Abstract
Upon integration into the host chromosome, retroviral gene expression requires transcription by the host RNA polymerase II, and viral messages are subject RNA processing events including 5′-end capping, pre-mRNA splicing, and polyadenylation. At a minimum, RNA splicing is required to generate the env mRNA, but viral replication requires substantial amounts of unspliced RNA to serve as mRNA and for incorporation into progeny virions as genomic RNA. Therefore, splicing has to be controlled to preserve the large unspliced RNA pool. Considering the current view that splicing and polyadenylation are coupled, the question arises as to how genome-length viral RNA is efficiently polyadenylated in the absence of splicing. Polyadenylation of many retroviral mRNAs is inefficient; in avian retroviruses, ∼15% of viral transcripts extend into and are polyadenylated at downstream host genes, which often has profound biological consequences. Retroviruses have served as important models to study RNA processing and this review summarizes a body of work using avian retroviruses that has led to the discovery of novel RNA splicing and polyadenylation control mechanisms.
Keywords: retroviruses, RNA splicing, Spliceosome, snRNP, SR protein, hnRNP H, Polyadenylation, Review
2. INTRODUCTION
2.1. Retroviruses and RNA processing
Retroviruses employ a unique replication scheme in which a long, single-stranded RNA genome is converted into a double-stranded DNA molecule that is inserted into and becomes a permanent resident of the host genome (reviewed in (1, 2)). From the chromosomal position, the integrated viral DNA (the provirus) is transcribed by the host RNA polymerase II (pol II) to generate genome-length viral RNA that has the same modifications as typical host mRNAs (a 5′ cap and a 3′ poly(A) tail). A portion of this full-length viral RNA is packaged into progeny virions, and an additional pool is translated into structural and enzymatic proteins that compose the virus particles. However, some viral proteins are synthesized from spliced transcripts, so the primary transcript also serves as a substrate for RNA splicing. The number of spliced mRNA species can be quite large, as is the case for complex retroviruses like human immunodeficiency virus (HIV) (see (3) and a review by M. McLaren, K. Marsh, and A. Cochrane in this series). Clearly, the extent of splicing must necessarily be controlled to preserve the genome-length RNA, which typically represents ∼50% or greater of the total. Another issue raised by the recent appreciation that splicing and polyadenylation are coupled is how the full-length viral RNA is efficiently polyadenylated in the absence of splicing. This review focuses on the progress made in understanding RNA processing control in the simple retrovirus, Rous sarcoma virus (RSV), and the implications for the processing of cellular mRNAs.
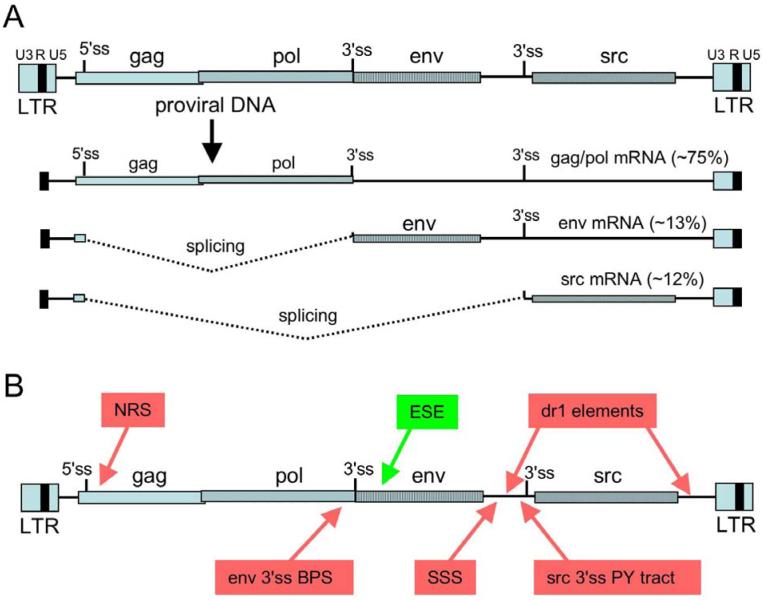
RSV is a member of the avian sarcoma/leucosis virus (ASLV) family and contains the gag, pol, and env genes common to all retroviruses (Figure 1A). The gag gene encodes a polyprotein that is processed into virion core components, pol encodes the reverse transcriptase and integrase enzymes, and the env gene codes for the envelope glycoprotein that is embedded in the virion membrane. In addition, RSV harbors the src gene that is responsible for cell transformation and is thus a replication-competent oncovirus. Upon entering the cell, the reverse transcriptase that enters with the infecting virion converts the RNA genome into a dsDNA and upon accessing the host DNA, the integrase enzyme inserts the viral DNA into the host genome. The reverse transcription process duplicates the U3 and U5 regions such that long terminal repeats (LTRs) are formed at each end of the provirus in the order U3-RU5 (Figure 1A). Transcription factors recognize promoter elements within U3 and initiate pol II transcription at the beginning of R within the 5′ LTR, and full-length RNA results from 3′-end processing at the end of R in the 3′ LTR (discussed in more detail below). Much of the RNA remains completely unspliced and is exported to the cytoplasm where it serves as an mRNA for the gag/pol genes, and is incorporated into progeny virions. Within the nucleus, the primary transcript is also a substrate for splicing to generate the sub-genomic env and src mRNAs (Figure 1A), which are generated in approximately equal amounts (10% to 15% each). The accumulation of appropriate quantities of genome-length RNA is crucial for replication success and therefore, understanding how splicing is controlled has been the subject of much work. Below I review the mechanisms of splicing control in RSV and the current view of how 3′-end formation and splicing are integrated to ensure the proper processing of full-length viral RNA.
Figure 1.
Elements that influence RSV RNA processing. A) Structure of the RSV provirus and mRNA species. Shown are the LTRs at the termini of the provirus with the U3, R (black box), and U5 regions indicated. The gag, pol, env, and src gene coding regions are shown with different patterned boxes, and the 5′ and 3′ splice sites are indicated. Transcription starts at the beginning of the 5′ R region, and RNAs are cleaved and polyadenylated at the end of the 3′ R region. The relative percentages of unspliced, env, and src mRNA are indicated, and splicing is depicted by dotted lines. B) Summary of cis elements that control splicing in RSV. The schematic of the proviral DNA is as in Figure 1. Red boxes indicate negative-acting elements and include: the negative regulator of splicing, or NRS; the suboptimal BPS associated with the env 3′ ss; the suppressor of src splicing (SSS) that specifically represses src splicing; dr1, the direct repeat elements that flanks the src gene and specifically repress src splicing; the suboptimal pyrimidine tract associated with the src 3′ ss. Splicing to env is also controlled by a positive-acting exonic splicing enhancer (green). Adapted and updated from (20).
2.2. pre-mRNA splicing pathways
Most genes in higher eukaryotes are interrupted by sequences (introns) that must be removed from precursor-mRNA (pre-mRNA), and the functional portions (exons) spliced together to form the mature mRNA (reviewed in (4, 5)). RNA splicing is accomplished by a large macromolecular machine termed the spliceosome that recognizes the exon/intron boundaries and executes the splicing reaction. Typically, metazoan genes contain multiple introns and the fact that a large number of host mRNAs undergo alternative splicing (the differential inclusion / exclusion of exons) (5) increases the challenge of understanding how splice junctions are recognized and paired to generate various spliced isoforms. Since viral RNAs are spliced by the host spliceosome and several retroviruses utilize alternative splicing for production of mRNA (3), these issues are relevant for understanding retroviral splicing control.
