W, Nakanishi G, Arata J, Jetten AM. Differential expression of human cornifin alpha and beta in squamous differentiating epithelial tissues and several skin lesions. J Invest Dermatol. 1997;108:200–204. doi: 10.1111/1523-1747.ep12334240. [DOI] [PubMed] [Google Scholar]
- Green H, Djian P. Consecutive actions of different gene-altering mechanisms in the evolution of involucrin. Mol Biol Evol. 1992;9:977–1017. doi: 10.1093/oxfordjournals.molbev.a040775. [DOI] [PubMed] [Google Scholar]
- Green KJ, Parry DAD, Steinert PM, Virata MLA, Wagner RM, Angst BD, Niles LA. Structure of the human desmoplakins: implications for function in the desmosomal plaque. J Biol Chem. 1990;265:2603–2612. [PubMed] [Google Scholar]
- Greenberg CS, Birckbichler PJ, Rice RH. Transglutaminases: multifunctional crosslinking enzymes that stabilize tissues. FASEB J. 1991;5:3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- Hohl D, Lichti U, Turner ML, Roop DR, Steinert PM. Characterization of human loricrin: structure and function of a new class of epidermal cell envelope proteins. J Biol Chem. 1991;266:6626–6636. [PubMed] [Google Scholar]
- Hohl H, de Viragh PA, Amiguett-Barras F, Gibbs S, Backendorf C, Huber M. The small proline-rich proteins constitute a multigene family of differentially regulated cornified cell envelope precursor proteins. J Invest Dermatol. 1995;104:902–905. doi: 10.1111/1523-1747.ep12606176. [DOI] [PubMed] [Google Scholar]
- Holbrook KA, Wolff K. The structure and development of the skin. In: Fitzpatrick TB, editor. Dermatology in General Medicine. New York: McGraw- Hill; 1993. pp. 97–145. [Google Scholar]
- Ishida-Yamamoto A, Eady RA, Watt FM, Roop DR, Hohl D, Iizuka H. Immunoelectron microscopic analysis of cornified cell envelope formation in normal and psoriatic epidermis. J Histochem Cytochem. 1996;44:167–175. doi: 10.1177/44.2.8609373. [DOI] [PubMed] [Google Scholar]
- Ishida-Yamamoto A, Kartasova T, Matsuo S, Kuroki T, Iizuka H. Involucrin and SPRR are synthesized sequentially in differentiating cultured epidermal. J Invest Dermatol. 1997;108:12–16. doi: 10.1111/1523-1747.ep12285611. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Elias PM. Skin as an organ of protection. In: Fitzpatrick TB, Eisen AZ, Wolff K, Freedberg IM, Austen KF, editors. Dermatology in General Medicine. 4th ed. New York: McGraw-Hill; 1993. pp. 241–253. [Google Scholar]
- Jarnik M, Kartasova T, Steinert PM, Lichti U, Steven AC. Differential expression and cell envelope incorporation of small proline rich protein 1 indifferent cornified epithelia. J Cell Sci. 1996;109:1381–1391. doi: 10.1242/jcs.109.6.1381. [DOI] [PubMed] [Google Scholar]
- Jarnik M, Simon MN, Steven AC. Cornified cell envelope assembly: a model based on electron microscopic determinations of thickness and projected density. J Cell Sci. 1998;111:1051–1060. doi: 10.1242/jcs.111.8.1051. [DOI] [PubMed] [Google Scholar]
- Jetten AM, De Luca LM, Nelson K, Schroeder W, Burlingame S, Fujimoto W. Regulation of cornifin alpha expression in the vaginal and uterine epithelium by estrogen and retinoic acid. Mol Cell Endocrinol. 1996;14:7–15. doi: 10.1016/0303-7207(96)03871-3. [DOI] [PubMed] [Google Scholar]
- Kartasova T, Kohno Y, Koizumi H, Osada S, Huh N, Lichti U, Steinert PM, Kuroki T. Sequence and expression patterns of mouse SPR1: correlation of expression with epithelial function. J Invest Dermatol. 1996;106:294–304. doi: 10.1111/1523-1747.ep12340741. [DOI] [PubMed] [Google Scholar]
- Kim S-Y, Chung S-I, Steinert PM. Highly active soluble processed forms of the transglutaminase 1 enzyme in epidermal keratinocytes. J Biol Chem. 1995;270:18026–18035. doi: 10.1074/jbc.270.30.18026. [DOI] [PubMed] [Google Scholar]
- Lee S-C, Lee J-B, Kook J-P, Seo J-J, Nam K-I, Park S-S, Kim Y-P. Expression of differentiation markers during fetal skin development in humans: immunohistochemical studies on the precursor proteins forming the cornified cell envelope. J Invest Dermatol. 1999;112:882–886. doi: 10.1046/j.1523-1747.1999.00602.x. [DOI] [PubMed] [Google Scholar]
