Abstract
The proper localization of resident membrane proteins to the trans-Golgi network (TGN) involves mechanisms for both TGN retention and retrieval from post-TGN compartments. In this study we report identification of a new gene, GRD20, involved in protein sorting in the TGN/endosomal system of Saccharomyces cerevisiae. A strain carrying a transposon insertion allele of GRD20 exhibited rapid vacuolar degradation of the resident TGN endoprotease Kex2p and aberrantly secreted ∼50% of the soluble vacuolar hydrolase carboxypeptidase Y. The Kex2p mislocalization and carboxypeptidase Y missorting phenotypes were exhibited rapidly after loss of Grd20p function in grd20 temperature-sensitive mutant strains, indicating that Grd20p plays a direct role in these processes. Surprisingly, little if any vacuolar degradation was observed for the TGN membrane proteins A-ALP and Vps10p, underscoring a difference in trafficking patterns for these proteins compared with that of Kex2p. A grd20 null mutant strain exhibited extremely slow growth and a defect in polarization of the actin cytoskeleton, and these two phenotypes were invariably linked in a collection of randomly mutagenized grd20 alleles. GRD20 encodes a hydrophilic protein that partially associates with the TGN. The discovery of GRD20 suggests a link between the cytoskeleton and function of the yeast TGN.
INTRODUCTION
The secretory and endocytic pathways of eukaryotic cells consist of a series of membrane-enclosed compartments that communicate using transport vesicles or by direct fusion. In vesicular transport, a vesicle forms from a donor membrane and fuses with a membrane from an acceptor compartment (Rothman and Wieland, 1996; Schekman and Orci, 1996). A protein present in the donor compartment can be either actively sequestered within a forming vesicle or actively excluded from the vesicle or can diffuse into the vesicle at its prevailing concentration in the donor compartment. Once formed, the vesicle and associated protein cargo are targeted to the appropriate acceptor membrane via the interaction of SNARE proteins associated with the vesicle and target membranes. The processes by which proteins are actively sequestered within or excluded from forming vesicles are important determinants in maintenance of the correct functional and structural organization of the donor compartment. Therefore, the mechanisms that govern these processes have attracted considerable interest.
In addition to its role in proteolytic processing, the trans-Golgi network (TGN) is an intersection point of the endocytic and secretory membrane trafficking pathways and mediates many protein-sorting events. For example, at least four different types of vesicles form from the yeast TGN for transport to other destinations. Soluble vacuolar hydrolases such as carboxypeptidase Y (CPY) and the membrane protein carboxypeptidase S are packaged into one class of vesicles that enter a pathway, leading to the lysosome-like vacuole (Vida et al., 1993; Marcusson et al., 1994). The membrane protein alkaline phosphatase (ALP) enters another class of TGN-derived vesicles targeted to the vacuole via a pathway distinct from that used by CPY (Cowles et al., 1997b; Piper et al., 1997) and dependent on the AP-3 adaptor complex (Cowles et al., 1997a; Stepp et al., 1997; Vowels and Payne, 1998). Vesicles that carry exocytic cargo to the plasma membrane also form from the TGN, and there is strong evidence that multiple types of exocytic vesicles are generated (Harsay and Bretscher, 1995; Mulholland et al., 1997). Finally, yet another class of vesicles may form from the yeast TGN and transport proteins back to earlier secretory pathway compartments (Harris and Waters, 1996). The yeast TGN also serves as an acceptor compartment for vesicles derived from endosomes (Bryant et al., 1998) and possibly earlier Golgi compartments.
The mechanism by which soluble vacuolar hydrolases such as CPY are sorted at the yeast TGN has been intensely investigated using genetic and biochemical approaches (for reviews, see Stack et al., 1995; Conibear and Stevens, 1998). CPY contains a sorting signal that is recognized by the lumenal domain of the CPY receptor, Vps10p, a type I integral membrane protein (Marcusson et al., 1994; Cooper and Stevens, 1996). Receptor–ligand complexes at the TGN then enter vesicles that are targeted to a prevacuolar endosome where the ligand and receptor are thought to dissociate. CPY is then transported on to the vacuole, whereas Vps10p is recycled back to the TGN via a retrograde pathway.
Related to the trafficking of Vps10p between the TGN and prevacuolar endosome are mechanisms that ensure that the TGN retains its resident membrane proteins. The yeast TGN contains three membrane-bound enzymes, Kex1p, Kex2p, and dipeptidyl aminopeptidase (DPAP) A, that are involved in proteolytic maturation of the α-factor pheromone (Fuller et al., 1988). Signals that specify TGN localization are contained within the 100- to 120-amino-acid cytoplasmic domains of all three of these resident enzymes (Cooper and Bussey, 1992; Wilcox et al., 1992; Nothwehr et al., 1993). Kex2p and DPAP A each contain an aromatic amino acid-based signal involved in mediating retrieval from a post-Golgi compartment, probably the same endosomal compartment that Vps10p is retrieved from (Brickner and Fuller, 1997; Bryant and Stevens, 1997). Both proteins also contain “static” retention signals that effect a slow rate of transport from the TGN to the prevacuolar endosome. The cumulative action of the two independent localization mechanisms results in very efficient localization to the TGN.
Several genetic screens have been used to identify genes necessary for proper trafficking and localization of TGN resident proteins and of Vps10p. For example, loss of function of >50 VPS (vacuolar protein-sorting) and PEP (peptidase-deficient) genes are known to cause aberrant secretion of CPY, and some of these have been shown to directly affect trafficking of Vps10p (Stack et al., 1995; Conibear and Stevens, 1998). In addition, the GRD and SOI genes were identified based on alterations in the localization of model TGN membrane proteins A-ALP and Kex2p, respectively (Nothwehr et al., 1996; Redding et al., 1996). A-ALP consists of the cytoplasmic domain of DPAP A fused to the transmembrane and lumenal domains of ALP (Nothwehr et al., 1993). Several gene products are known to mediate retrieval of Vps10p, Kex2p, and A-ALP from an endosomal compartment, although some differences in the retrieval machinery used by each cargo protein have been observed. Retrieval of Vps10p from the prevacuolar endosome has been shown to require a multisubunit “retromer complex” that has been proposed to serve as vesicle coat for sorting of Vps10p into vesicles that form from the endosome (Seaman et al., 1998). Loss of function of several of the retromer subunits including Vps35p and Vps5p have been shown to cause defects in TGN localization of A-ALP and Kex2p (Nothwehr and Hindes, 1997; Nothwehr et al., 1999). Moreover, Vps35p has been shown to have a direct role in retrieval of A-ALP, and the structural features on Vps35p that mediate retrieval of A-ALP are distinct from features required for Vps10p retrieval (Nothwehr et al., 1999). Grd19p is required for retrieval of A-ALP and Kex2p and has little if any role in Vps10p retrieval (Voos and Stevens, 1998), but its relationship to the retromer complex is unknown. In contrast, the machinery necessary for reducing the rate of exit of resident membrane proteins from the TGN has not yet been identified.
In this study we report the extensive phenotypic characterization of a newly identified gene involved in TGN localization, GRD20. We find that rapidly after loss of Grd20p function, a severe defect in TGN localization of Kex2p occurs as well as missorting of CPY. A complete loss of GRD20 function also results in a severe growth defect; however, the growth defect appears to be independent of the TGN sorting defect. Rather, the growth defect in grd20 mutants appears to be related to a defect in polarization of the actin cytoskeleton. The Grd20 protein localizes in part to the TGN. GRD20 is the first example of a gene involved in both actin cytoskeleton organization and in TGN function in yeast.
MATERIALS AND METHODS
grd Mutant Screen and Cloning of GRD20
Yeast strains SNY89 and SNY90 (Table 1) were mutagenized by integrating a library of yeast genomic fragments containing Tn3-based transposons inserted at random locations (Burns et al., 1994). The yeast genomic fragments containing Tn3 transposons engineered with the Escherichia coli LacZ and yeast LEU2 genes were digested with NotI before transformation into yeast. The mutant Leu+ transformants were propagated at 30°C, and A-ALP activity of colonies on plates was assessed using a variation of a previously described method (Chapman and Munro, 1994; Nothwehr et al., 1996). Each plate was overlaid with 5 ml of 50°C solution containing 0.35% (wt/vol) agar, 0.5 M Tris, pH 9.0, 5 mM MgSO4, 1.0% Triton X-100, 5 mg/ml Fast Red dye, and 1 mg/ml napthol AS phosphate, and the color change was observed. Clones consistently exhibiting elevated A-ALP activity were assessed for proteolytic processing of A-ALP, CPY secretion, and pro-α-factor (MATα parent only). Mutants that secreted CPY were used for complementation analysis with the vps and grd mutant collections (Rothman and Stevens, 1986; Robinson et al., 1988; Raymond et al., 1992; Nothwehr et al., 1996). Diploids were analyzed either for CPY secretion or processing of A-ALP, depending on the severity of each phenotype. Mutants chosen for further analysis were back-crossed against the parental wild-type strain of the opposite mating type (either SNY89 or SNY90) and sporulated, and the resulting tetrads were analyzed for linkage of the grd phenotype with a single transposon. A plasmid from a YCp50-based yeast genomic library (Rose et al., 1987) called pC-5 was found that complemented the grd phenotype of yeast mutant 8923.176. Further analysis showed that open reading frame (ORF) YER157w/GRD20 was sufficient for complementation. Furthermore, the transposon in 8923.176 was shown by PCR to be inserted around the Met-183 codon of the GRD20 ORF.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SNY36-9A | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 pho8Δ∷ADE2 | Nothwehr et al., 1995 |
| SNY17 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷LEU2 | Nothwehr et al., 1995 |
| KWY1 | MATa/α ura3-52/ura2-52 leu2-3,112/leu2-3,112 his3-Δ200/his3-Δ200 trp1-901/trp1-901 LYS2/lys2-801 suc2-Δ9/suc2-Δ9 pho8Δ∷LEU2/phos8Δ∷ADE2 | This study |
| SNY89 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8∷ste13-pho8 | This study |
| SNY90 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 pho8∷ste13-pho8 | This study |
| 8923.176 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8∷ste13-pho8 grd20∷Tn3-LEU2 | This study |
| SNY97-14D | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8∷ste13-pho8 grd20∷Tn3-LEU2 | This study |
| SBY4-10A | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 grd20-Δ1 | This study |
| SBY7-7A | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ∷ADE2 grd20∷Tn3-LEU2 | This study |
| SBY6 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pep4Δ∷TRP1 pho8Δ∷ADE2 grd20∷Tn3-LEU2 | This study |
| LSY2 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 pho8Δ∷ADE2 pep4Δ∷TRP1 | This study |
| AHY47 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 kex2Δ∷LEU2 | This study |
| SBY9-8A | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 grd20∷tn3-LEU2 kex2Δ∷LEU2 | This study |
| SNY108-1A | MATα ura3-52 leu2-3,112 lys2-801 (suc2-Δ9 or SUC2) grd20∷tn-LEU2 sec1-1 | This study |
| LCY7 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 vps1-100 (ts) | T.H. Stevens |
| SNY2 | MATα ura3-52 leu2-3,112 gal2 sec1-1 vps1-100 (ts) | This study |
| SNY121-12A | MATα ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 pho8Δ∷ADE2 can1-100 grd20Δ∷URA3 | This study |
| SNY44-13B | MATα ura3-52 leu2-3,112 his4-519 ade6 BAR1 or bar1-1 pho8-ΔX end4-1 | Nothwehr et al., 1995 |
Plasmids and Yeast Strain Construction
Most of the yeast strains and plasmids used in this study are described in Tables 1 and 2, respectively. A centromeric (CEN) plasmid called pSB1 containing the GRD20 gene was constructed by subcloning a 2.69-kbp PvuII–HindIII fragment from pC-5 into the SmaI and HindIII sites of pRS316 (Sikorski and Hieter, 1989). The influenza hemagglutinin (HA) epitope-tagged GRD20 allele was constructed by first introducing a BamHI site by site-directed mutagenesis just upstream of the GRD20 stop codon in a plasmid consisting of Bluescript KS+ (Stratagene, La Jolla, CA) containing the 0.45-kbp BglII–HindIII fragment from GRD20. The resulting plasmid (pSB8) was digested with BamHI and ligated to a ∼120-bp BglII linker containing three copies of the HA epitope (YPYDVPDYA), resulting in plasmid pSB9. To reconstruct the full-length GRD20::HA allele in pRS316 the 0.5-kbp BglII–HindIII fragment from pSB1 was replaced with the 0.6-kbp BglII–HindIII fragment from pSB9.