The initial function of the spliceosome is to identify the splice junctions that are to be joined together. This is accomplished through the identification of conserved but degenerate splicing signals at the exon/intron boundaries, the 5′ and 3′ splice sites. The 5′ splice site (ss) has a short consensus sequence whereas recognition of the 3′ ss requires the branchpoint sequence (BPS) and pyrimidine (PY) tract. The vast majority of cellular introns are spliced by a spliceosome that that contains five small nuclear ribonucleoprotein (snRNP) particles (U1, U2, U4, U5 and U6) and a large number of proteins (4, 6). The small nuclear RNA (snRNA) components of the snRNPs make extensive base pairing interactions with the premRNA as well as each other. The 5′ ss is initially recognized by U1, and the PY tract associated with the 3′ ss is bound by U2 auxiliary factor (U2AF), which is required for subsequent binding of U2 to the BPS. It is thought that splice site pairing is determined at this early point and that regulating alternative splicing involves modulating these early steps (7). This pre-spliceosome is then joined by the U4/U6.U5 tri-snRNP to form a mature complex in which complex rearrangements occur between the snRNA and substrate, and between the snRNAs, to deliver the splice sites to the active site of the spliceosome. Crucial to this process is a switch in 5′ ss base pairing interaction from U1 to U6 snRNA, and a structure formed between U6 and U2 snRNP is thought to carry out catalysis (4, 8, 9). The splicing reaction itself involves two sequential transesterification reactions that cleave the splice sites, join the exons, and release the intron as a lariat. A second spliceosome was described in 1996 that excises a rare class of introns whose splice sites are highly conserved and deviate substantially from conventional introns (10, 11). Removal of these introns is remarkably similar to the ‘major’ pathway but utilizes a unique set of snRNPs (U11, U12, U4atac, U6atac) that perform analogous functions to the snRNPs involved in conventional splicing. U5 snRNP and numerous other splicing factors are shared between the two splicing pathways (12). As will be discussed below, both pathways influence RSV splicing control.
3. CONTROL OF RSV RNA SPLICING: THE SPLICE SITES
Splicing control in RSV involves the maintenance of suboptimal splice sites and the regulation of splice site use by a positive element and several negative-acting elements. These control regions are summarized in Figure 1B and discussed below.
3.1. The env 3′ ss is suboptimal
One of the mechanisms by which splicing is controlled in RSV is the maintenance of suboptimal splicing signals. Presumably, a population of viral RNAs is able to escape splicing by going unrecognized by the splicing apparatus. While it appears that the 5′ ss is not involved in splicing control (13, 14), evidence for a suboptimal env 3′ ss stemmed from the examination of a virus in which a 24 nt oligonucleotide was inserted 12 nt upstream of the env 3′ ss (15). This virus showed a marked delay in replication that correlated with substantial oversplicing to the env 3′ ss. Upon longer-term passage, a class of phenotypic revertants was isolated that had mutations in the original insert that lowered splicing to wild-type levels. The results were consistent with the replication defect stemming from a paucity of unspliced RNA. A second class of revertants was identified that had deletions downstream of the env 3′ ss in the env exon (14). In the context of a wild-type virus, the same deletions caused replication defects and resulted in very little env splicing, which suggested that the deleted sequences played a positive role in env splicing. It is now known that the deletion eliminated an RNA splicing enhancer (Figure 1B).
Examination of the mutations in an in vitro splicing assay showed that env RNA substrates containing the 24 nt insertion were spliced ∼5-fold better than the wild-type and revertant RNAs, consistent with what was observed in the virus experiments (16). The effect of the insertion was explained by the finding that the insertion created a new, efficiently used BPS that increased env splicing; point mutation suppressors acted directly on this BPS to either reduce splicing or decrease the second step of splicing, whereas the exon deletions blocked splicing before the first step and most likely at an early splice site recognition step, consistent with the deletion of a splicing enhancer. The latter point was confirmed with the in vitro demonstration that env splicing substrates lacking the splicing enhancer failed to form any spliceosomal complexes (17). Interestingly, suppressors defective in the second splicing step in vitro showed a similar pattern in vivo, and the suggestion was made that splicing intermediates play a role in viral splicing regulation through the differential binding of U2AF to the PY tract, and the spliceosomal protein SAP49 to the BPS region (17, 18). Collectively, the work suggests that in the wild-type virus, the PY tract associated with the env 3′ ss is functional and that the efficiency of env 3′ ss use is controlled by the competing activities of poor BPS recognition and a splicing enhancer located a short distance downstream (Figure 1B).
3.2. The src 3′ ss is suboptimal
Splicing to the src 3′ ss in RSV is also very inefficient and considerable work supports the notion that, like env, the src 3′ ss is suboptimal. However, in contrast to the env 3′ ss, it is the PY tract associated with the src 3′ ss that accounts in part for its inefficient use (Figure 1B). The 14 nt PY tract is interrupted by five purines, which were postulated to play a role in the regulation of src splicing (19). When the src PY tract was mutated to an uninterrupted stretch of 14 pyrimidines, splicing to the src 3′ ss increased to ∼54%, compared to ∼16% for wild type, and there was a corresponding decrease in unspliced RNA. The improved splicing also activated a cryptic 5′ ss within the env gene. The increased splicing correlated with slower replication kinetics for the mutant virus, consistent with the unspliced RNA being limiting for replication. Some viruses that arose upon continued passage of the oversplicing mutants had deletions of the src 3′ ss or the src gene that restored high levels of genome-length RNA. However, an additional population of revertants appeared to produce full-length unspliced RNA and upon examination, they contained small deletions in the putative BPS or in the improved PY tract, and they showed decreased splicing to src and normal replication kinetics (20). It was also shown in vivo that the src 3′ ss is inefficiently used when paired with a quality 5′ ss (13). Thus, in addition to the weak env 3′ ss, RSV replication also requires the maintenance of a weak src 3′ ss.
4. CONTROL OF RSV RNA SPLICING: NEGATIVE ELEMENTS DISTINCT FROM THE SPLICE SITES
4.1. The suppressor of src splicing (SSS)
In addition to suboptimal 3′ splice sites, RSV harbors an element upstream of the src 3′ ss that regulates splicing specifically at that site (Figure 1B). The presence of such an element was originally suggested from experiments to determine the reason for different levels of src splicing observed between different ALV strains (21). It was found that a 262 bp region between the env and src genes could confer the low src splicing efficiency of the PrC strain on a PrA strain, suggesting the presence of a negative element in this region. Surprisingly, only 4 differences were noted between strains, two in the intron and two in the src exon. When unidirectional deletions from the 5′ end were made into this region, src splicing increased approximately two-fold with a concomitant decrease in unspliced RNA (22). There was little change in env splicing, indicating that the repressive sequence is specific to the src 3′ ss. This sequence, called the suppressor of src splicing (SSS), was also shown to function additively with but independently of the suboptimal src PY tract (20) and to block splicing of a heterologous intron in a position-dependent manner (13).
RSV RNA splicing in mammalian cells is quite different than in chicken cells. In NIH3T3 or human fibroblast cells, very little env splicing was detected whereas ∼50-60% of the RNA represented spliced src transcripts (23, 24). These results suggested that mammalian cells might lack a negative factor required for proper splicing control, which could in part explain the nonpermissive nature of these cells for RSV replication. Subsequent work with wild-type and SSS mutant viruses showed a similar, high level of src splicing in human fibroblasts, consistent with a lack of SSS function in those cells (24). Furthermore, using an in vitro splicing system derived from HeLa cells, there was no difference in splicing of src minigenes containing or lacking the SSS, but SSS-specific repression was observed upon addition of chicken cell extract. These data suggested that an inhibitory factor present in chicken extracts, but not mammalian extracts, is responsible for splicing regulation by the SSS. However, the identity of the factor and the mechanism of action remain to be elucidated.
4.2. Negative effects of direct repeat elements on src splicing
A second cis element that may repress src splicing was identified as the dr1 direct repeat element that flanks the src gene (25) (Figure 1B). These elements also act as constitutive transport elements (CTEs) and promote the accumulation of unspliced viral RNA in the cytoplasm (26-28). Deletion of both elements causes severe replication defects characterized by increased turnover of unspliced RNA, reduced unspliced RNA in the cytoplasm, and poor particle production (29). Deletion of either repeat causes a delayed replication phenotype (27). Curiously, mutation of either dr1 element caused an increase in src mRNA but not env mRNA, which suggested that both direct repeats contribute to a specific repressive effect on src splicing (25, 29). An indirect effect of the mutations on splicing by compromising CTE activity to increase the nuclear pool of unspliced RNA splicing substrate is possible but seems unlikely given that env mRNA was not elevated. It was speculated that factors that bind to the repeats might loop out the RNA and influence splicing factor binding at the src 3′ ss (25). Understanding the mechanism by which the direct repeats repress src splicing deserves additional attention.