- Marekov LN, Steinert PM. Ceramides are bound to structural proteins of the human foreskin epidermal cornified cell envelope. J Biol Chem. 1998;273:17763–17770. doi: 10.1074/jbc.273.28.17763. [DOI] [PubMed] [Google Scholar]
- Mehrel T, et al. Identification of a novel gene encoding a major cell envelope protein, loricrin. Cell. 1990;60:1103–1112. doi: 10.1016/0092-8674(90)90073-n. [DOI] [PubMed] [Google Scholar]
- Melino, G., Candi, E., and Steinert, P.M. (1999). Transglutaminases in cell death. Methods Enzymol. (in press). [DOI] [PubMed]
- Nemes Z, Marekov LN, Fesus L, Steinert PM. A novel function for transglutaminase 1: attachment of long chain ω-hydroxyceramides to involucrin by ester bond formation. Proc Natl Acad Sci USA. 1999a;96:8402–8407. doi: 10.1073/pnas.96.15.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes Z, Marekov LN, Steinert PM. Involucrin crosslinking-linking by transglutaminase 1: binding to anionic membranes directs residue specificity. J Biol Chem. 1999b;274:11013–11021. doi: 10.1074/jbc.274.16.11013. [DOI] [PubMed] [Google Scholar]
- Nemes Z, Steinert PM. Bricks and mortar of the epidermal barrier. Exp Mol Med. 1999;31:5–19. doi: 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- Pillai S, Bickle DD. Role of intracellular free calcium in the cornified envelope formation of keratinocytes: differences in the mode of action of extracellular calcium and 1,25-dihydroxvitamin D3. J Cell Physiol. 1991;146:94–100. doi: 10.1002/jcp.1041460113. [DOI] [PubMed] [Google Scholar]
- Reichert U, Michel S, Schmidt R. The cornified envelope: a key structure of terminally differentiating keratinocytes. In: Darmon M, Blumenberg M, editors. Molecular Biology of the Skin. New York: Academic Press; 1993. pp. 107–150. [Google Scholar]
- Rezniczek GA, de Pereda JM, Reipert S, Wiche G. Linking integrin alpha6beta4-based cell adhesion to the intermediate filament cytoskeleton: direct interaction between the beta4 subunit and plectin at multiple molecular sites. J Cell Biol. 1998;141:209–225. doi: 10.1083/jcb.141.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice RH, Green H. The cornified envelope of terminally differentiated human epidermal keratinocytes consists of cross-linked protein. Cell. 1977;11:417–422. doi: 10.1016/0092-8674(77)90059-9. [DOI] [PubMed] [Google Scholar]
- Rice RH, Green H. Presence in human epidermal cells of a soluble protein precursor of the cross-linked envelope: activation of the crosslinking by calcium ions. Cell. 1979;18:681–694. doi: 10.1016/0092-8674(79)90123-5. [DOI] [PubMed] [Google Scholar]
- Robinson NA, Eckert RL. Identification of transglutaminase-reactive residues in S100A11. J Biol Chem. 1998;273:2721–2728. doi: 10.1074/jbc.273.5.2721. [DOI] [PubMed] [Google Scholar]
- Robinson NA, Lapec S, Welter JF, Eckert RL. S100A11, S100A10, annexin I, desmosomal proteins, small proline rich proteins, plasminogen activator inhibitor-2, and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J Biol Chem. 1997;272:12035–12036. doi: 10.1074/jbc.272.18.12035. [DOI] [PubMed] [Google Scholar]
- Roop DR, Cheng CK, Titterington L, Meyers CA, Stanley JR, Steinert PM, Yuspa SH. Synthetic peptides corresponding to keratin subunits elicit highly specific antibodies. J Biol Chem. 1984;259:8037–8043. [PubMed] [Google Scholar]
- Ruhrberg C, Hajibagheri MA, Parry DAD, Watt FM. Periplakin, a novel component of cornified envelopes and desmosomes that belongs to the plakin family and forms complexes with envoplakin. J Cell Biol. 1997;139:1835–1849. doi: 10.1083/jcb.139.7.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhrberg C, Hajibagheri MA, Simon M, Dooley TP, Watt FM. Envoplakin, a novel precursor of the cornified envelope that has homology to desmoplakin. J Cell Biol. 1996;134:715–729. doi: 10.1083/jcb.134.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhrberg C, Watt FM. The plakin family: versatile organizers of cytoskeletal architecture. Curr Opin Genet Dev. 1997;7:392–397. doi: 10.1016/s0959-437x(97)80154-2. [DOI] [PubMed] [Google Scholar]