Table 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pSN55 | STE13-PHO8 gene fusion encoding A-ALP in pRS316 | Nothwehr et al., 1993 |
| pSN92 | PHO8 gene in pRS316 | Nothwehr et al., 1993 |
| pAH16 | STE13-PHO8 gene fusion encoding A-ALP in pRS314 | Nothwehr et al., 1999 |
| pSB1 | pRS316 containing the GRD20 gene | This study |
| pSB1-125 | pRS316 containing the grd20-1 temperature-sensitive allele | This study |
| pSB1-1212 | pRS316 containing the grd20-2 temperature-sensitive allele | This study |
| pSB1-139 | pRS316 containing the grd20-3 temperature-sensitive allele | This study |
| pSB1-23 | pRS316 containing the grd20-4 temperature-sensitive allele | This study |
| pSB1-65 | pRS316 containing the grd20-5 temperature-sensitive allele | This study |
| pSB10 | pRS316 containing the GRD20 gene with 3 copies of the HA epitope inserted at the C-terminus of the ORF | This study |
| pSB18 | pRS313 containing the grd20-1 temperature-sensitive allele | This study |
| pSB19 | pRS313 containing the SUC2 gene | This study |
| pSN179-A | pRS316 containing the KEX2 gene | This study |
| pSL2099 | pRS315 containing a GAL1-STE3∷myc fusion construct | Davis et al., 1993 |
A construct for generation of yeast strains lacking all but the first 7 codons of the 801-codon GRD20 ORF was made by first subcloning the 1.8-kbp PstI–EcoRI fragment from the 5′ untranslated region of GRD20 into Bluescript KS+, resulting in plasmid pSN313. Next, a pRS306 (Sikorski and Hieter, 1989) derivative lacking a region of the polylinker was made by digesting pRS306 with SacI–SpeI, filling in with Klenow and deoxynucleotides, and self-ligating, resulting in pSN316. Finally, the 1.8-kbp BamHI–EcoRI fragment from pSN313 and the 1.6-kbp HindIII fragment from pC-5 corresponding to the 3′ untranslated region of GRD20 were successively subcloned into the BamHI–EcoRI and HindIII sites of pSN316, respectively, generating pSB12. pSB12 was linearized with SacI and transformed into the diploid strain KWY1. Ura+ transformants were grown nonselectively and then grown in the presence of 5-fluoroorotic acid to select for Ura− loop-outs. The Ura− strains were screened by PCR for the presence of grd20-Δ1 allele. A GRD20/grd20-Δ1 heterozygous diploid was then sporulated, and tetrads were dissected giving rise to the haploid strain SBY4-10A that contains the grd20-Δ1 allele. Replacement of the entire GRD20 coding sequence with the URA3 gene was also performed. DNA containing the grd20Δ::URA3 allele was generated by PCR amplifying the URA3 gene from pRS306 using primers that contained regions of homology to the beginning and end of the GRD20 ORF. The grd20Δ::URA3 was first introduced into a diploid strain, which was then transformed with a GRD20-containing plasmid and sporulated giving rise to strain SNY121-12A.
A URA3-based CEN plasmid harboring the KEX2 gene was constructed by subcloning a 3.9-kbp fragment from pKX9 (a gift from Robert Fuller, University of Michigan) into the BamHI site of pRS316, resulting in plasmid pSN179-A. Finally, the pho8::ste13-pho8 allele that expresses the A-ALP fusion protein was introduced using construct pSN288 (Nothwehr et al., 1999) into yeast strains SEY6210 (Robinson et al., 1988) and SNY36–9A (Nothwehr et al., 1995), resulting in strains SNY89 and SNY90, respectively.
Generation of grd20 Temperature-sensitive Mutants
The GRD20 gene was subjected to random PCR mutagenesis using an in vivo gap repair method. Plasmid pSB17 consists of the 2.69-kbp PvuII–HindIII GRD20 fragment cloned into the SmaI and HindIII sites of pRS315 (Sikorski and Hieter, 1989). Primers 270 (5′-CCGGCTCCTATGTTGTGTGG-3′) and 271 (5′-GGATGTGCTGC-AAGGCGATTA-3′), which hybridize just outside of the polylinker and extend toward it, were used to amplify a 3.0-kbp PCR product from template pSB17 under mutagenic conditions (Cadwell and Joyce, 1992). The PCR fragment was cotransformed into yeast strain SBY4–10A along with a gel-purified fragment of pSB1 in which the ORF region had been removed by digestion with EcoRI–AatII. Yeast transformants containing circular plasmids generated via homologous recombination were selected on minimal media lacking uracil. A total of 13,000 transformants were screened at 22 and 36°C both for defects in CPY sorting using a colony-blotting assay and for growth defects. Plasmids were rescued from mutants exhibiting temperature-sensitive phenotypes, and the linkage of the phenotype to the plasmid was verified.
Antibody Generation
A vector for expression of the Kex2p cytosolic domain fused to glutathione S-transferase was constructed by subcloning the 0.38-kbp EcoRI–SnaBI fragment from the KEX2 gene into the EcoRI and SmaI sites of pGEX-5X-1 (Pharmacia Biotech, Piscataway, NJ) resulting in pAH39. To generate a plasmid expressing the Kex2p cytosolic domain tagged with 6×His, the 0.38-kbp EcoRI–SalI fragment from pAH39 was subcloned into the EcoRI and SalI sites of pET-28a(+) (Novagen, Madison, WI), resulting in pSB15. Induction of E. coli carrying pAH39 with isopropyl-1-thio-β-d-galactopyranoside produced a fusion protein that was purified using gluthathione-agarose chromatography and injected into New Zealand White rabbits. For affinity purification of rabbit antibodies against Kex2p, the 6×His-tagged Kex2p cytosolic domain was purified from isopropyl-1-thio-β-d-galactopyranoside-induced E. coli carrying pSB15 using a Ni-nitrilotetraacetic acid-agarose column (Qiagen, Valencia, CA). The purified protein was covalently attached to cyanogen bromide-activated Sepharose (Sigma, St. Louis, MO), and the resulting column was used for affinity purification.
A Grd20p antigen construct for expression in and purification from E. coli was made by subcloning the 0.73-kbp EcoRI–SalI fragment from GRD20 into the EcoRI and SalI sites of pGEX-5X-3 (Pharmacia Biotech). Rabbit antiserum raised against the resulting purified antigen (corresponding to residues 7–251 of Grd20p fused downstream of glutathione S-transferase) was then affinity purified against a protein corresponding to the same region of Grd20p fused to maltose-binding protein.
Radiolabeling, Immunoprecipitation, and Immunoblot Analysis
The procedure used for immunoprecipitation of CPY was performed using a rabbit antibody against CPY as previously described (Vater et al., 1992).
For immunoprecipitation of A-ALP, Vps10p, and Kex2p, cultures were grown in media lacking methionine and cysteine, and 35S-Express label (New England Nuclear, Boston, MA) was added to start a pulse. To initiate a chase, 50 μg/ml unlabeled methionine and cysteine were then added. At the end of the chase 0.5 OD600 units of culture were adjusted to 10 mM NaN3 on ice, pelleted, and spheroplasted at 30°C for 30 min in a solution containing 50 mM Tris, pH 7.5, 1.4 M sorbitol, 2 mM MgCl2, 10 mM NaN3, 0.3% β-mercaptoethanol, and 11 μg/ml oxalyticase (Enzogenetics, Corvallis, OR). The spheroplasts were then pelleted and lysed in 50 μl of 1% SDS, 8 M urea, 0.5 mM PMSF, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A at 100°C for 5 min. Immunoprecipitation was then carried out for 2 h on ice using rabbit antibodies against Kex2p, ALP (Nothwehr et al., 1996), or Vps10p (a generous gift from T.H. Stevens, University of Oregon) in a 1-ml volume containing 10 mM Tris, pH 8.0, 0.1% Triton X-100, 0.05% SDS, 0.4 M urea, and 2 mM EDTA. Immune complexes were precipitated by adding 50 μl of IgGsorb (The Enzyme Center, Malden, MA) and incubating for 1 h on ice. The precipitates were washed twice with 10 mM Tris, pH 8.0, 0.1% SDS, 0.1% Triton X-100, and 2 mM EDTA. The samples were analyzed by SDS-PAGE and fluorography as described previously (Stevens et al., 1986). Radiolabeled proteins were quantified from gels using a Phosphorimager system (Fuji Photo Film, Tokyo, Japan).
Immunoprecipitation of invertase was performed essentially as previously described (Gaynor and Emr, 1997). Briefly, strains carrying plasmid pSB19 were spheroplasted and then incubated in minimal media containing low glucose (0.1%), 1 M sorbitol, 1 mg/ml BSA, and 50 mM potassium phosphate, pH 5.7, for 30 min before the pulse and chase. After the chase, the cells were pelleted (I fraction), and the media were removed (E fraction). The I and E fractions were trichloroacetic acid precipitated and denatured at 100°C in 100 μl of 50 mM Tris, pH 7.5, 1 mM EDTA, 1% SDS, and 6 M urea, diluted with 900 μl of 100 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Tween 20, and 0.1 mM EDTA, and rabbit anti-invertase serum (a generous gift from S. Emr, University of California, San Diego) was added. The immune complexes were precipitated using IgGsorb, washed, denatured, and separated on SDS-PAGE gels.
Immunoblots of secreted pro-α-factor were performed by applying freshly growing cells to the surface of a YEPD agar plate and immediately overlaying with a nitrocellulose filter. After incubating for 16 h at 30°C the cells were washed from the filter, and the filter was blocked in 0.75% nonfat dry milk in TTBS (20 mM Tris, pH 7.5, 500 mM NaCl, and 0.1% Triton X-100) and incubated with anti-α-factor serum (a gift from R. Schekman, University of California, Berkeley). Detection was carried out using an anti-rabbit HRP secondary antibody and the Super Signal chemiluminescence substrate (Pierce, Rockford, IL). A similar approach was used for CPY colony blots, except a mouse anti-CPY antibody (Molecular Probes, Eugene, OR) and an anti-mouse HRP secondary antibody were used.
For Western blotting of Grd20p, yeast cells were incubated at 70°C for 10 min in the presence of 50 mM Tris, pH 6.8, containing 8 M urea, 5% SDS, and 5% β-mercaptoethanol and were lysed using glass beads; 0.30 OD equivalents of cell extract per lane were run on an SDS-PAGE gel, and the gel was electroblotted onto nitrocellulose as previously described (Towbin et al., 1979). The filter was blocked and probed with a rabbit anti-Grd20p antibody, and immune complexes were detected by chemiluminescence as described above.