4.3. The negative regulator of splicing (NRS)
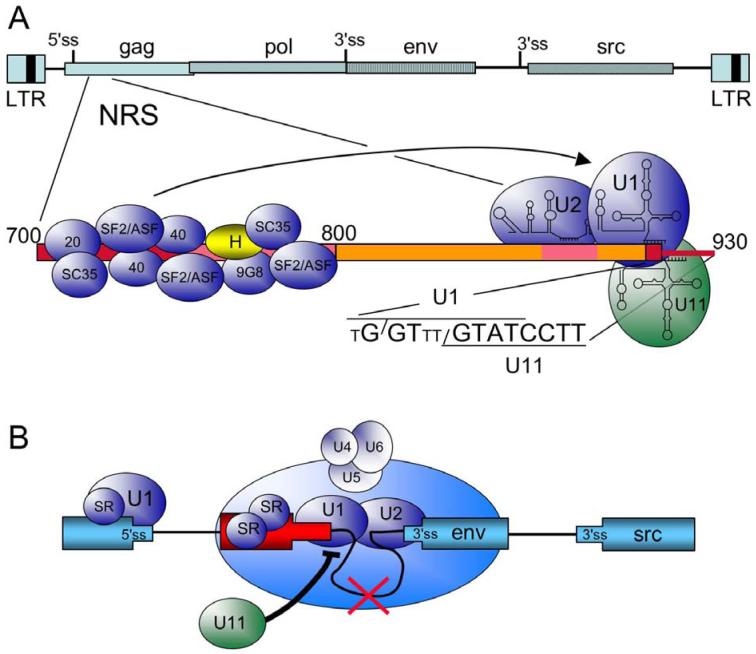
A third element in RSV that is distinct from the splice sites and serves to repress splicing is the well-characterized negative regulator of splicing (NRS) (Figures 1B and 2A). It was originally observed that deletions in the gag gene caused elevated env and src mRNA levels (22, 30, 31) and a concomitant decrease in unspliced RNA, although a more recent report suggested a predominant effect on src splicing (32). These results suggested that a splicing suppressor resided in the gag gene. The repressive element was also able to block splicing of a heterologous myc intron (30) and was localized to a ∼230 nt region between nts 700 and 930 in the RSV genome (33). It is important to note that the element is ∼300 nt from the 5′ ss and ∼4,000- and ∼6,000 nt from the env and src 3′ splice sites, respectively. Additional mapping studies indicated a bipartite nature to the NRS; splicing inhibition required an upstream purine rich region (nts 700 to 800, referred to as NRS5′) and a discrete sequence located ∼120 nt away in the downstream region (NRS3′, nts 801-930) (Figure 2A) (33). The presence of splice site-like sequences in NRS3′ suggested that components of the splicing machinery were involved in the inhibition process.
Figure 2.
Summary of cis elements and trans factors important for NRS splicing control. A) At top is a schematic of the RSV proviral DNA (as in Figure 1) and below is an expansion of the NRS (nt 700 to 930). Regions important for NRS splicing inhibition are boxed in red and pink. NRS5′, from nt 700 to 800, binds the indicated SR proteins (blue) and hnRNP H (yellow). U1, U2, and U11 snRNPs bind to the distal end of NRS3′ (nts 800 to 930). The sequence of the degenerate U1 binding site (small T represents non-consensus residues) and the perfect U11 binding site are shown. Slashes indicate the splice site for each sequence. U2 snRNP most likely binds to an upstream BPS-like sequence (not shown). The arrow indicates that the SR protein promote U1 binding to NRS3′. B) Model for NRS splicing inhibition. In the schematic, the RSV exons are in blue, and the NRS is in red. SR proteins promote U1 binding to the authentic 5′ ss and the NRS, setting up a competition for interactions with the 3′ splice sites (env in this example). As detailed in the text, NRS-bound U1 interacts with the 3′ ss to form an early splicing complex, which then matures into a non-catalytic spliceosome-like complex upon the addition of the U4/U6.U5 tri-snRNP, which is not correctly integrated into the complex. Splicing from the NRS fails (red X), and the complex sequesters the 3′ ss from a productive interaction with the authentic 5′ ss. U11 binding would block U1 binding to the overlapping site (thick black line), preventing assembly of the NRS-3′ss complex, and allowing normal splicing to occur.
4.3.1. snRNPs bind to NRS3′
In vitro approaches have provided important insights into the mechanism of NRS-mediated splicing inhibition. The NRS blocked splicing of an adenovirus splicing substrate when located in the intron (34) but not when inserted upstream or downstream of either exon (M.T. McNally, unpublished data). Inhibition occurred before the first step of splicing, but larger than normal splicing complexes were detected that contained the full complement of U2-dependent snRNPs plus two additional snRNPs (34). However, integration of the U4/U6.U5 tri-snRNP was faulty in that it could be dissociated by heparin treatment (34, 35). This aberrant incorporation of the tri-snRNP likely explains why splicing does not occur from the NRS (discussed below). It was subsequently shown that the NRS itself binds U1 and U2 snRNPs, and further characterization identified U11 and U12 snRNPs as NRS-binding factors (Figure 2A). As discussed above, U11 recognizes 5′ splice sites within U12-dependent introns, and early work suggested that U11 was directly involved in NRS splicing inhibition (34). However, it was later shown that U11 is dispensable for splicing inhibition but that U1 binding to a degenerate, overlapping site within NRS3′ was essential for inhibition (36, 37). A clear role for U2 snRNP in NRS activity remains to be established. Collectively, the data suggested that the NRS might represent a novel decoy 5′ ss that somehow subverts the splicing machinery to block splicing. As detailed below, this appears to be the case.
4.3.2. SR proteins bind to NRS5′
While splice site-like sequences were an indication that snRNPs might interact with NRS3′, no obvious clues to the nature of factors that might interact with NRS5′ were evident from the sequence. Cross-linking analyses showed that several members of the SR protein family of splicing factors interacted with NRS5′, including SRp20, ASF/SF2, and proteins whose size was consistent with SRp40 and SRp55 (38) (Figure 2A). A yeast 3-hybrid screen also identified 9G8 and SC35 as NRS-binding proteins (39). These findings were consistent with the observations that SR proteins often interact with purine rich sequences, of which NRS5′ is ∼70% purines. SR proteins are a family of structurally and functionally related proteins that contribute to splice site selection and many steps in spliceosome assembly (40, 41). One well-documented property of SR proteins is that they bind exonic splicing enhancers (ESE) and promote splicing through recruitment of components of the splicing machinery, including snRNPs, to splice sites (42, 43). As would be expected of an extensive SR protein binding platform, NRS5′ functioned as a potent splicing enhancer in an ESE assay, and bona fide ESEs could replace NRS5′ and support NRS splicing inhibition (44). These observation pointed toward a role for the SR proteins in recruitment of U1 to the degenerate 5′ ss in NRS3′, an idea that was confirmed in vitro (36) and which further suggested that the NRS functions as a pseudo 5′ ss to block splicing. This model predicted that the NRS (through U1 snRNP) would interact with and form a complex with a 3′ splice site.
4.3.3. Interaction of the NRS with a 3′ ss; the NRS complex
The findings that SR proteins, U1 snRNP, and the U1 binding site are required for NRS splicing inhibition strengthened the notion that the NRS is recognized as a pseudo 5′ ss and predicted that the NRS itself would assemble into complexes similar to those formed on authentic 5′ splice sites. Using gel filtration chromatography, it was demonstrated that the NRS alone assembles into a complex that is indistinguishable from the early (E′) complex that forms on isolated 5′ splice sites (45, 46). Furthermore, the complex appeared functionally relevant since there was a correlation between the sequence requirements for assembly in vitro and splicing inhibition in vivo, and assembly of the complex required SR proteins, the U1 binding site, and U1 snRNP (47). It was subsequently shown that a chimeric substrate composed of the NRS fused to an adenovirus 3′ ss (i.e., a substrate designed to detect an NRS - 3′ ss interaction) formed a large ATP-independent complex whose assembly required the NRS U1 binding site and the branchpoint and pyrimidine tracts associated with the 3′ ss (48). These data were consistent with an interaction between the NRS pseudo 5′ ss and the adenovirus 3′ ss, and with formation of a complex with characteristics of a bona fide spliceosomal E complex.