- Simon M, Green H. Participation of membrane-associated proteins in the formation of the cross-linked envelope of the keratinocyte. Cell. 1984;36:827–834. doi: 10.1016/0092-8674(84)90032-1. [DOI] [PubMed] [Google Scholar]
- Simon M. The epidermal cornified cell envelope and its precursors. In: Leigh IM, Lane EB, Watt FM, editors. The Keratinocyte Handbook. Cambridge, United Kingdom: Cambridge University Press; 1994. pp. 275–292. 275–292. [Google Scholar]
- Simon M, Green H. Enzymatic crosslinking of involucrin and other proteins by keratinocyte particulates in vitro. Cell. 1985;40:677–683. doi: 10.1016/0092-8674(85)90216-8. [DOI] [PubMed] [Google Scholar]
- Simon M, Green H. The glutamine residues reactive in transglutaminase-catalyzed crosslinking of involucrin. J Biol Chem. 1988;263:18093–18098. [PubMed] [Google Scholar]
- Steinert PM. A model for the hierarchical structure of the human epidermal cornified cell envelope. Cell Death & Differ. 1995;2:23–31. [PubMed] [Google Scholar]
- Steinert PM, Candi E, Kartasova T, Marekov LN. Small proline rich proteins are cross-bridging proteins in the cornified cell envelopes of stratified squamous epithelia. J Struct Biol. 1998b;122:76–85. doi: 10.1006/jsbi.1998.3957. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Kartasova T, Marekov LN. Biochemical evidence that small proline rich proteins and trichohyalin function in epithelia to modulate the biomechanical properties of their cornified cell envelopes. J Biol Chem. 1998a;273:11758–11769. doi: 10.1074/jbc.273.19.11758. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin and SPRs are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J Biol Chem. 1995;270:17702–17711. doi: 10.1074/jbc.270.30.17702. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN. Involucrin is an early component in the assembly of the epidermal cornified cell envelope. J Biol Chem. 1997;272:2021–2030. doi: 10.1074/jbc.272.3.2021. [DOI] [PubMed] [Google Scholar]
- Steven AC, Bisher ME, Roop DR, Steinert PM. Biosynthetic pathways of filaggrin and loricrin elucidated by immuno-labeling of newborn mouse epidermis. J Struct Biol. 1990;104:150–162. doi: 10.1016/1047-8477(90)90071-j. [DOI] [PubMed] [Google Scholar]
- Steven AC, Steinert PM. Protein composition of cornified cell envelopes of epidermal keratinocytes. J Cell Sci. 1994;107:693–700. [PubMed] [Google Scholar]
- Swartzendruber DC, Wertz PM, Madison KC, Downing DT. Evidence that the corneocyte has a chemically bound lipid envelope. J Invest Dermatol. 1987;88:709–715. doi: 10.1111/1523-1747.ep12470383. [DOI] [PubMed] [Google Scholar]
- Watt F, Green H. Involucrin synthesis is correlated with cell size in human epidermal cell culture. J Cell Biol. 1981;90:738–742. doi: 10.1083/jcb.90.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Keeble S, Fisher C, Hudson DL, Codd J, Salisbury JR. Onset of expression of peanut lectin-binding glycoproteins is correlated with stratification of keratinocytes during human epidermal development in vivo and vitro. J Cell Sci. 1989;94:355–359. doi: 10.1242/jcs.94.2.355. [DOI] [PubMed] [Google Scholar]
- Wertz PW, Madison KC, Downing DT. Covalently bound lipids of human stratum corneum. J Invest Dermatol. 1989;92:109–111. doi: 10.1111/1523-1747.ep13071317. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Beegen H, Eckert RL. Biophysical characterization of involucrin reveals a molecule ideally suited to function as an intermolecular cross-bridge of the keratinocyte cornified envelope. J Biol Chem. 1992;267:12233–12238. [PubMed] [Google Scholar]
- Yaffe MB, Murthy S, Eckert RL. Evidence that involucrin is a covalently linked constituent of highly purified cultured keratinocyte cornified envelopes. J Invest Dermatol. 1993;100:3–9. doi: 10.1111/1523-1747.ep12349857. [DOI] [PubMed] [Google Scholar]
- Zahn H, Gattner H-G. Hair sulfur amino acid analysis. In: Jolles P, Zahn H, Hocker H, editors. Formation and Structure of Human Hair. Basel: Birkhauser Verlag; 1997. pp. 239–258. [DOI] [PubMed] [Google Scholar]