For Western blotting of Ste3p-myc, cells were propagated for several generations at 22°C in minimal media containing 2% each of raffinose and galactose. Cultures with a density of 1 × 107 cells/ml were then shifted to 36°C, and 15 min later glucose was added to a final concentration of 2%. At 0, 30, 60, and 90 min after glucose addition, 12 ml of culture were withdrawn, adjusted to 10 mM NaN3, and were immediately centrifuged at 4°C to pellet the cells. The cells were then washed once with 1 ml of ice-cold distilled H2O, and the cell pellets were stored at −80°C. The cell pellets were thawed on ice and resuspended in 100 μl of a solution containing 8 M urea, 5% SDS, 50 mM Tris, pH 6.8, 5% β-mercaptothanol, 0.5 mM PMSF, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A and were incubated at 65°C for 10 min. The cells were lysed using glass beads, and lysates were centrifuged for 5 min. Eight percent of the supernatant from each sample was loaded on 10% SDS-PAGE gels and blotted to nitrocellulose (Towbin et al., 1979). Ponceau S staining of total protein on the Western blots confirmed that similar amounts of protein were loaded at each time point (our unpublished results). Ste3-myc was detected using the mouse anti-c-myc monoclonal antibody 9E10 (Babco, Richmond, CA), and processing for chemiluminescent detection was carried out (described above). The blots were imaged using the LAS-1000 Luminescent Image Analyzer (Fuji Photo Film).
Fluorescence Microscopy
The procedures for preparation of fixed spheroplasted yeast cells, attachment to microscope slides, and costaining of the A-ALP fusion protein and Vma2p using an anti-ALP polyclonal antibody and anti-Vma2p monoclonal antibody 13D11-B2 (Molecular Probes) were previously described (Roberts et al., 1991; Nothwehr et al., 1995). All secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA) and were used at a 1:500 dilution unless otherwise indicated.
Simultaneous detection of single-copy Kex2p and Vma2p was carried out using solutions containing the following reagents: 1) affinity-purified Kex2p antibody and a mouse anti-Vma2p antibody (Molecular Probes), 2) biotin-conjugated goat anti-rabbit immunoglobulin G (IgG; heavy and light chain [H+L]), and 3) goat anti-mouse IgG (H+L) antibody conjugated to Texas Red and streptavidin conjugated to FITC.
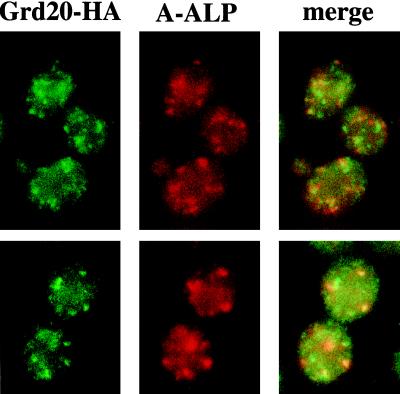
Simultaneous detection of Grd20-HA and A-ALP was accomplished by incubating with the following reagents: 1) mouse anti-HA monoclonal antibody HA.11 (Babco) and rabbit anti-ALP antibody, 2) biotin-conjugated goat anti-rabbit IgG (H+L), and 3) goat anti-mouse IgG (H+L) antibody conjugated to alexa 488 (Molecular Probes) and Texas Red conjugated to streptavidin. The extent of colocalization of Grd20-HA and A-ALP was determined by analyzing the punctate structures stained in 30 cells from a single experiment. The cells were chosen on the criteria that they stained well for each antigen and had clearly defined signals. The analysis involved scoring a total of 229 Grd20-HA-postive punctate structures and 217 A-ALP structures. For the 30 cells analyzed, the number of Grd20-HA and A-ALP staining punctate structures per cell was about the same (7.6 and 7.2, respectively), and 51% of the Grd20-HA structures also stained for A-ALP.
For detection of actin, 10-ml YEPD cultures of yeast in log phase were fixed by adding 1 ml of 37% formaldehyde and were incubated at room temperature for 1 h. The cells were then pelleted and washed twice with 1-ml volumes of distilled H2O and then twice with PBS. The cells were pelleted and then permeablized by incubating in 1 ml of 0.2% Triton X-100 in PBS for 10 min at room temperature followed by two washes in PBS. Approximately 2 OD600 units of cells were stained by incubating for 60 min in 40 μl of PBS containing 0.15 μM Texas Red-phalloidin (Molecular Probes). The cells were washed three times with 1-ml volumes of PBS, and cells were analyzed by microscopy.
Yeast cells were photographed using an Olympus BX-60 epifluorescence microscope (Olympus, Lake Success, NY). Film negatives were digitized using a Polaroid SprintScan 35 scanner and adjusted using Adobe Photoshop 3.0 (Adobe Systems, Mountain View, CA).
RESULTS
Identification of the GRD20 Gene and Its Product
We previously carried out a genetic screen of yeast mutagenized with ethylmethane sulfonate and UV irradiation, resulting in the identification of 18 complementation groups of mutants defective in retention of resident TGN membrane proteins (Nothwehr et al., 1996). This screen was based on the observation that strains defective for Golgi retention mislocalized a model TGN resident protein, A-ALP, to the vacuole where it was activated by proteolytic processing. Strains that correctly localized A-ALP to the TGN exhibited little or no processing of A-ALP and therefore had low levels of enzyme activity.
To facilitate cloning of the affected genes in the grd mutants, in this study we repeated the GRD screen using strains that were randomly mutagenized by transposon insertion mutagenesis. New grd mutant alleles were generated by transforming yeast with a library of plasmids harboring random yeast fragments into which Tn3 bacterial transposons had been inserted (Burns et al., 1994). The transposons were engineered to contain the yeast LEU2 gene; thus chromosomal integration of the transposons was selected for by growing transformants on media lacking leucine.
Mutants shown by a colorimetric ALP activity assay to have elevated levels of activity were then screened for other phenotypes associated with TGN retention such as a defect in α-factor pheromone processing and aberrant secretion of the vacuolar hydrolase CPY. The obtained mutant alleles were complementation tested against previously identified grd and vps mutants to determine whether they fell into already identified complementation groups. Selected mutants were then back-crossed with the wild-type parent of the opposite mating type to confirm linkage of the mutant phenotype with the transposon insertion (LEU2 marker) and to determine whether the strain contained single or multiple transposons.
One recessive mutant (8923.176) had low but detectable A-ALP activity, moderate CPY missorting, and a strong defect in processing of α-factor. The 8923.176 mutant strain complemented all of the grd and vps mutants and thus represented a new grd complementation group named grd20. The GRD20 gene was cloned from a library of yeast genomic fragments carried in the CEN plasmid YCp50 (Rose et al., 1987) by complementation of a growth defect associated with the grd20::Tn3 allele (see below). The insert from a clone, pC-5, that complemented both the growth and grd phenotypes was sequenced and found by comparison with the yeast genome database to be derived from chromosome 5. By subcloning segments of the insert into the low-copy-number plasmid pRS316, the complementing ORF was found to be YER157w, a gene with no previously assigned function. PCR was used to confirm (see MATERIALS AND METHODS) that the sole Tn3 transposon in the 8923.176 mutant strain was inserted into the YER157w ORF around codon 183, demonstrating that YER157w was in fact GRD20.
GRD20 encodes an 801-amino-acid protein of predicted size 92.5 kDa and pI of 5.2. In reasonable agreement with the predicted size, a rabbit anti-Grd20p antibody detected a band of ∼109 kDa on a Western blot of total proteins from a wild-type yeast strain that was absent in a grd20 null strain (Figure 1A). Analysis of the sequence using various structure and domain-predicting computer programs (e.g., SMART; Schultz et al., 1998) predicted that Grd20p contains a region (amino acids 87–114) with a strong propensity for adopting coiled-coil structure (Figure 1B). No other well-characterized protein motifs such as signal sequence or transmembrane domains were found. BLAST database searches using the Grd20p amino acid sequence identified two ORFs with significant similarity to Grd20p: SBC1539.05 from Schizosaccharomyces pombe and Y71F9A_290.a from Caenorhabditis elegans (GenBank accession numbers AC006893 and CAB51337, respectively). A region of Y71F9A_290.a including nearly the entire 428-residue ORF was found to be 23.5% identical to a central region of Grd20p corresponding to residues 122–552 (Figure 1B). In addition, two human expressed sequence tags (GenBank accession numbers AA603511 and AA280321) also exhibit extensive similarity to residues 531–658 of Grd20p: 29 and 30% identity, respectively. However, the complete human sequences were not available, precluding a more extensive comparison. The significant conservation between the yeast, C. elegans, and human proteins suggests that Grd20p has homologues in higher eukaryotes that carry out functions similar to that carried out by Grd20p in yeast.
Figure 1.
The GRD20 encodes a conserved protein with a predicted coiled-coil domain. (A) Whole-cell protein extracts from strains SNY17 (GRD20), SBY7-7A (grd20::Tn3), and SBY4-10A (grd20-Δ1) were separated by SDS-PAGE and analyzed by Western blot using a rabbit anti-Grd20p antibody. (B) Drawings representing the Grd20p and Y71F9A-290.a protein sequences. The shaded boxes denote regions of sequence similarity between the two proteins (23.5% identity using the ALIGN program with default settings). The black box in Grd20p corresponds to a region (residues 87–114) having a high propensity for forming coiled-coil structure (p = 0.931 using COILS 2.1 program [Lupas et al., 1991] with a window of 28). The arrow indicates the approximate position of the Tn3 transposon insertion in the grd20::Tn3 allele.
grd20 Mutants Exhibit Dramatic Growth Defects
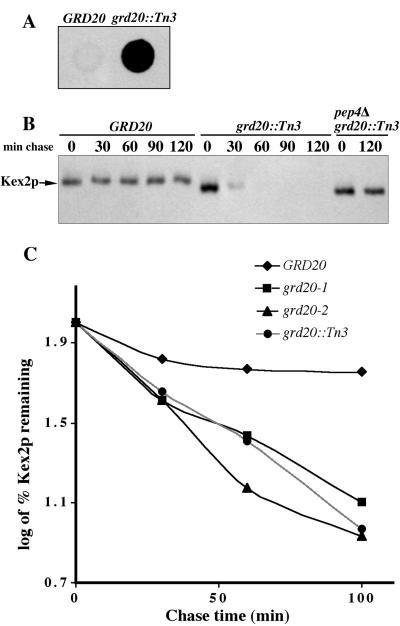
To initiate a more thorough phenotypic analysis of GRD20, a strain containing a complete deletion of the gene was generated. Whereas strains carrying the transposon insertion allele (grd20::Tn3) showed modest growth defects at 24 and 30°C and somewhat stronger defects at 36°C, strains carrying the null allele grd20-Δ1 grew extremely poorly at all temperatures tested (Figure 2). Loss-of-function mutations in several other genes in yeast have been shown to have dramatic effects on localization of Golgi membrane proteins but little or no effect on growth (Nothwehr et al., 1996, 1999; Brickner and Fuller, 1997; Voos and Stevens, 1998). Therefore, these results suggested that Grd20p might be involved in another process in addition to TGN localization. These data also indicated that the grd20::Tn3 allele must encode a partially functional protein despite being recessive to wild type. Consistent with this, a protein product of ∼145 kDa was detected in the grd20::Tn3 strain using a rabbit antibody generated against residues 7–251 of Grd20p (Figure 1A). PCR mapping indicated that the transposon is oriented so that GRD20 and the LacZ gene are transcribed in the same direction. In addition, grd20::Tn3 strains exhibit LacZ activity, indicating that the fusion is in-frame (our unpublished results). Thus the protein is derived from a fusion of GRD20 (first ∼183 codons) with LacZ. The predicted molecular mass of such a protein (130 kDa) also agrees reasonably well with the observed size (145 kDa) of the protein expressed from the grd20::Tn3 allele.
Figure 2.
A loss of Grd20p function causes a severe growth defect. The following strains were spotted onto YEPD plates from left to right: SNY17, SBY4-10A, SBY7-7A, SBY4-10A carrying pSB1, SBY4-10A carrying pSB1-125, and SBY4-10A carrying pSB1-1212 (see Table 2 for descriptions of plasmids). Approximately 5000, 500, and 50 viable cells were applied at the three positions (from top to bottom) for each strain. The plates were incubated at the indicated temperatures for 36 h before being photographed.