Despite initiating spliceosome assembly, the NRS-Ad3′ substrate is not catalytically active in vitro ((35) and M.T. McNally, unpublished data), suggesting that events subsequent to E complex formation are defective. This idea is supported by a recent publication providing evidence that under splicing conditions, the NRS-Ad3′ substrate assembles a 50S splicing complex with the full complement of spliceosomal snRNPs (plus U11), but the U4/U6.U5 tri-snRNP is not stably integrated into the complex (35). This result is similar to what was observed by Gontarek et al. (34) when the NRS was inserted into the adenovirus intron. It is this stalled spliceosome, formed between the NRS and a viral 3′ ss, that presumably accounts for splicing inhibition by sequestering the 3′ss and preventing it from interacting with the authentic viral 5′ ss (Figure 2B).
4.3.4. The NRS pseudo 5′ ss
It is still not understood why splicing fails to occur from the NRS U1 binding site, despite the binding of U1 snRNP and assembly of a spliceosome-like complex. One possibility is that, despite the poor 5′ ss consensus sequence, U1 snRNP binds the NRS irreversibly. This would block subsequent base pairing of U6 to the pseudo 5′ ss and prevent catalysis. However, this is not supported by evidence that U1 binding to the NRS is reversible in vitro (M.T. McNally, unpublished data). A second possible mechanism stems from the observation of a 12/13 nt complementarity between the NRS and the region of U6 snRNA that normally binds to the 5′ ss. This potentially hyperstable interaction is predicted to block subsequent U6/U2 contacts required for catalysis. While attractive, the hyperstable model is not supported by the finding that mutations that would disrupt the extensive U6/NRS interaction have no effect on NRS splicing inhibition (e.g., (36) and M.T. McNally, unpublished data). Currently, the thought is that the sequence of the NRS pseudo 5′ ss itself, or its structure (49), accounts for the lack of splicing from the NRS and splicing inhibition. The sequence of the pseudo 5′ ss is highly underrepresented among human 5′ splice sites ((50), and M.T. McNally and B. Tian, unpublished data) and it has a poor score as assessed by the MaxEntScan software for 5′ ss quality (51). Interestingly, in support of the importance of the unique sequence of the NRS 5′ ss in splicing inhibition are the observations that mutation of the non-consensus uridines (Figure 2A) to consensus adenosine converts the NRS to a functional U2-dependent intron, whereas mutations to other bases largely inactivate the NRS (36, 50).
The finding that U1 is not irreversibly bound to the NRS pseudo 5′ ss suggests that the lack of splicing is explained by a post-U1 binding event. A role for U6 snRNP in splicing inhibition is suggested from experiments showing that NRS mutations in the +5 G (which affects U1 and U6 binding) that inactivate the NRS can be partially rescued by expression of compensatory U6 snRNA alone, and rescue is increased with both U1 and U6 compensatory RNA. Likewise, splicing from an NRS that is mutated toward the 5′ ss consensus can be partially suppressed by compensatory U6 snRNA (M.T. McNally, unpublished data). Future experimentation is required to more fully understand how the NRS assembles into a spliceosome in which aberrant snRNA interactions lead to a non-catalytic complex.
4.3.5. U11/U12 snRNP binding to the NRS
One of the more surprising initial findings was that the NRS efficiently binds U11 and U12 snRNPs (the factors that recognize the 5′ and 3′ splice sites of U12-dependent introns, respectively, despite being ∼100-fold less abundant than U1 snRNP (34, 52). While U11 can bind the NRS independently of U12 (53) and it is clear that U11 is not directly involved in splicing suppression, U11 still plays a regulatory role in NRS splicing control by modulating U1 binding. It was observed that mutations in the U11 site abolish U11 binding in vitro but increase U1 binding ∼4 fold, and such NRS mutants block splicing in cells more efficiently than wild type (36, 37). Since the U1 and U11 binding sites overlap (Figure 2A), U11 binding would prevent the U1 interaction and transcripts would not enter into the inhibitory NRS complex. Curiously, the NRS binds U11 much more efficiently in vitro than authentic U12-dependent splicing substrates (53), which prompted studies to understand why this is so. In contrast to U1, the high-affinity SR protein binding sites in NRS5′ are not required for U11 binding (M.T. McNally, unpublished data).
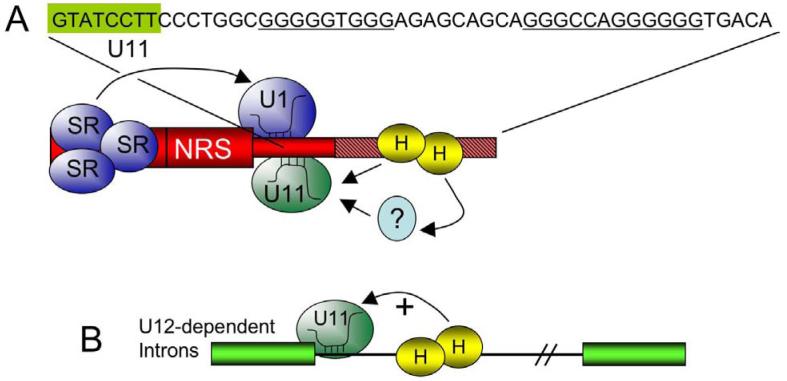
Subsequent efforts to identify NRS features that account for the high efficiency U11 binding showed a prominent role for a G-rich element just downstream of the U11 site (Figure 3A) (53). Mutations in the G tracts strongly reduced U11 binding in vitro as measured in RNA pull-down experiments, and also in vivo as measured with a heterologous splicing assay. In this assay, the NRS is fused to a U12-dependent 3′ss where the NRS U11 5′ ss site is used efficiently and accurately; splicing thus serves as a reporter for U11 binding in vivo (36). The G tracts were shown to bind heteronuclear ribonucleoprotein (hnRNP) H, and two lines of evidence suggested that hnRNP H mediates U11 binding via the G tracts. First, U11 binding was decreased in vitro when hnRNP H was depleted from extracts, and U11 binding could be partially rescued upon addition of recombinant hnRNP H (54). Second, NRS splicing to a U12-dependent 3′ ss in cells was abolished when the hnRNP H sites were deleted, but splicing could be restored by tethering hnRNP H (but not other proteins) to the RNA substrates as an MS2-fusion protein (54). These studies clearly demonstrated that hnRNP H mediates the efficient binding of U11 to the NRS and suggested that hnRNP H might more broadly influence U12-dependent splicing of host genes. This proved to be the case for two introns that contain hnRNP H bindings sites just downstream of the U11 5′ ss (P120 and SCN4A introns), and examination of the 404 U12-dependent introns known at the time showed that ∼18% harbor potential hnRNP H sites downstream (Figure 3B). Thus, RSV has usurped hnRNP H to ensure efficient U11 binding to the NRS. It should be noted that hnRNP H also regulates alternative splicing of numerous other viral and cellular U2-dependent introns, often by counteracting the effects of negative-acting factors such as hnRNP A1 (55-60).
Figure 3.
hnRNP H promotes U11 binding to the NRS. A) The sequence of the U11 binding site (boxed green) and the downstream G-rich region (underlined) is shown above a schematic of the NRS. The NRS, shown in red, is depicted with SR proteins promoting U1 binding (arrow). The G-rich region (hatched pink) binds hnRNP H (yellow H) that in turn promotes U11 binding (arrows). The interaction might direct or through another protein (?). B) The observation that hnRNP H promotes U11 binding to the NRS led to the finding that a subset of U12-dependent cellular introns harbor G tracts requires hnRNP H for U11 binding (arrow) and splicing.
5. POLYADENYLATION OF RSV RNA
5.1. Retroviruses and polyadenylation
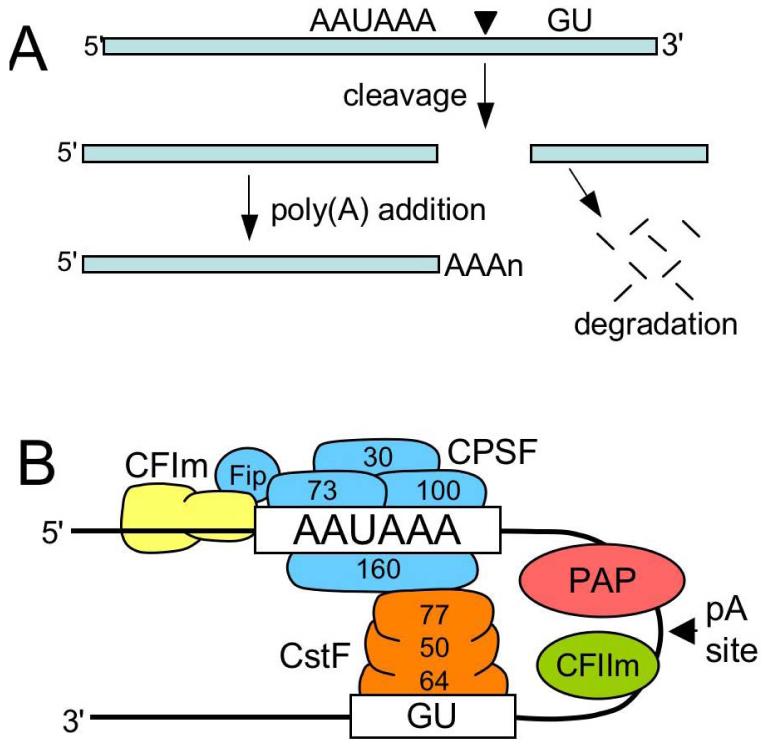
Most cellular mRNAs undergo 3′-end processing reactions involving cleavage of the RNA at its eventual 3′ terminus and addition of a poly(A) tail (Figure 4A). The polyadenylation process is intimately coupled to other RNA processing reactions, including 5′-end capping and splicing, which ensures efficient and faithful processing of the mRNA (61). Retroviral mRNA is also polyadenylated by the host cell machinery but because the poly(A) signals are present within the LTRs at each end of the provirus, use of the 5′ poly(A) site must be suppressed (1, 3). In addition, polyadenylation in many retroviruses is inefficient and viral transcripts terminate at poly(A) sites within downstream cellular genes. This is important for the incorporation of cellular sequences into retroviral genomes and for activation of downstream cellular genes, which in the case of oncogenes results in tumorigenesis (1). The issue of poly(A) site choice is trivial in avian retroviruses because the major poly(A) signal (AAUAAA) is upstream of the repeat sequence (transcription start site) and thus is only present at the 3′ end of the transcript. Polyadenylation efficiency is also low in RSV compared to cellular genes in that ∼15% of the viral transcripts represent read-through RNA (62), and the extent of read-through can have profound consequences on tumorigenesis in infected animals (63, 64). Recent studies have led to a new understanding of polyadenylation control in RSV and have revealed a novel mechanism of coupled splicing-polyadenylation control that involves long-distance interactions between the NRS, the env 3′ss, and the poly(A) site.
Figure 4.
Polyadenylation pathway and factors. A) Poly(A) addition occurs by endonucleolytic cleavage of the RNA between the AAUAAA and GU-rich elements at the eventual poly(A) addition site (black triangle), and subsequent addition of the poly(A) tail to the 5′ cleavage product. The downstream fragment is degraded. B) Schematic of polyadenylation factors. The multi-subunit CPSF (blue, molecular size of subunits indicated) is shown interacting with the AAUAAA signal, as are components of CstF (orange) bound to the GU-rich element. CFIm assists in CPSF binding via hFip. PAP, and CFIIm are also shown.
5.2. Polyadenylation and coupling to splicing
For a simple cleavage and poly(A) addition mechanism of 3′-end formation, an unexpectedly large number of proteins are dedicated to polyadenylation of mRNA (65) (Figure 4B), which likely reflects the fact that polyadenylation is often regulated. The primary signals for poly(A) site use are the AAUAAA signal present ∼10-30 nt upstream of the cleavage site (site of poly(A) addition) and a U- or GU-rich element ∼30 nt downstream that is referred to as the downstream element (DSE) (Figure 4B). In addition, numerous studies have identified upstream elements (USEs) that can contribute to polyadenylation efficiency, and recent bioinformatics analyses revealed a variety of conserved elements associated with poly(A) sites that might be involved in constitutive or regulated polyadenylation (66). The AAUAAA signal is recognized by a multi-subunit complex called the cleavage and polyadenylation specificity factor (CPSF), which binds cooperatively with another multi-subunit factor (cleavage stimulatory factor, or CstF) that interacts with the downstream DSE (Figure 4B). Another factor called cleavage factor I (CFIm) binds to discrete sequences near the poly(A) site and contributes, via Fip1, to CPSF binding and recruitment of poly(A) polymerase (PAP)(67). With the help of additional cleavage factors, the RNA is cleaved at the poly(A) site by a component of CPSF and the poly(A) tail is added by PAP.
Early studies by Niwa and Berget (68, 69) showed that polyadenylation in vitro is stimulated by an upstream intron, and that mutation of the poly(A) signal depresses splicing of a terminal intron. These studies suggested a link between splicing and polyadenylation, and it has become quite clear in recent years that the splicing and poly(A) machineries are functionally linked. Numerous interactions between splicing and polyadenylation factors have been described that are thought to mediate the coupling process. These include interactions between the U1 snRNP and CFIm (70), the U1 snRNP A (U1A) protein and CPSF (71), U2 snRNP-associated SF3b proteins and CPSF (72), SRm160 and CPSF (73), and U2AF65 and both PAP and CFIm (74). In the latter case, it was shown that the RS-like domain of U2AF65 directly mediates coupling via an interaction with a similar alternating charge domain present in the CFIm 59-kD subunit. Given the benefits of splicing on polyadenylation of cellular mRNAs, the question arises as to how genome-length (i.e., unspliced) retroviral mRNA is efficiently polyadenylated in the absence of splicing. Recent work has revealed an interesting solution to this problem.
5.3. A role for the NRS in RSV polyadenylation
5.3.1. The RSV poly(A) site is inherently weak
The observation that ∼15% of RSV transcripts fail to use the viral poly(A) site suggests that the viral site is weak. To address this issue, the RSV poly(A) site was characterized in an in vitro polyadenylation assay and found to be poorly used compared to the well-characterized and efficiently used SV40 late poly(A) site (75, 76). Examination of the sequences surrounding the RSV poly(A) suggested that the poor in vitro activity stemmed in part from a lack of quality USE and DSEs. The RSV AAUAAA and cleavage site was shown to be functional when either the RSV USE or DSE regions were replaced by the analogous regions from SV40, and vice versa (N.L. Maciolek and M.T. McNally, submitted). These results suggested that the RSV elements were functional but suboptimal and also indicated that the poor poly(A) activity observed for RSV was not due to the presence of an inhibitory element. Further work showed that one of the major deficiencies of the RSV substrate is the lack of a quality DSE and very inefficient binding of CstF. The RSV substrate could be activated when the downstream region was replaced with MS2 binding sites and an MS2-CstF fusion protein (but not control MS2 fusions) was provided (N.L. Maciolek and M.T. McNally, submitted). These results in part explain the relatively high level of read-through transcripts in RSV, but also suggest that additional cis elements are present within the RSV RNA that stimulate use of the weak poly(A) site.
5.3.2. The NRS is required for optimal RSV polyadenylation
The first indication that distant cis elements were required for RSV polyadenylation came from the work of Miller and Stoltzfus (77) who showed that two distinct upstream regions were required for wild-type poly(A) levels. Deletions that encompassed the env 3′ ss increased read-through transcripts, which suggested that conventional coupling via 3′ ss-binding factors might promote 3′-end processing. However, because the src 3′ ss was intact in these mutants, and deletion of the src 3′ ss was without effect, the mere presence of a 3′ ss was insufficient to promote polyadenylation. The second region was located in the gag gene and encompassed the NRS, and subsequent mutagenesis work directly implicated the NRS in poly(A) control since specific NRS mutations led to read-through transcripts (32, 39). Thus, it appeared that the NRS and the env 3′ ss region were both required for proper polyadenylation in RSV. Fogel and McNally (39) also showed that deletions in the SR protein binding region or mutation of the snRNP binding sites decreased poly(A) efficiency, which suggested that no single NRS-binding factor was responsible for NRS-mediated polyadenylation stimulation. This led to a model in which the NRS complex (formed through the interaction of the NRS with a viral 3′ ss) stabilized the binding of one or more splicing factors to the unspliced RNA, and these factors mediated coupling via one of the mechanisms described above (39).