A Loss of Grd20p Function Results in Missorting of a Vacuolar Hydrolase
Newly synthesized CPY is modified from a core glycosylated endoplasmic reticulum (ER) form (p1CPY) to an outer chain modified form (p2CPY) because of the action of mannosyltransferases in the Golgi apparatus before being proteolytically processed to the mature form (mCPY) in the vacuole (Stevens et al., 1982). A loss of retention of TGN membrane proteins is often associated with a failure to properly sort the vacuolar hydrolase CPY, a process carried out in the TGN by a sorting receptor, Vps10p (Vida et al., 1993; Marcusson et al., 1994). The ability of the grd20 null and transposon insertion mutants to sort CPY was assessed by pulse labeling cultures with [35S]methionine and cysteine for 10 min and chasing for 45 min with unlabeled amino acids. CPY was then immunoprecipitated from extracellular and intracellular fractions to assess the amount of CPY that was aberrantly secreted. Whereas >90% of the newly synthesized CPY in a wild-type strain was transported to the vacuole and processed to the mature form, 52% of the CPY in the grd20-Δ1 strain was secreted in the Golgi modified p2CPY form (Figure 3A). The almost identical severity of the CPY missorting phenotype in the null and transposon allele strains (52 vs. 56% secretion) is in contrast to the obvious difference in growth rates (Figure 2). The uncoupling of these phenotypes suggests either that Grd20p contains multiple domains having distinct functions in the cell or that the minimum level of Grd20p function necessary to mediate normal growth is lower than that necessary for CPY sorting. Finally, the data demonstrate that CPY missorting is not a secondary consequence of poor growth.
Figure 3.
Grd20p is required for efficient sorting of the vacuolar hydrolase CPY. (A) Strains SNY17 (GRD20), SBY4-10A (grd20-Δ1), SNY97-14D (grd20::Tn3), and SBY4-10A/pSB10 (grd20-Δ1 + pSB10) were pulsed with [35S]methionine and cysteine for 10 min and chased with unlabeled amino acids for 45 min. The strains were incubated at 30°C throughout the experiment. CPY was immunoprecipitated from the intracellular (I) and extracellular (E) fractions and analyzed by SDS-PAGE and fluorography. The left panel was overexposed to allow detection of CPY in the grd20-Δ1 strain that incorporated label poorly. (B) The following strain/plasmid combinations were propagated for several doublings at 22°C: SBY4-10A/pSB1 (GRD20), SBY4-10A/pSB1-125 (grd20-1), and SBY4-10A/pSB1-1212 (grd20-2). The strains were then shifted to 36°C for 5 min or incubated at 22°C as indicated. After a 10-min pulse and 0- and 45-min chase, CPY was immunoprecipitated and analyzed as in A. (C) Strain SBY4-10A transformed with plasmids pSB1 + pSB18 (GRD20 + GRD20), pSB1 + pSN335 (GRD20 + grd20-1), pSB1 + pSN336 (GRD20 + grd20-2), and pSB1-125 + pSN336 (grd20-1 + grd20-2) was subjected to CPY immunoprecipitation as described in B after a 45-min chase. For both A and B the percentage of CPY secreted from each strain after a 45-min chase as determined by phosphorimager analysis is indicated below each E lane. The CPY secretion values in B represent the average of two experiments, and the values from each of those experiments are shown in Table 3 for strains incubated at 36°C. The positions of ER (p1) and Golgi (p2) precursor forms and vacuole localized mature form (m) of CPY indicated were assigned based on the migration of CPY expressed in wild-type strains.
Generation of grd20 Temperature-sensitive Mutants
Although multiple phenotypes were associated with loss of Grd20p function, it was possible that some of these phenotypes were indirectly related to Grd20p. Such indirect phenotypes would only appear after a prolonged absence of Grd20p function. To address this issue we generated a panel of temperature-sensitive grd20 mutants and analyzed their phenotypes in addition to the nonconditional alleles.
To generate temperature-sensitive mutants, yeast transformants carrying randomly mutagenized grd20 alleles were screened for growth and aberrant CPY secretion at the permissive and nonpermissive temperatures of 23 and 36°C, respectively (see MATERIALS AND METHODS). Based on the growth and CPY secretion phenotypes at 36°C, two general classes of mutants were found, as exemplified by the grd20-1 and grd20-2 alleles. Although both alleles exhibit near normal growth at 23°C, the grd20-1 strain exhibited little or no growth at 36°C (Figure 2). In contrast, the grd20-2 allele did exhibit growth at 36°C, albeit at a reduced rate.
To determine whether the CPY missorting phenotype was exhibited rapidly after the loss of Grd20p function, strains carrying the grd20-1 and grd20-2 alleles were propagated overnight at 22°C and then shifted to 36°C 5 min before a 10-min pulse and 45-min chase. Although strains carrying either of the alleles exhibited near normal CPY sorting at 22°C, the grd20-1 and grd20-2 strains exhibited substantial CPY missorting at 36°C (Figure 3B, lanes 3, 4, 11, and 12). The grd20-1 strain aberrantly secreted 47% of its CPY after just a 5-min preincubation at the nonpermissive temperature before the pulse. This level of missorting approaches that of the null strain (52%); thus the CPY sorting defect occurs very rapidly after a loss of Grd20p function. The extent of the CPY sorting defect in the grd20-2 strain was somewhat less than that of the other mutants (33% secretion), suggesting that under these conditions it still retains some residual CPY sorting function.
The p1, p2, and mature forms of CPY can be clearly observed at the 0-min chase time in the wild-type strain (Figure 3B, lane 5). However, the grd20-1 cells fail to convert p1CPY to the p2 form, although the CPY secreted from grd20-1 cells appears slightly larger than bona-fide p1CPY (Figure 3B, compare lanes 4 and 5). In the grd20-2 cells there appears to be a conversion of p1CPY to a partially glycosylated “pseudo p2” form (Figure 3B, compare lanes 5 and 9); thus the glycosylation defect is less severe for this mutant. Clearly, the underglycosylation of CPY is not due to a trafficking defect that would prevent CPY from reaching the Golgi mannosyltransferases, because the underglycosylated CPY is secreted. Rather, these data imply that either the localization or activity of one or more mannosyltransferases in the Golgi is affected in the mutants.
The extent of aberrant CPY secretion at 36°C and growth rate at 22 and 36°C were determined for five temperature-sensitive mutants representing the two phenotypic classes (Table 3). Unexpectedly, the majority of the CPY that remained intracellular after a 45-min chase in three of the mutants (grd20-1, grd20-3, and grd20-4) was not proteolytically processed to the mature form (Figure 3B, lane 3, and Table 3). These three mutants all exhibited little or no growth at 36°C. In contrast, wild-type and temperature-sensitive strains that grew at 36°C processed most of their intracellular CPY to the mature form. These results hint at a trafficking defect in grd20-1, grd20-3, and grd20-4 cells that prevents sorted CPY from reaching the vacuole or missorted CPY from reaching the cell surface. Surprisingly, the grd20 null mutant does not exhibit the marked intracellular accumulation of unprocessed CPY and does not appear to have a CPY glycosylation defect (Figure 3A). The grd20-1 and grd20-2 alleles do not appear to encode proteins that interfere with function of wild-type Grd20p (Figure 3C). However, in terms of the accumulation of unprocessed CPY and the glycosylation defect, the grd20-1 allele does dominate over grd20-2 in a haploid strain carrying both alleles (Figure 3, compare single mutant allele strains in B with the double mutant allele strain in C). Thus it is possible that grd20-1 encodes a protein that in the absence of wild-type Grd20p somehow interferes with trafficking of CPY. Alternatively, the results could be explained by the ability of grd20 null strains over a long period to somehow compensate for a complete loss of Grd20p.
Table 3.
Relationship between various phenotypes in grd20 temperature-sensitive strains
| Allele | Growth rate at 22°Ca | Growth rate at 36°C | % CPY secretion at 36°Cb | % intracellular CPY in pro formb | % cells with actin cablesc |
|---|---|---|---|---|---|
| GRD20 | ++++ | ++++ | <10, <10 | <5, <5 | 81 |
| grd20-1 | +++ | − | 47, 46 | 78, 64 | 0 |
| grd20-2 | +++ | ++ | 45, 20 | 43, 10 | 59 |
| grd20-3 | +++ | − | 34 | 70 | 0 |
| grd20-4 | +++ | − | 36 | 73 | 0 |
| grd20-5 | +++ | ++ | 32 | 29 | 43 |
The strains listed from the top to bottom correspond to SBY4-10A transformed with the following plasmids, respectively: pSB1, pSB1-125, pSB1-1212, pSB1-139, pSB1-23, and pSB1-65.
For determination of the growth rate, cells were spotted onto YEPD plates and incubated at the indicated temperatures as described in the legend to Figure 2. The relative rates of growth were qualitatively scored after 48 hr of growth with ++++ indicating maximal growth and − indicating little or no growth detected (i.e., refer to the data for the grd20-1 strain at 36°C).
The percent CPY secretion was determined as described in the legend to Figure 3B. The percent intracellular CPY in the nonvacuolar processed pro form was determined by dividing the amount of pro-CPY in the intracellular fraction by the total CPY in the intracellular fraction. The two percent secretion and percent pro-CPY values given for GRD20, grd20-1, and grd20-2 are the result of two independent determinations, whereas the result of a single experiment is given for the other strains.
The yeast strains were grown and stained with Texas Red-conjugated phalloidin according to the legend for Figure 9. A total of 100–150 cells from each strain were then scored for the presence of actin cables (see MATERIALS AND METHODS). The analysis was limited to cells with small buds and a single nucleus, because in wild-type cells actin cables in this phase of the cell cycle were easier to identify than at other phases of the cell cycle.
grd20 Mutants Exhibit Selective Effects on TGN Membrane Protein Localization
The α-factor processing defect in several yeast mutants has been previously shown to be due to mislocalization of the Kex2p endoprotease to the vacuole where it undergoes rapid degradation (Payne and Schekman, 1989; Nothwehr et al., 1996; Redding et al., 1996). Colony blot analysis demonstrates that the grd20::Tn3 strain secretes a substantial amount of unprocessed α-factor (Figure 4A). Therefore, we analyzed Kex2p turnover by pulse labeling yeast with [35S]methionine and cysteine, chasing for various times with unlabeled amino acids, and immunoprecipitating Kex2p. Kex2p was rapidly degraded in the grd20::Tn3 mutant strain with a half-time of ∼30 min (Figure 4B). In contrast, no degradation of Kex2p was detected in the wild-type strain even after a 120-min chase. The degradation of Kex2p in the grd20::Tn3 mutant was dependent on the vacuolar protease Pep4p and thus likely reflects mislocalization to the vacuole. Although the grd20 null allele mutants would be expected to be similarly defective for Kex2p retention, we were unable to characterize the Kex2p degradation phenotype of such strains because of poor incorporation of radioactive label apparently caused by slow growth.
Figure 4.
Kex2p is rapidly degraded by vacuolar proteases in grd20 mutants. (A) Colony immunoblot of strains AHY47/pSN179-A (GRD20) and SBY9-8A/pSN179-A (grd20::Tn3) in which secreted, unprocessed α-factor was detected using a rabbit anti-α-factor antibody. (B) Strains SNY17 (GRD20), SBY7-7A (grd20::Tn3), and SBY6 (pep4Δ grd20::Tn3) were pulsed for 10 min and chased for the indicated times at 30°C. Kex2p was then immunoprecipitated and analyzed by SDS-PAGE and fluorography. (C) The following strain/plasmid combinations were propagated for several doublings at 22°C, shifted to 36°C for 5 min, and then pulsed for 10 min and chased for the indicated times: SBY4-10A/pSB1 (GRD20), SBY4-10A/pSB1-125 (grd20-1), SBY4-10A/pSB1-1212 (grd20-2), and SBY7-7A. After immunoprecipitation and separation by SDS-PAGE, the percentage of Kex2p remaining at each time point was determined and normalized to the amount of Kex2p present at 0 min (arbitrarily set at 100%).