Evidence of an important role for the NRS in splicing and polyadenylation control also comes from animal studies that examined the effects of NRS mutations on infection and disease. It was observed that a recombinant, non-acute ALV (EU-8) caused rapid-onset B-cell lymphomas in infected chickens, and tumors had proviral integrations upstream of or within the first intron of the c-myb gene (78). It was further shown that chimeric mRNA was produced by read-through transcription and splicing from the viral 5′ ss to the 3′ ss of c-myb exon 2. Production of a truncated c-myb protein strongly correlated with rapid-onset tumors, and aberrant splicing of read-through transcripts was required for the effect. The determinant for tumorigenicity in EU-8 was shown to be a 42 nt deletion within the SR protein-binding region of the NRS, and the Δ42 NRS was partially compromised in a splicing inhibition assay (64). A subsequent study showed that impaired NRS function rather than matrix protein alterations caused the increase in tumors, since viruses harboring silent point mutations that preserved the gag coding region but inactivated the NRS showed an increase in short-latency lymphomas (63). Significantly, these viruses also demonstrated an increase in read-through transcripts. These studies highlight the important biological significance of a functional NRS in ALV splicing and polyadenylation control in an animal setting.
5.3.3. The NRS alone stimulates RSV polyadenylation in vitro
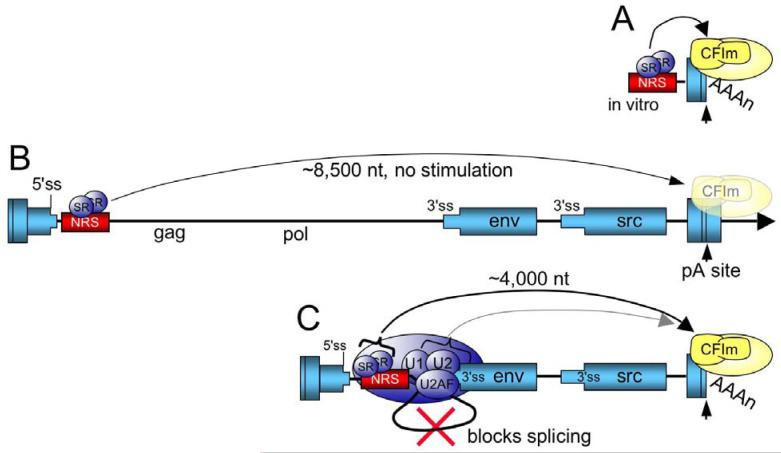
Two recent reports used an in vitro approach to investigate the mechanism by which the NRS stimulates RSV polyadenylation (75, 76). In support of the above model, it was shown that a strong adenovirus 3′ ss and to a lesser extent, the weaker src 3′ ss, could stimulate use of a RSV poly(A) substrate (75). Thus, conventional coupling interactions (presumably through U2AF) can activate RSV polyadenylation in vitro. However, in contrast to what was predicted in the above model, it was shown that the NRS alone could stimulate RSV polyadenylation in vitro and neither the env or src 3′ ss were required for the effect (75, 76). Furthermore, the U1 binding site, whose mutation caused increased read-through in the viral context, was also not required for NRS-stimulated polyadenylation. Using a panel of NRS mutants that eliminate binding of SR proteins, hnRNP H, U2 snRNP, and U1/U11 snRNPs, the poly(A) stimulatory activity was mapped to NRS5′ where SR proteins bind, and an hnRNP H binding site present in that region was not required. These results suggested that SR proteins might promote RSV polyadenylation, and two lines of evidence supported this idea (75). First, addition of purified SR proteins to an extract that lacked these factors stimulated RSV polyadenylation, but not poly(A) addition to an SV40 substrate whose activity does not require SR proteins. Second, RSV substrates in which the SR protein-binding region was replaced with high-affinity binding sites for three different SR proteins were also polyadenylated. These results strongly indicate that SR proteins promote RSV polyadenylation in vitro and represent the first direct evidence that SR proteins can act as polyadenylation factors (Figure 5A). While SR proteins can boost RSV polyadenylation in vitro, where the binding sites are close to the poly(A) site, the question remained whether SR proteins alone also stimulate RSV polyadenylation from their native location in the virus, which is ∼8,500 upstream of the poly(A) site.
Figure 5.
Model for SR protein stimulation of RSV polyadenylation. A) For in vitro substrates, the NRS is in close proximity to the poly(A) site where SR proteins stimulate polyadenylation, perhaps through an interaction the RS domain with a similar domain within CFIm. CFIm and the polyadenylation machinery are in yellow. B) In the viral context where the NRS-3′ss complex cannot form, SR proteins associated with the NRS are too far away from the poly(A) site to stimulate polyadenylation (washed out poly(A) factors) and read-through transcripts increase (arrow). The RSV ‘exons’ are represented as blue boxes and the intron as a line. C) SR proteins promote NRS complex assembly with the env 3′ ss, which blocks splicing. The NRS-3′ss complex repositions the SR proteins closer to the poly(A) site where they promote poly(A) complex formation (arrow), perhaps through an interaction with CFIm. It is also possible that additional NRS complex factors contribute to poly(A) efficiency through conventional coupling interactions (light shaded arrow).
5.3.4. SR proteins, the NRS complex, and proximity to the poly(A) site in vivo
The activity of SR proteins in splicing often shows a distance constraint. For example, in mediating the activity of ESEs and in exon definition, SR proteins fail to function at distances much more than 300 nt (79, 80). Therefore, experiments were designed to determine if SR proteins bound to the NRS also stimulate polyadenylation in the virus from the native NRS position ∼8,500 nt away. In contrast to the in vitro results, the high-affinity SR protein binding sites did not promote polyadenylation when inserted in place of the NRS (75) (Figure 5B). Significantly, the high-affinity sites did partially rescue the read-through and splicing defects of a provirus that harbored a deletion of the SR protein-binding region, i.e., a virus in which the NRS-3′ ss inhibitory complex could still form. These results again point out the importance of SR proteins in splicing control and suggested their importance in polyadenylation. However, a role for the SR proteins in promoting polyadenylation was ambiguous since it was not possible to discount the possibility that the SR proteins simply promoted assembly of the NRS-3′ss complex, and that other factors within the complex mediated conventional coupling interactions to boost polyadenylation effect. Reasoning that the SR proteins might only stimulate polyadenylation when close to the poly(A) site (as was the case for the in vitro substrates) and that the NRS-3′ss complex might serve to bring the SR proteins closer to the poly(A) site, deletion viruses were constructed in which the high-affinity SR protein binding sites were place at a position similar to that which would occur from an NRS - env 3′ ss interaction. These viruses lacked the NRS and env 3′ss, so the authentic NRS complex could not form, yet now the high-affinity sites partially rescued the poly(A) defect. These data support a model in which SR proteins provide two functions: they promote U1 binding to the NRS and assembly of the NRS-3′ss complex that blocks splicing, and this complex repositions the SR proteins closer to the poly(A) site such that they promote polyadenylation (Figure 5C). These data also reconcile the early observations of Miller and Stoltzfus (77) that sequences within gag and surrounding the env 3′ ss are both required for optimal RSV polyadenylation; the NRS must interact with the env 3′ ss to reposition the SR proteins closer to the poly(A) site, and loss of either element disrupts this process. Given that the RS-domain of U2AF interacts with a similar domain in CFIm to mediate coupling (74), and the finding that several SR proteins interact with CFIm (81), an attractive idea is that SR proteins within the NRS-3′ss complex promote polyadenylation by recruiting CFIm via RS domain interactions. The fact that only partial rescue of the poly(A) defect was observed might suggest that the activity of the SR proteins is augmented by other factors in the NRS-3′ss complex (Figure 5C). These ideas are currently being explored in our laboratory.
5.3.5. A role for hnRNP H in RSV polyadenylation control?
hnRNP H binds to G-rich elements associated with a number of cellular and viral poly(A) sites and stimulates polyadenylation (82, 83). There are two hnRNP H binding regions in the NRS: an upstream site that is embedded in the SR protein binding region, and a strong downstream site that is required for efficient U11 binding (see 4.3.5). It was anticipated that hnRNP H might also promote NRS-mediated RSV polyadenylation. In contrast to a positive role in RSV polyadenylation, Wilusz and Beemon (76) showed that sequestering hnRNP H by adding a G-rich oligonucleotide to reactions increased use of the RSV poly(A) site in vitro, which led to a model whereby hnRNP H binding to the NRS (presumably the upstream site) might out compete SR protein binding. This is not supported by data from Maciolek and McNally (75) where no effect on in vitro polyadenylation was observed when the upstream hnRNP H binding site was mutated (the NRS in this poly(A) substrate lacked the downstream sites). Furthermore, there was no effect on RSV polyadenylation in vivo when either the upstream or downstream hnRNP H binding sites were mutated (39, 75). A virus with both regions mutated has yet to be tested. It is possible that the in vitro inhibition of RSV polyadenylation is an artifact of placing strong hnRNP H sites at an inappropriate position relative to the poly(A) site. Such artifacts were observed for the SV40 substrate where positioning hnRNP H binding sites upstream of the poly(A) site abolished polyadenylation (84). Additional work is required to determine if hnRNP H plays a regulatory role in RSV polyadenylation.
6. CONCLUSIONS AND PERSPECTIVES
As this “Protein-RNA Interactions in Viral RNA Processing” review series demonstrates, viral systems have proven invaluable for discovering and dissecting cellular processes, including RNA processing. A very prominent example is the discovery of RNA splicing, which stemmed from work on adenovirus (85, 86). Retroviruses have also been quite useful for studying the regulation of splicing, and the avian viruses have proved fruitful in revealing novel mechanisms by which splicing and polyadenylation are controlled and integrated. Early work by Katz and Skalka (14) using an avian sarcoma virus provided one of the first example of exonic sequences that promote splicing, and this element turned out to be one of the first splicing enhancers described. Since that time, the exon has transitioned to a focal point for the binding of positive and negative splicing regulatory factors in cellular and viral genes. Additional work on splicing control identified the NRS, and contributions from several labs have culminated in a detailed understanding of a novel mechanism of splicing control. The NRS functions as an elaborate pseudo 5′ ss that, with the assistance of SR proteins, binds U1 snRNP and initiates the assembly of a non-productive splicing complex with the viral 3′ splice sites. The viral 3′ ss is thus sequestered from a productive interaction with the authentic viral 5′ ss. The observations and conclusions of Miller and Stoltzfus (77) concerning polyadenylation control in RSV stimulated recent work that has uncovered a novel mechanism of coupling between the splicing and polyadenylation machineries that takes placed in the absence of splicing. A significant finding was that SR proteins promote NRS-mediated polyadenylation, which was the first demonstration of this activity for SR protein family members. Still, much remains to be learned about splicing and polyadenylation control in avian retroviruses.
With respect to the NRS, a number of fundamental question remain to be answered. Of prominence, why the NRS-3′ss complex is non-productive remains to be explained. What is it about the sequence of the NRS U1-type 5′ ss that allows assembly of a spliceosome that is unable to undergo catalysis? What role does the structure of the NRS play in the inhibition process? What features prevent the stable incorporation of the U4/U6.U5 tri-snRNP into the NRS-3′ss complex? What roles do other sequences within the NRS that have received less attention, like the branchpoint-pyrimidine tract element that appears to bind U2 snRNP, play in splicing control? Additionally, does U11 binding simply fine-tune splicing control by modulating U1 binding, or does U11 play another unexpected role in RSV biology? With regard to polyadenylation control, do SR proteins present within the NRS-3′ss complex act alone in poly(A) stimulation, or is there cooperation with other factors within the complex. While the SR proteins appear to be repositioned closer to the poly(A) site via the NRS-3′ss complex, they are still more than 4000 nts from the poly(A) site in linear terms. How do the SR proteins promote polyadenylation over such a long distance? The details of what poly(A) factors are influenced by the NRS and SR proteins and how coupling occurs at the molecular level remain to be elucidated. Future work will also address if and how hnRNP H influences RSV polyadenylation. There is also little known about how splicing to the src 3′ ss is controlled by the suppressor of src splicing (SSS) and the dr1 sequences. What factors do they bind, and how do they interface with the splicing machinery? Clearly, while we have a fairly sophisticated understanding of several aspects of RNA processing control in avian retroviruses, numerous mysteries remain to be solved. As has been true to date, gaining an understanding these remaining problems in viral RNA processing regulation will likely improve our appreciation of cellular RNA processing as well.
7. ACKNOWLEDGMENTS
We are grateful to members of the McNally lab, C. Martin Stoltzfus, and an anonymous reviewer for helpful comments on the manuscript. This work was supported by Public Health Service Grant R01 CA78709 from the National Cancer Institute.
8. REFERENCES
- 1.Goff SP. Retroviridae: The Retroviruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams and Wilkins; Philadelphia, Pa: 2007. [Google Scholar]
- 2.Coffin JM, Hughes S, Varmus H. Retroviruses. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- 3.Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3(1):18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burge CB, Tushchl TH, Sharp PA. Splicing of precursors to mRNAs by the Spliceosome. In: Gesteland RF, Cech T, Atkins JF, editors. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. [Google Scholar]
- 5.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 6.Jurica MS, Moore MJ. Pre-mRNA Splicing. Awash in a Sea of Proteins. Mol Cell. 2003;12(1):5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 7.Lim SR, Hertel KJ. Commitment to splice site pairing coincides with A complex formation. Mol Cell. 2004;15(3):477–83. doi: 10.1016/j.molcel.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 8.Valadkhan S, Manley JL. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413(6857):701–7. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- 9.Valadkhan S. The spliceosome: a ribozyme at heart? Biol Chem. 2007;388(7):693–7. doi: 10.1515/BC.2007.080. [DOI] [PubMed] [Google Scholar]
- 10.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84(5):801–11. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 11.Hall SL, Padgett RA. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271(5256):1716–8. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 12.Tarn WY, Steitz JA. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci. 1997;22(4):132–7. doi: 10.1016/s0968-0004(97)01018-9. [DOI] [PubMed] [Google Scholar]
- 13.McNally MT, Beemon K. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J Virol. 1992;66(1):6–11. doi: 10.1128/jvi.66.1.6-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz RA, Skalka AM. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990;10(2):696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz RA, Kotler M, Skalka AM. cis-acting intron mutations that affect the efficiency of avian retroviral RNA splicing: implication for mechanisms of control. J Virol. 1988;62(8):2686–95. doi: 10.1128/jvi.62.8.2686-2695.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu X-D, Katz RA, Skalka AM, Maniatis T. The role of branchpoint and 3′ exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991:211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- 17.Bouck J, Fu XD, Skalka AM, Katz RA. Role of the constitutive splicing factors U2AF65 and SAP49 in suboptimal RNA splicing of novel retroviral mutants. J Biol Chem. 1998;273(24):15169–76. doi: 10.1074/jbc.273.24.15169. [DOI] [PubMed] [Google Scholar]
- 18.Bouck J, Fu XD, Skalka AM, Katz RA. Genetic selection for balanced retroviral splicing: novel regulation involving the second step can be mediated by transitions in the polypyrimidine tract. Mol Cell Biol. 1995;15(5):2663–71. doi: 10.1128/mcb.15.5.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Stoltzfus CM. A suboptimal src 3′ splice site is necessary for efficient replication of Rous sarcoma virus. Virology. 1995;206(2):1099–1107. doi: 10.1006/viro.1995.1033. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Simpson SB, Stoltzfus CM. Selection and characterization of replication-competent revertants of a Rous sarcoma virus src gene oversplicing mutant. J Virol. 1996;70(6):3636–44. doi: 10.1128/jvi.70.6.3636-3644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoltzfus CM, Lorenzen SK, Berberich SL. Noncoding region between the env and src genes of Rous sarcoma virus influences splicing efficiency at the src gene 3′ splice site. J Virol. 1987;61(1):177–84. doi: 10.1128/jvi.61.1.177-184.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berberich SL, Stoltzfus CM. Mutations in the regions of the Rous sarcoma virus 3′ splice sites: implications for regulation of alternative splicing. J Virol. 1991;65(5):2640–6. doi: 10.1128/jvi.65.5.2640-2646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berberich SL, Macias M, Zhang L, Turek LP, Stoltzfus CM. Comparison of Rous sarcoma virus RNA processing in chicken and mouse fibroblasts: evidence for double-spliced RNA in nonpermissive mouse cells [published erratum appears in J Virol 1990 Dec;64(12):6360] J Virol. 1990;64(9):4313–20. doi: 10.1128/jvi.64.9.4313-4320.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amendt BA, Simpson SB, Stoltzfus CM. Inhibition of RNA splicing at the Rous sarcoma virus src 3′ splice site is mediated by an interaction between a negative cis element and a chicken embryo fibroblast nuclear factor. J Virol. 1995;69(8):5068–76. doi: 10.1128/jvi.69.8.5068-5076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo W, Winistorfer SC, Stoltzfus CM. Selective inhibition of splicing at the avian sarcoma virus src 3′ splice site by direct-repeat posttranscriptional cis elements. J Virol. 2000;74(18):8513–23. doi: 10.1128/jvi.74.18.8513-8523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogert RA, Lee LH, Beemon KL. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70(6):3834–43. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson SB, Guo W, Winistorfer SC, Craven RC, Stoltzfus CM. The upstream, direct repeat sequence of Prague A Rous sarcoma virus is deficient in mediating efficient Gag assembly and particle release. Virology. 1998;247(1):86–96. doi: 10.1006/viro.1998.9233. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Cullen BR. Structural and functional analysis of the avian leukemia virus constitutive transport element. RNA. 1999;5(12):1645–55. doi: 10.1017/s1355838299991616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson SB, Zhang L, Craven RC, Stoltzfus CM. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J Virol. 1997;71(12):9150–6. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arrigo S, Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988;8:4858–67. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoltzfus CM, Fogarty SJ. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J Virol. 1989;63(4):1669–76. doi: 10.1128/jvi.63.4.1669-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Sullivan CT, Polony TS, Paca RE, Beemon KL. Rous sarcoma virus negative regulator of splicing selectively suppresses SRC mRNA splicing and promotes polyadenylation. Virology. 2002;302(2):405–12. doi: 10.1006/viro.2002.1616. [DOI] [PubMed] [Google Scholar]