Kex2p turnover was also analyzed in strains carrying the temperature-sensitive grd20-1 and grd20-2 alleles that had been preincubated at the nonpermissive temperature for 5 min before carrying out the pulse and chase (Figure 4C). As has been previously described (Wilcox et al., 1992), we observed that Kex2p was more stable in wild-type strains at 30 than at 36°C (Figure 4, compare B and C). Strains carrying the temperature-sensitive alleles exhibited a severe Kex2p degradation defect, similar to that of the grd20::Tn3 strain (Figure 4C). Because both the Kex2p turnover and CPY missorting phenotypes occurred rapidly after a loss of function of Grd20p, it is likely that Grd20p carries out a function directly involved in protein sorting in the TGN and/or endosomal structures.
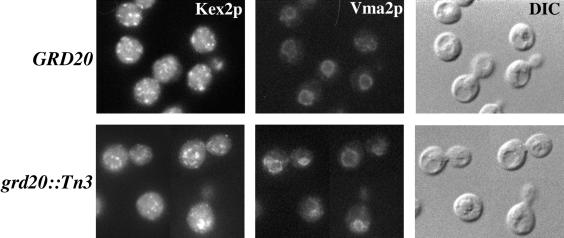
The subcellular location of Kex2p in the grd20::Tn3 mutant was investigated using immunofluorescence microscopy. In the experiment shown in Figure 5, both the GRD20 (wild-type) and grd20::Tn3 strains contained a mutation in the gene for the vacuolar protease Pep4p to prevent degradation of any aberrantly localized Kex2p. In wild-type cells localization of Kex2p expressed at normal levels from the chromosomal KEX2 allele exhibited a cytoplasmic punctate staining pattern typical of yeast TGN membrane proteins (Redding et al., 1991; Roberts et al., 1992). In the grd20::Tn3 strain, the staining was also punctate to some degree but generally tended to occur in patches clustered to a discrete region of the cell coinciding with the position of the vacuole. Vacuoles were observed in the cells by colocalization with the vacuolar membrane marker Vma2p and were indicated by the crater-like structures apparent using differential interference contrast optics (Figure 5, right panels). In some cases the Kex2p structures coincided with the vacuolar membrane, and in other cases they appeared to localize to the lumen of the vacuole. Interestingly, the membrane proteins carboxypeptidase S and Ste2p were recently shown to be targeted to the vacuolar lumen (Odorizzi et al., 1998), and it is possible that Kex2p uses a similar pathway, at least in grd20 cells. In the wild-type strain the Kex2p structures were scattered more randomly around the cell and were not associated with the vacuole. Taken together, these results indicate that in grd20 mutants Kex2p rapidly is mislocalized to the vacuole where it is degraded by vacuolar proteases.
Figure 5.
Kex2p is mislocalized to the vacuole in grd20 mutants. GRD20 pep4Δ (LSY2) and grd20::Tn3 pep4Δ (SBY6) strains propagated at 30°C were fixed, spheroplasted, and costained with antibodies against Kex2p and Vma2p. After subsequent treatment with fluorochrome-conjugated antibodies, the cells were viewed by differential interference contrast optics and by epifluorescence through filters specific for FITC and Texas Red.
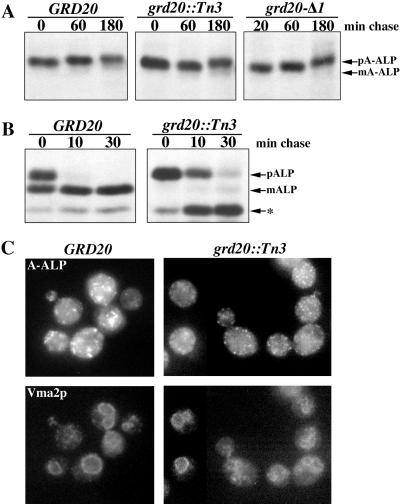
To determine whether localization of TGN membrane proteins other than Kex2p was affected in grd20 mutants, we analyzed the localization of A-ALP. Wild-type, grd20::Tn3, and grd20-Δ1 mutant cells were pulse labeled for 10 min and chased, and A-ALP was immunoprecipitated after each time point to determine whether it had undergone vacuolar processing. Surprisingly, the stability of A-ALP in the grd20 mutants was similar to that observed in the wild-type strain, and little or no A-ALP exhibited vacuolar processing within the chase period. For comparison, A-ALP is processed with a half-time of ∼60 min in several other grd mutants (Nothwehr et al., 1996). Vacuole-localized A-ALP and ALP under certain conditions are processed from the mature form to a faster-running form (discussed below), but no alternatively processed forms of A-ALP were observed within the 180-min chase period. Consistent with the view that A-ALP is not mislocalized to the vacuole in grd20 mutants, indirect immunofluorescence staining of A-ALP in grd20::Tn3 cells revealed a nonvacuolar staining pattern (Figure 6C). However, in the grd20::Tn3 mutant A-ALP appeared to be localized to punctate structures that were reduced in size and more numerous than in wild-type cells. It is interesting to note that the molecular mass of A-ALP increased slightly over the 180-min chase period in both wild-type and grd20 cells (Figure 6A), suggesting that A-ALP experiences prolonged exposure to Golgi mannosyltransferases in the mutant as well as wild-type strains. The low but detectable A-ALP activity that allowed us to identify the grd20::Tn3 mutant strain in our genetic screen does suggest, however, that in the steady state there is some low level of processed A-ALP in the vacuole.
Figure 6.
Localization and transport of A-ALP and ALP in grd20 mutants. Strains in A and B were propagated at 30°C, pulsed for 10 min, and chased for the indicated times, and either A-ALP or ALP was immunoprecipitated and analyzed by SDS-PAGE and fluorography. (A) Analysis of strains SNY17 (GRD20), SBY7-7A (grd20::Tn3), and SBY4-10A (grd20-Δ1) carrying a CEN plasmid directing expression of A-ALP (pSN55). (B) Analysis of strains SNY17 (GRD20) and SBY7-7A (grd20::Tn3) carrying a CEN plasmid directing expression of ALP (pSN92). The positions at which precursor A-ALP (pA-ALP), mature A-ALP (mA-ALP), precursor ALP (pALP), mature ALP (mALP), and a breakdown product of mature ALP (*) migrate are indicated. (C) Strains LSY2 (GRD20 pep4Δ) and SBY6 (grd20::Tn3 pep4Δ) carrying pSN55 were fixed, spheroplasted, and costained with antibodies against A-ALP (upper panels) and Vma2p (lower panels). After subsequent treatment with fluorochrome-conjugated antibodies, the cells were viewed by differential interference contrast optics and by epifluorescence through filters specific for FITC and Texas Red.
The CPY sorting defect in grd20 mutants hinted at a possible defect in trafficking of the CPY receptor Vps10p, because mutants such as vps35 fail to retrieve Vps10p from the prevacuolar compartment, leading to its default transport to the vacuole (Seaman et al., 1997; Nothwehr et al., 1999). Therefore, the degradation of Vps10p in wild-type and grd20::Tn3 cells was analyzed by pulsing for 10 min, chasing, and immunoprecipitation of Vps10p. In contrast to Kex2p (Figure 4B), Vps10p exhibited only minor degradation in the grd20::Tn3 strain even after a 180-min chase (Figure 7). Vps10p exhibited an immunofluorescence staining pattern similar to A-ALP in grd20::Tn3 cells (our unpublished results), indicating that Vps10p experiences very little mislocalization to the vacuole. These data suggest that A-ALP and Vps10p may be localized to a highly fragmented TGN or transport vesicles.
Figure 7.
The CPY receptor Vps10p is only slightly unstable in grd20 mutants. Strains SNY17 (GRD20) and SNY97–14D (grd20::Tn3) were pulsed for 10 min and chased for the indicated times at 30°C. Vps10p was then immunoprecipitated and analyzed by SDS-PAGE and fluorography. The positions of intact Vps10p and a form (*) generated by proteolytic cleavage by vacuolar proteases (Cereghino et al., 1995) are indicated.
Kex2p Is Mislocalized to the Vacuole in grd20 Cells Independent of the Plasma Membrane
The clathrin heavy chain and the dynamin family member Vps1p are thought to have a role in formation of vesicles from the TGN that carry cargo for eventual delivery to the vacuole (Conibear and Stevens, 1998). In strains lacking function of these proteins, resident TGN membrane proteins have been shown to be mislocalized to the plasma membrane (Seeger and Payne, 1992; Nothwehr et al., 1995). However, in strains defective for retrieval of TGN membrane proteins from the prevacuolar compartment, these proteins are mislocalized to the vacuole independent of the plasma membrane (Nothwehr and Hindes, 1997; Nothwehr et al., 1999; Seaman et al., 1997; Voos and Stevens, 1998). Therefore, we investigated whether the pathway used by Kex2p to reach the vacuole in grd20 mutants involved an initial mislocalization to the plasma membrane followed by endocytosis. If transport involved an initial mislocalization to the plasma membrane, then Kex2p would be packaged into secretory vesicles at the TGN, which would later fuse with the plasma membrane. To test this mechanism we combined the sec1-1 mutation, which blocks secretory vesicle fusion with the plasma membrane (Novick and Schekman, 1979), with the grd20::Tn3 mutation and analyzed the rate of Kex2p turnover at the nonpermissive temperature for sec1-1 (Table 4). Kex2p in the sec1-1 grd20::Tn3 double mutant was turned over with rapid kinetics, similar to the grd20::Tn3 single mutant (18- vs. 21-min half-life). A strain carrying the temperature-sensitive vps1-100 allele also exhibited rapid turnover of Kex2p (16 min), but in a sec1-1 vps1-100 strain Kex2p was very stable (>180 min). These data indicate that under the conditions of the experiment that a sec1-1 block in secretory vesicle fusion with the plasma membrane had been imposed. Taken together, the results support a model in which Kex2p reaches the vacuole independent of the plasma membrane in grd20 mutant cells; thus the role of Grd20p in Kex2p localization is clearly distinct from that of clathrin heavy chain and Vps1p.
Table 4.
Half-life of Kex2p expressed in a sec1-1 grd20 double mutant strain
| Relevant genotype | Half-life (min) at 36°C |
|---|---|
| Wild-type | 150 |
| grd20∷Tn3 | 21 |
| sec1-1 grd20∷Tn3 | 18 |
| vps1-100 | 16 |
| sec1-1 vps1-100 | >180 |
Strains SNY17 (wild-type), SBY7-7A (grd20∷Tn3), SNY108-1A (sec1-1 grd20∷Tn3), SNY2 (vps1-100), and LCY7 (sec1-1 vps1-100) were propagated at 22°C for several doublings and then shifted to 36°C for 30 min before pulsing for 10 min and chasing for various time points. Kex2p was immunoprecipitated and separated by SDS-PAGE, and the amount of Kex2p in each sample was determined (see MATERIALS AND METHODS). Plots of the log of the amount of Kex2p versus time were subjected to linear regression analysis to determine the half-time of Kex2p turnover.
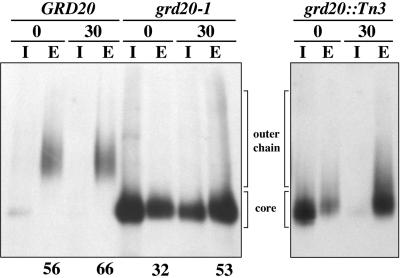
grd20 Mutants Exhibit Near Normal Secretion of Invertase and Trafficking of ALP to the Vacuole
The observation that the defective growth phenotype could be uncoupled from the Kex2p and CPY missorting phenotypes suggested that Grd20p was required for some process other than TGN membrane protein sorting. We initially explored the possibility that grd20 mutants may be defective in transport through the secretory pathway, because such a defect would be expected to affect growth dramatically. Wild-type, grd20::Tn3, and grd20-1 strains were analyzed for transport of the secretory protein invertase (Figure 8). The strains were propagated overnight at 22°C before being shifted to 36°C for 15 min, radioactively pulsed for 10 min, chased for 0 and 30 min, and subjected to immunprecipitation of invertase from the intracellular and extracellular fractions. In wild-type cells the core glycosylated ER form of invertase seen in the intracellular fraction after a 0-min chase completely received outer chain glycosylation by 30 min. The amount of invertase secreted from the grd20::Tn3 strain after 30 min was similar to that of the wild-type strain; however, the amount of secretion after 30 min in grd20-1 (53%) was somewhat less than that of wild type (66%). The results indicate mutations in GRD20 can cause a modest reduction in the rate of invertase secretion; however, it is unlikely that Grd20p plays a major role in trafficking of secretory proteins, because CPY is secreted from grd20 mutants, and the invertase secretion rate is not dramatically affected.
Figure 8.
Invertase secretion kinetics and carbohydrate processing in grd20 mutants. Strains SNY17 (GRD20), SBY4-10A/pSB1-125 (grd20-1), and SBY7-7A (grd20::Tn3) carrying plasmid pSB19 were propagated at 22°C and then shifted to 36°C for 15 min before a 10-min pulse and 0- or 30-min chase. Invertase immunoprecipitated from intracellular (I) and extracellular (E) samples was then analyzed by SDS-PAGE and fluorography. The positions of the ER glycosylated and early Golgi modified forms of invertase (core) and the medial and late Golgi forms (outer chain) are indicated. The percent invertase secretion at each time point as determined by phosphorimager analysis is indicated below the E lanes for strains GRD20 and grd20-1. The values for percent secretion represent the average of two independent experiments.
Similar to the results for glycosylation of CPY, invertase secreted from grd20 mutants was only partially converted from the core glycosylated ER form to the outer chain modified form, an event that involves mannose addition via mannosyltransferases in the Golgi. Furthermore, comparison of the migration of Kex2p from wild-type and grd20::Tn3 strains also suggests that Kex2p may be underglycosylated in the grd20::Tn3 mutant (Figure 4B).
Newly synthesized ALP is transported through the ER and Golgi before being packaged into TGN-derived vesicles for eventual delivery to the vacuole via a pathway distinct from that used by CPY, A-ALP, and Kex2p. As a further test of the integrity of the early secretory pathway and to test whether the ALP pathway is functional in grd20 mutants, we analyzed the rate of vacuole delivery of ALP in wild-type and grd20::Tn3 cells (Figure 6B). Immunoprecipitation of ALP from cells radioactively pulsed for 10 min and chased for various times revealed that by the end of the pulse period 54% of the ALP was delivered to the vacuole, as judged by its vacuolar proteolytic processing. This rate of vacuolar delivery is consistent with the previously reported half-time of ∼5 min (Klionsky and Emr, 1989). In grd20::Tn3 cells the rate of ALP delivery to the vacuole was delayed slightly (56% processing at 10 min vs. 89% for wild type) but by the 30-min time point 81% of the ALP had reached the vacuole. These results are consistent with the idea that trafficking through the ER and Golgi as well as in the ALP pathway between the TGN and the vacuole is not extensively perturbed in the absence of Grd20p function.
We did, however, note a difference in the manner in which ALP was processed in the vacuole of grd20::Tn3 cells (Figure 6B). In wild-type cells precursor ALP is converted to mature ALP, and then a breakdown product that exhibits greater SDS-PAGE mobility than the mature form gradually appears. This breakdown product is apparently generated in the vacuole, because its appearance is dependent on vacuolar proteases (our unpublished results). In the grd20::Tn3 mutant it appears that the mature protein is converted to the faster-running form much more rapidly. It is possible that this is caused by an imbalance of vacuolar hydrolases in the vacuole. Alternatively, underglycosylation of the ALP lumenal domain could cause increased exposure of the secondary cleavage site to proteolysis.
The Growth Defect in grd20 Mutants Correlates with a Defect in the Actin Cytoskeleton
Recent data have indicated a role for the actin cytoskeleton and associated proteins in membrane trafficking in animal cells (Stow et al., 1998) and in the yeast endocytic pathway (Kubler and Riezman, 1993; Munn et al., 1995). The yeast actin cytoskeleton consists of cables and filamentous actin patches that are polarized during most of the cell cycle. Actin patches are normally concentrated in regions of active secretion, such as a newly forming bud, and cables are oriented toward patch clusters (Kilmartin and Adams, 1984). Loss of function of a variety of genes disrupts the normal polarized localization of actin, and such defects are often linked with poor growth or lethality (Botstein et al., 1997).
To test whether grd20 mutants exhibited defects in the actin cytoskeleton, wild-type and grd20-1 cells were propagated at 22°C and then shifted to 36°C for 120 min. In wild-type strains actin is known to be transiently depolarized by sudden changes in temperature (Lillie and Brown, 1994); therefore, incubation for a full 120 min at 36°C was necessary to allow such perturbations to subside. After incubation at 36°C the cells were fixed and stained with Texas Red-conjugated phalloidin to label filamentous actin (Figure 9). In contrast to the wild-type strain, the grd20-1 cells exhibited a random distribution of actin patches between the mother cell and bud and an absence of actin cables. An essentially normal actin cytoskeleton was observed in the grd20-1 cells at 22°C (our unpublished results).
Figure 9.
Grd20p is necessary for normal actin polarization and organization. Strains SBY4-10A/pSB1 (GRD20) and SBY4-10A/pSB1-125 (grd20-1) were grown for several doublings at 22°C and then shifted to 36°C for 2 h, whereas strain SBY4-10A (grd20-Δ1) was propagated at 30°C before fixation. The cells were then fixed, stained with phalloidin conjugated to Texas Red, washed, mounted on slides, and visualized using differential interference contrast optics and by epifluorescence through a filter specific for Texas Red.
The actin polarization defects were quantified in all five grd20 temperature-sensitive mutants by determining the percentage of cells having visible actin cables (Table 3). All three alleles of the class that exhibited essentially no growth at 36°C (grd20-1, grd20-3, and grd20-4) exhibited strong actin depolarization defects, whereas the alleles that grew slowly at 36°C (grd20-2 and grd20-5) exhibited a nearly normal actin cytoskeleton. All of the mutants exhibited similar CPY sorting defects (Table 3), and grd20-1 and grd20-2 exhibited similar Kex2p localization defects (Figure 4C). However, the mutants can clearly be divided into two groups: 1) a group with essentially no growth at 36°C, intracellular accumulation of unprocessed CPY, and actin polarization defects; and 2) a group that grows slowly at 36°C, accumulates very little unprocessed CPY, and has near normal actin organization.
The effect of a complete loss of Grd20p function on the actin cytoskeleton was assessed by phalloidin staining of grd20-Δ1 cells that had been grown at 30°C. As was the case of the grd20-1 allele incubated at the nonpermissive temperature, actin polarization in the null allele strain was dramatically altered (Figure 9). Wild-type cells propagated at 30°C exhibited a phalloidin staining pattern (our unpublished results) that was indistinguishable from that of wild-type cells incubated for 2 h at 36°C (Figure 9). In contrast to the wild-type strain most of the grd20-Δ1 cells with small buds exhibited little if any polarization of actin patches, and only occasionally was very weak staining of cables observed. We noted a somewhat more extensive actin polarization defect in the grd20-2 and grd20-5 strains incubated at the nonpermissive temperature than in the grd20-Δ1 strain at 30°C, but nevertheless these results confirm that GRD20 is required for normal actin polarization. In addition, the grd20-Δ1 cells tended to clump together and exhibited altered budding morphology, with cells often showing elongated buds and in some cases multiple buds as well as abnormally wide bud necks (Figure 9). Such morphological defects have been observed in many other yeast mutants with defects in the actin cytoskeleton (Botstein et al., 1997). The grd20-1 temperature-sensitive mutant did not exhibit obvious morphological defects (Figure 9), suggesting that they only appear after a prolonged absence of Grd20p function.
Endocytosis of the a-Factor Receptor Is Defective in grd20 Mutants
The a-factor pheromone receptor Ste3p is subject to two modes of endocytosis: a ligand-induced process and a constitutive process that occurs in the absence of ligand (Davis et al., 1993). In both cases the receptor is internalized and delivered to the vacuole, where it is degraded in a PEP4-dependent manner. The rapid constitutive turnover of Ste3p (half-life of ∼20 min) is mediated by a signal in the cytoplasmic domain of Ste3p and is dependent on genes that mediate trafficking through the endocytic pathway. The actin cytoskeleton has been shown to be necessary for the internalization step of endocytosis of the α-factor pheromone receptor (Kubler and Riezman, 1993; Geli and Riezman, 1996). Because the actin cytoskeleton is perturbed in grd20 mutant cells, we assessed whether the grd20-1 mutant was defective for endocytic turnover of Ste3p.
To assess the half-life of Ste3p in wild-type and grd20-1 cells in the absence of ligand, the strains were transformed with a construct expressing Ste3p tagged at the C terminus with the c-myc epitope. This tagged STE3 allele has been shown to complement ste3 mutations, and the Ste3-myc protein exhibits turnover similar to wild-type Ste3p (Davis et al., 1993). The STE3::myc construct was under the control of the glucose-repressible GAL1 promoter. Wild-type and grd20-1 strains carrying the GAL1-STE3::myc construct were propagated for several generations at the permissive temperature of 22°C in media containing galactose as a carbon source. The cultures were then shifted to the nonpermissive temperature of 36°C, and glucose was added to shut off expression of STE3-myc. The amount of Ste3-myc protein present 0, 30, 60, and 90 min after addition of glucose was assessed by Western blot analysis using an antibody against the c-myc epitope (Evan et al., 1985). Consistent with the previously observed rapid constitutive turnover of Ste3p, in the wild-type strain the majority of Ste3-myc was degraded by 30 min, and it was undetectable by 90 min (Figure 10). In contrast, Ste3-myc was turned over much more slowly in the grd20-1 strain with substantial amounts of protein remaining after 90 min. For comparison, we also analyzed Ste3-myc turnover in a strain containing a temperature-sensitive mutation in the END4 gene, which is known to be necessary for internalization of both the α-factor and a-factor receptors from the cell surface (Raths et al., 1993; Roth and Davis, 1996). The block in turnover of Ste3-myc in the grd20-1 strain was nearly as extensive as in the end4-1 strain (Figure 10). These results indicate that delivery of Ste3-myc from the cell surface to the vacuole is blocked in grd20-1 cells and thus suggest a role for Grd20p in mediating transport of Ste3p through the endocytic pathway.
Figure 10.
Grd20p is required for rapid endocytic delivery of Ste3p to the vacuole. The following strain/plasmid combinations were grown for several generations at 22°C in minimal media containing galactose: SNY121-12A/pSB18 + pSL2099 (GRD20 END4), SBY4-10A/pSB1-125 + pSL2099 (grd20-1 END4), and SNY44-13B/pSL2099 (GRD20 end4-1). The cultures were then shifted to nonpermissive temperature (36°C), and 15 min later glucose was added to shut off expression of Ste3-myc. Equivalent volumes of culture were withdrawn at 0, 30, 60, and 90 min after addition of glucose. Protein extracts were immediately prepared from the cells, separated by SDS-PAGE, and analyzed by Western blotting using a mouse antic-myc antibody. The blots were developed as described in MATERIALS AND METHODS.