- 33.McNally MT, Gontarek RR, Beemon K. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology. 1991;185(1):99–108. doi: 10.1016/0042-6822(91)90758-4. [DOI] [PubMed] [Google Scholar]
- 34.Gontarek RR, McNally MT, Beemon K. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 1993;7(10):1926–36. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- 35.Giles KE, Beemon KL. Retroviral splicing suppressor sequesters a 3′ splice site in a 50S aberrant splicing complex. Mol Cell Biol. 2005;25(11):4397–405. doi: 10.1128/MCB.25.11.4397-4405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNally LM, McNally MT. U1 small nuclear ribonucleoprotein and splicing inhibition by the rous sarcoma virus negative regulator of splicing element. J Virol. 1999;73(3):2385–93. doi: 10.1128/jvi.73.3.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hibbert CS, Gontarek RR, Beemon KL. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA. 1999;5(3):333–43. doi: 10.1017/s1355838299981347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNally LM, McNally MT. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J Virol. 1996;70(2):1163–72. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fogel BL, McNally LM, McNally MT. Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acids Res. 2002;30(3):810–7. doi: 10.1093/nar/30.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6(9):1197–211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6(5):837–47. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 42.Jamison SF, Pasman Z, Wang J, Will C, Luhrmann R, Manley JL, Garcia-Blanco MA. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res. 1995;23(16):3260–7. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25(3):106–10. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 44.McNally LM, McNally MT. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol Cell Biol. 1998;18(6):3103–3111. doi: 10.1128/mcb.18.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staknis D, Reed R. SR proteins promote the first specific recognition of Pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14(11):7670–82. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cook CR, McNally MT. Characterization of an RNP complex that assembles on the Rous sarcoma virus negative regulator of splicing element. Nucleic Acids Res. 1996;24(24):4962–4968. doi: 10.1093/nar/24.24.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cook CR, McNally MT. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- 48.Cook CR, McNally MT. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J Virol. 1999;73(3):2394–400. doi: 10.1128/jvi.73.3.2394-2400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabello-Villegas J, Giles KE, Soto AM, Yu P, Mougin A, Beemon KL, Wang YX. Solution structure of the pseudo-5′ splice site of a retroviral splicing suppressor. RNA. 2004;10(9):1388–98. doi: 10.1261/rna.7020804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paca RE, Hibbert CS, O'Sullivan CT, Beemon KL. Retroviral splicing suppressor requires three nonconsensus uridines in a 5′ splice site-like sequence. J Virol. 2001;75(16):7763–8. doi: 10.1128/JVI.75.16.7763-7768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11(23):377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 52.Montzka KA, Steitz JA. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc Natl Acad Sci U S A. 1988;85(23):8885–9. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNally LM, Yee L, McNally MT. Two regions promote U11 snRNP binding to a retroviral splicing inhibitor element (NRS) J Biol Chem. 2004;279(37):38201–8. doi: 10.1074/jbc.M407073200. [DOI] [PubMed] [Google Scholar]
- 54.McNally LM, Yee L, McNally MT. hnRNP H is required for optimal U11 snRNP binding to a retroviral RNA processing control element: Implications for U12-dependent RNA splicing. J Biol Chem. 2006;281(5):2478–88. doi: 10.1074/jbc.M511215200. [DOI] [PubMed] [Google Scholar]
- 55.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3(5):e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous Nuclear Ribonucleoprotein F/H Proteins Modulate the Alternative Splicing of the Apoptotic Mediator Bcl-x. J Biol Chem. 2005;280(24):22641–50. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 57.Schaub MC, Lopez SR, Caputi M. Members of the heterogeneous nuclear ribonucleoprotein H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J Biol Chem. 2007;282(18):13617–26. doi: 10.1074/jbc.M700774200. [DOI] [PubMed] [Google Scholar]
- 58.Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate premRNA splicing. PLoS Biol. 2006;4(2):e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13(5):593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacquenet S, Mereau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlant C. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J Biol Chem. 2001;276(44):40464–75. doi: 10.1074/jbc.M104070200. [DOI] [PubMed] [Google Scholar]
- 61.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416(6880):499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 62.Herman SA, Coffin JM. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J Virol. 1986;60(2):497–505. doi: 10.1128/jvi.60.2.497-505.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polony TS, Bowers SJ, Neiman PE, Beemon KL. Silent point mutation in an avian retrovirus RNA processing element promotes c-myb-associated short-latency lymphomas. J Virol. 2003;77(17):9378–87. doi: 10.1128/JVI.77.17.9378-9387.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith MR, Smith RE, Dunkel I, Hou V, Beemon KL, Hayward WS. Genetic determinant of rapid-onset B-cell lymphoma by avian leukosis virus. J Virol. 1997;71(9):6534–40. doi: 10.1128/jvi.71.9.6534-6540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63(2):405–45. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11(10):1485–93. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19(11):1315–27. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niwa M, Berget SM. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 1991;5(11):2086–95. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 69.Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4(9):1552–9. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 70.Awasthi S, Alwine JC. Association of polyadenylation cleavage factor I with U1 snRNP. RNA. 2003;9(11):1400–9. doi: 10.1261/rna.5104603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lutz CS, Murthy KG, Schek N, O'Connor JP, Manley JL, Alwine JC. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10(3):325–37. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 72.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23(2):195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 73.McCracken S, Lambermon M, Blencowe BJ. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol Cell Biol. 2002;22(1):148–60. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25(20):4854–64. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maciolek NL, McNally MT. Serine/Arginine-Rich Proteins Contribute to Negative Regulator of Splicing Element-Stimulated Polyadenylation in Rous Sarcoma Virus. J Virol. 2007;81(20):11208–17. doi: 10.1128/JVI.00919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilusz JE, Beemon KL. The negative regulator of splicing element of rous sarcoma virus promotes polyadenylation. J Virol. 2006;80(19):9634–40. doi: 10.1128/JVI.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller JT, Stoltzfus CM. Two distant upstream regions containing cis-acting signals regulating splicing facilitate 3′-end processing of avian sarcoma virus RNA. J Virol. 1992;66(7):4242–51. doi: 10.1128/jvi.66.7.4242-4251.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang W, Kanter MR, Dunkel I, Ramsay RG, Beemon KL, Hayward WS. Minimal truncation of the c-myb gene product in rapid-onset B-cell lymphoma. J Virol. 1997;71(9):6526–33. doi: 10.1128/jvi.71.9.6526-6533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270(6):2411–4. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 80.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8(14):1703–12. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 81.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279(34):35788–97. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 82.Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30(8):1842–50. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bagga PS, Arhin GK, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26(23):5343–50. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bagga PS, Ford LP, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end premRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23(9):1625–31. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 86.Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977;74(8):3171–5. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]