Grd20p Partially Colocalizes with the TGN Marker A-ALP
The requirement of Grd20p for actin organization and protein localization in the TGN suggested that Grd20p might associate with Golgi or endosomal compartments and/or with the actin cytoskeleton. To investigate localization of Grd20p, we introduced an HA epitope tag to its C terminus. The HA-tagged Grd20p was fully functional when expressed using the low-copy-number plasmid pSB10, as judged by complementation of the CPY sorting defect in a grd20 null strain (Figure 3A). The staining pattern of Grd20-HA as determined using fluorescence microscopy was consistent with a partially cytosolic localization, because cells exhibited a weak diffuse background staining (Figure 11). In addition to this diffuse staining pattern, however, was a punctate staining pattern reminiscent of that of Golgi membrane markers. The punctate structures were randomly distributed around cytoplasm of mother cells and buds and thus are unlikely to correspond to actin patches that are usually observed adjacent to the plasma membrane and are concentrated in the bud (Figure 9). When cells expressing untagged Grd20p were analyzed using the same anti-HA staining procedure, little or no staining was observed, demonstrating the specificity of this technique (our unpublished results). Costaining of cells for both Grd20-HA and the TGN marker A-ALP was carried out to test whether Grd20-HA associated with the TGN (Figure 11). Quantitative analysis (see MATERIALS AND METHODS) revealed that 51% of the Grd20-HA-positive structures also stained for A-ALP. Thus Grd20-HA associates with the TGN and some other unidentified structures, possibly early or medial Golgi compartments. For structures that stained for both proteins we sometimes observed subtle differences in the shape of the structure as viewed by Grd20-HA staining as compared with that seen by A-ALP staining. These data may indicate that Grd20p and A-ALP associate with different subregions of TGN compartments or could reflect a loose association of Grd20-HA with the TGN, possibly as a component of a Golgi-associated cytoskeletal complex.
Figure 11.
Grd20-HA partially colocalizes with the TGN marker A-ALP. Strain SBY4-10A carrying plasmids pSB10 and pAH16 was fixed, spheroplasted, and costained with anti-HA and anti-ALP antibodies. The anti-HA antibody was detected with an alexa 488-conjugated secondary antibody, whereas the anti-ALP antibody was detected with a Texas Red-conjugated secondary antibody. The cells were viewed by epifluorescence using filters specific for each fluorochrome. Two fields stained for each antigen are shown, with the panels on the right representing images derived from a merge of the images for each antigen.
DISCUSSION
The localization of membrane proteins to the TGN of eukaryotic cells is a complex dynamic process involving both static retention and retrieval from a post-Golgi compartment. Our studies on GRD20 underscore differences in the trafficking pathways for various TGN membrane proteins and suggest a link between the actin cytoskeleton and protein sorting in the TGN/endosomal system.
Role of Grd20p in Localization and Trafficking of CPY and TGN Membrane Proteins
The loss of Grd20p function severely affects localization of Kex2p but has a much different effect on localization and trafficking of the TGN membrane proteins A-ALP and Vps10p. We observed rapid vacuolar degradation of Kex2p and missorting of CPY in the grd20::Tn3 mutant, and these phenotypes were exhibited shortly after loss of Grd20p function in grd20 temperature-sensitive strains. In contrast to Kex2p, A-ALP and Vps10p exhibited little or no mislocalization to the vacuole in the grd20::Tn3 strain. Rather, these proteins exhibited diffuse cytoplasmic staining patterns. Similar results were obtained when A-ALP localization was analyzed in the grd20 null strain. As discussed in INTRODUCTION, the machinery for retrieval of Kex2p, Vps10p, and A-ALP from the prevacuolar endosome is highly overlapping. Thus, a novel aspect of this work is the apparent specificity of Grd20p for localization of Kex2p.
How do we explain the differences in the fate of Kex2p compared with Vps10p and A-ALP in grd20 mutant cells? If in fact A-ALP and Vps10p are localized to a fragmented TGN in grd20 cells, there are at least two possible models capable of explaining our data. First, it is possible that Grd20p is required for retrieval of Kex2p but not for Vps10p and A-ALP. Vps10p and A-ALP could then cycle between the TGN and the prevacuolar endosome, whereas nonretrieved Kex2p is rapidly transported to the vacuole. Alternatively, the normally slow rate of Kex2p transport from the TGN to the prevacuolar endosome could be accelerated in grd20 mutants with no affect on the rate of transport of Vps10p and A-ALP. The fact that CPY sorting is partially defective in grd20 mutants indicates that some aspect of Vps10p function has been affected. One possibility is that Vps10p has nearly normal trafficking between a fragmented TGN and the prevacuolar endosome in grd20 mutants, but some aspect of Vps10p function at the TGN has been affected. For example, ionic conditions within the TGN might be altered, preventing efficient binding of the CPY to its receptor. Sufficient levels of Ca2+ and Mn2+ ions are known to be necessary for efficient CPY sorting (Dürr et al., 1998).
The staining patterns for A-ALP and Vps10p in the grd20::Tn3 mutant could instead reflect entrapment of these proteins in transport vesicles. These vesicles would be more likely to originate from the prevacuolar endosome rather than from the TGN, because anterograde trafficking between the TGN and vacuole does not seem to be generally impaired in grd20 mutants. Kex2p is mislocalized to the vacuole in the grd20::Tn3 mutant; thus the entrapment of A-ALP and Vps10p in vesicles in this mutant would imply that in wild-type cells Kex2p uses a different set of retrograde vesicles than are used by Vps10p and A-ALP. This leads then to a complex model in which Grd20p is required for consumption of certain vesicles at the TGN and is also required at a separate step for retention of Kex2p, such as in retrieval from the prevacuolar compartment.
Although it seems clear that Kex2p, A-ALP, and Vps10p pass through the prevacuolar endosome during their cycling itinerary (Cereghino et al. 1995; Piper et al., 1995; Rieder et al., 1996; Voos and Stevens, 1998; Nothwehr et al., 1999), it is possible that Kex2p may initially be transported from the TGN to a third compartment such as an early endosome before being transported to the prevacuolar endosome. The specificity of Grd20p for Kex2p retention could then be explained by a defect in transport of Kex2p from an early to prevacuolar endosome, resulting in aberrant trafficking to the vacuole. Additional analysis of the trafficking of these proteins in wild-type and grd20 mutant cells will be needed to distinguish between these models.
Grd20p is required for optimal function of the carbohydrate processing machinery in the Golgi apparatus, as judged by the failure of CPY and invertase to be converted from their ER to Golgi modified forms. Thus it is likely that Grd20p performs a role at Golgi compartments proximal to the TGN as well as at the TGN itself and possibly endosomes. Consistent with this idea, we find using fluorescence microscopy that Grd20p exhibits a punctate distribution that partially colocalizes with the TGN membrane protein A-ALP. Given the grd20 defects in glycosylation reactions known to occur in Golgi compartments proximal to the TGN, an intriguing possibility is that Grd20p localizes to both the TGN and earlier Golgi compartments and is required for localization of resident membrane proteins throughout the Golgi. Alternatively, alterations in the levels of Ca2+ and Mn2+ ions could explain the glycosylation defect, because these ions are necessary for efficient glycosylation in the Golgi (Nakajima and Sharma et al., 1974; Ballou, 1975; Parodi, 1979; Haselbeck and Schekman, 1986). Levels of these ions in the yeast Golgi are regulated by the ion pump Pmr1p (Dürr et al., 1998). Thus alteration in Pmr1p levels in the Golgi could explain several phenotypes associated with grd20. Exploration of these models should represent fruitful directions for future studies.
We were surprised to find that certain grd20 temperature-sensitive alleles such as grd20-1 exhibited a marked intracellular accumulation of unprocessed CPY in contrast to the grd20 null allele. The basis of the buildup of unprocessed CPY is most likely due to a trafficking defect rather than a vacuolar protease defect, because the phenotype was observed rapidly after shifting of grd20-1 cells to the nonpermissive temperature. The vacuoles of such cells incubated at permissive temperature process CPY normally (Figure 3B), indicating that before shift they contain normal processing activity. It seems very unlikely that the temperature shift could rapidly inactivate the CPY processing enzymes protease A and B (Vandenhazel et al., 1996) in grd20-1 cells. In addition, grd20-1 vacuoles do not appear to have a general proteolysis defect, because Kex2p is rapidly degraded in the vacuole.
If a trafficking defect reduces the rate of CPY to the vacuole of grd20-1 cells, the next question is where such a block would occur. A defect in ER-to-Golgi and/or intra-Golgi trafficking of CPY is possible but seems unlikely, because Kex2p is rapidly transported to and degraded in the vacuole in grd20-1 cells. Kex2p is transported to the vacuole independent of the plasma membrane in the grd20::Tn3 mutant, suggesting that it uses the TGN-to-late endosome-to-vacuole pathway also used by CPY, although as discussed above, a variation of this trafficking pathway is possible. A small but significant reduction in the rate of invertase secretion was observed in grd20-1 cells, and the same trafficking defect may be responsible for the buildup of unprocessed CPY. Taken together, we think our data are most consistent with a model in which the rate of transport of missorted CPY between the TGN and cell surface is reduced in certain mutant grd20 alleles such as grd20-1. Interestingly, mutations in several genes that affect the actin cytoskeleton also affect membrane trafficking in the late secretory pathway (Botstein et al., 1997), and there is a good correlation between the actin polarization and CPY trafficking defects in our panel of grd20 mutants. The grd20-1 allele was found to be dominant over an allele with a very small amount of accumulation of unprocessed CPY; thus it is likely that the protein product of grd20-1 interferes in some way with trafficking.
A Link between the Yeast Actin Cytoskeleton and TGN Function?
Like several other genes in yeast, GRD20 was found to be required for proper organization and polarization of the actin cytoskeleton. Analysis of a collection of randomly mutagenized temperature-sensitive alleles indicated that mutants that failed to grow at 36°C consistently exhibited the actin cytoskeleton defect, whereas mutants that grew at 36°C (albeit slowly) had normal actin organization. Thus the extremely slow growth of the grd20 null mutant strain appears to be due at least in part to a defect in actin polarization. Many of the mutations that have been described as causing actin polarization defects are in genes encoding proteins that clearly have a direct role in the actin cytoskeleton, such as actin itself (Novick and Botstein, 1985), capping protein (Armatruda et al., 1990), and tropomyosin (Liu and Bretscher, 1989). In contrast, mutations in other genes that cause actin organization defects such as the d-lactate dehydrogenase gene (Amberg et al., 1995) do not have an obvious role in the actin cytoskeleton. Although mutants that have actin defects usually grow poorly, it is clear that actin depolarization is a specific phenotype and not simply a secondary effect of poor growth. This was recently demonstrated by analyzing a large number of yeast mutants having poor growth. Only a small subset was defective for actin organization (Karpova et al., 1998). Therefore, although the actin polarization defect in grd20 mutants correlated with poor growth, it is unlikely that the actin defect is a consequence of the growth defect per se. Rather, it is more likely that both the growth and actin defects are directly related to a loss of some aspect of Grd20p function.
There is a growing body of evidence implicating the actin cytoskeleton in yeast vesicular trafficking pathways. For example, in both yeast and animal cells the actin cytoskeleton plays a role in endocytosis. Mutations in the yeast actin gene ACT1 and in a gene encoding a homologue to the mammalian actin-bundling protein fimbrin caused defects in internalization of the α-factor receptor Ste2p (Kubler and Riezman, 1993). In addition, the normal polarized distribution of actin is altered in several yeast endocytosis mutants (Bθnθdetti et al., 1994; Munn et al., 1995; Wendland et al., 1996). The defect in endocytic transport of Ste3-myc in the grd20-1 mutant strain reported in this study is thus likely to stem from a disorganized actin cytoskeleton. The actin cytoskeleton in yeast has also been shown to play a role in polarization of the late secretory pathway so that exocytic vesicles are directed toward the plasma membrane of the bud (Novick and Botstein, 1985). An actin-binding protein that might mediate interactions between secretory vesicles and actin is Myo2p, an unconventional myosin (Johnston et al., 1991; Govindan et al., 1995).
Recent data have also implicated a role for actin specifically in Golgi function in animal cells. An isoform of the actin-binding protein spectrin has recently been shown to associate with the Golgi apparatus (Beck et al., 1994; Devarajan et al., 1997). Moreover, the spectrin-associated protein ankyrin associates with the TGN and ER (Devarajan et al., 1996; Beck et al., 1997). The well-studied spectrin network on the erythrocyte membrane forms a meshwork attached to actin filaments (Hartwig, 1995). Thus, by analogy it is thought that association of spectrin and ankyrin with the Golgi apparatus may provide structural integrity and specify subdomains on Golgi membranes. Intriguingly, spectrin appears to also have a role in sorting and transport of cargo proteins (Devarajan et al., 1997).
Previous evidence for a connection between the actin cytoskeleton and the Golgi apparatus in yeast derives from the discovery of the SAC1 gene that is synthetically lethal with the act1-2 allele and is required for actin polarization (Novick et al., 1989). In addition, SAC1 genetically interacts with the SEC14 gene that encodes a phosphatidylinositol transfer protein necessary for membrane trafficking through the Golgi (Cleves et al., 1989). Sac1p is a membrane protein that is localized to both the Golgi apparatus and the ER and is required for ATP import into the ER and also acts in lipid metabolism as a polyphosphoinositide phosphatase (Whitters et al., 1993; Guo et al., 1999; Kochendorfer et al., 1999). Thus SAC1 influences the levels of polyphosphoinositides, which in turn have been proposed to regulate actin organization (Guo et al., 1999).
Does the actin cytoskeleton play a role in sorting and trafficking of TGN/endosomal membrane proteins in yeast? In the case of GRD20, a subset of recessive alleles that have defects in TGN retention and CPY sorting also have actin polarization defects. Grd20p does not appear to be a stably associated component of the actin cytoskeleton, because it does not seem to colocalize with actin patches or cables (Figure 11; our unpublished results). This observation coupled with the existence of yeast mutants that have altered actin organization but appear to have normal CPY sorting (Liu and Bretscher, 1992; Moreau et al., 1997; Srinivasan et al., 1997) would seem to argue against a direct role for actin in protein-sorting events at the TGN. However, an exciting possibility is that Grd20p may regulate or be a component of an unidentified yeast Golgi-associated cytoskeletal complex that is also associated in some way with actin. Ankyrin-like repeat-containing proteins have been identified in yeast, and least one of these, Akr1p, plays a role in protein trafficking (Givan and Sprague, 1997). Thus a putative Golgi-associated cytoskeletal complex could be analogous to the spectrin–ankyrin network that associates with the Golgi of animal cells. An indirect association of actin with TGN protein sorting would be more consistent with the existence of mutations that affect actin but not TGN sorting. The grd20 null mutant exhibited a substantial actin polarization defect but was somewhat less affected than the grd20-1 class of temperature-sensitive alleles. Thus it is possible that, to a certain degree, alleles such as grd20-1 may interfere with actin organization. The severity of the actin defect could reduce the rate of transport in the late secretory pathway, which could account for the accumulation of unprocessed intracellular CPY in such mutants.
The possibility that TGN localization of the Kex2p protease in yeast may depend on an actin-associated cytoskeletal components is suggested by the recent discovery that furin, the animal cell homologue of Kex2p, interacts with ABP-280, a nonmuscle filamen involved in actin cross-linking (Liu et al., 1997). ABP-280 interaction with the cytosolic domain of furin was shown to modulate the rate of internalization of furin from the cell surface and to mediate the retrieval of furin from early endosomes to the TGN. Thus it is possible that an actin-associated protein in yeast may perform a similar function. Our identification of Grd20p may serve as an entry point to the identification of components of the cytoskeleton that have a role in Golgi function in yeast.
ACKNOWLEDGMENTS
We thank John Cooper and Beverly Wendland for useful discussions and reagents for actin analysis. Scott Emr, Tom Stevens, Nick Davis, and Randy Schekman generously provided antibodies and/or plasmids. Anna Hindes provided excellent technical assistance, and David Eide and Liz Conibear provided valuable critical evaluation of the manuscript. This work was supported by a US Department of Education Graduate Assistance in Areas of Need fellowship to R.G.S. and National Institutes of Health grant GM-53449 to S.F.N.
Abbreviations used:
- ALP
alkaline phosphatase
- CEN
yeast centromere
- CPY
carboxypeptidase Y
- DPAP
dipeptidyl aminopeptidase
- ER
endoplasmic reticulum
- HA
hemagglutinin
- H+L
heavy and light chain
- IgG
immunoglobulin G
- ORF
open reading frame
- TGN
trans-Golgi network
REFERENCES
- Amberg D, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Armatruda JF, Cannon JF, Tatchell K, Hug C, Cooper JA. Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature. 1990;344:352–354. doi: 10.1038/344352a0. [DOI] [PubMed] [Google Scholar]
- Beck KA, Buchanan JA, Malhotra V, Nelson WJ. Golgi spectrin: identification of an erythroid beta-spectrin homolog associated with the Golgi complex. J Cell Biol. 1994;127:707–723. doi: 10.1083/jcb.127.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck KA, Buchanan JA, Nelson WJ. Golgi membrane skeleton: identification, localization and oligomerization of a 195 kDa isoform associated with the Golgi complex. J Cell Sci. 1997;110:1239–1249. doi: 10.1242/jcs.110.10.1239. [DOI] [PubMed] [Google Scholar]
- Bθnθdetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Amberg D, Mulholland J, Huffaker T, Adams A, Drubin D, Stearns T. The yeast cytoskeleton. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 1–92. [Google Scholar]
- Brickner JH, Fuller RS. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J Cell Biol. 1997;139:23–26. doi: 10.1083/jcb.139.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Piper RC, Weisman LS, Stevens TH. Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J Cell Biol. 1998;142:651–663. doi: 10.1083/jcb.142.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes & Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GG. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R, Munro S. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Cooper A, Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997a;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Snyder WB, Burd CG, Emr SD. Novel Golgi to vacuole delivery pathway in yeast—identification of a sorting determinant and required transport component. EMBO J. 1997b;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis NG, Horecka JL, Sprague GF., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan P, Stabach PR, De Matteis MA, Morrow JS. Na+, K+-ATPase transport from the endoplasmic reticulum to the Golgi requires the Golgi spectrin-ankyrin G119 skeleton in Madin Darby canine kidney cells. Proc Natl Acad Sci USA. 1997;94:10711–10716. doi: 10.1073/pnas.94.20.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan P, Stabach PR, Mann AS, Ardito T, Kashgarian M, Morrow JS. Identification of a small cytoplasmic ankyrin (AnkG119) in the kidney and muscle that binds βIΣ* spectrin and associates with the Golgi apparatus. J Cell Biol. 1996;133:819–830. doi: 10.1083/jcb.133.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr G, Strayle J, Plemper R, Elbs S, Klee SK, Catty P, Wolf DH, Rudolph HK. The medial Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum associated protein degradation. Mol Biol Cell. 1998;9:1149–1162. doi: 10.1091/mbc.9.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RS, Sterne RE, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport—cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Role of type I myosins in receptor-mediated endocytosis in yeast. Science. 1996;272:533–535. doi: 10.1126/science.272.5261.533. [DOI] [PubMed] [Google Scholar]
- Givan SA, Sprague GF. The ankyrin repeat-containing protein akr1p is required for the endocytosis of yeast pheromone receptors. Mol Biol Cell. 1997;8:1317–1327. doi: 10.1091/mbc.8.7.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SL, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay H, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig JH. Actin-binding proteins. 1: spectrin super family. Protein Profile. 1995;2:703–800. [PubMed] [Google Scholar]
- Haselbeck A, Schekman R. Interorganelle transfer and glycosylation of yeast invertase in vitro. Proc Natl Acad Sci USA. 1986;83:2017–2021. doi: 10.1073/pnas.83.7.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Moltz SL, Riles LE, Güldener U, Hegemann JH, Veronneau S, Bussey H, Cooper JA. Depolarization of the actin cytoskeleton is a specific phenotype in Saccharomyces cerevisiae. J Cell Sci. 1998;111:2689–2696. doi: 10.1242/jcs.111.17.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochendorfer KU, Then AR, Kearns BG, Bankaitis VA, Mayinger P. SacIp plays a crucial role in microsomal ATP transport, which is distinct from its function in Golgi phospholipid metabolism. EMBO J. 1999;18:1506–1515. doi: 10.1093/emboj/18.6.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GP, Thomas L, Warren RA, Enns CA, Cunningham CC, Hartwig JH, Thomas G. Cytoskeletal protein ABP-280 directs the intracellular trafficking of furin and modulates proprotein processing in the endocytic pathway. J Cell Biol. 1997;139:1719–1733. doi: 10.1083/jcb.139.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bretscher A. Characterization of TPM1 disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretory vesicle. Mol Biol Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Stevenson BJ, Geli MI, Riezman H. end5, end6, and end7—mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Ballou CE. Yeast manno-protein biosynthesis: solublization and selective assay of four mannosyltransferases. Proc Natl Acad Sci USA. 1975;72:3912–3916. doi: 10.1073/pnas.72.10.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bruinsma P, Strawn LS. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol Biol Cell. 1999;10:875–890. doi: 10.1091/mbc.10.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Hindes AE. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Novick P, Osmond BC, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Parodi AJ. Biosynthesis of yeast mannoproteins. Synthesis of mannan outer chain and of dolichol derivatives. J Biol Chem. 1979;254:8343–8352. [PubMed] [Google Scholar]
- Payne GS, Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Piper RC, Bryant NJ, Stevens TH. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the vps-dependent pathway. J Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Brickner JH, Marschall LG, Nichols JW, Fuller RS. Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol Cell Biol. 1996;16:6208–6217. doi: 10.1128/mcb.16.11.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Roth AF, Davis NG. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the Vps29, Vps30, and Vps35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CB, Babczinski P, Lehle L, Tanner W. The role of dolicholmonophosphate in glycoprotein biosynthesis in Saccharomyces cerevisiae. Eur J Biochem. 1974;46:35–41. doi: 10.1111/j.1432-1033.1974.tb03594.x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy SF, Emr S, Decamilli P, Nussbaum R. Disruption of three phosphatidylinositol-polyphosphate 5-phosphotose genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol. 1997;74:350–360. [PubMed] [Google Scholar]
- Stack JH, Horazdovsky BF, Emr SD. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- Stepp JD, Huang K, Lemmon SK. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Stevens TH, Rothman JH, Payne GS, Schekman R. Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J Cell Biol. 1986;102:1551–1557. doi: 10.1083/jcb.102.5.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow JL, Faith KR, Burgess DR. Budding roles for myosin II on the Golgi. Trends Cell Biol. 1998;8:138–141. doi: 10.1016/s0962-8924(98)01238-0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenhazel HB, Kiellandbrandt MC, Winther JR. Biosynthesis and function of yeast vacuolar proteases—review. Yeast. 1996;12:1–16. doi: 10.1002/(sici)1097-0061(199601)12:1<1::aid-yea902>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald SI, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Huyer G, Emr SD. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol. 1993;121:1245–1256. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Stevens TH. Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J Cell Biol. 1998;140:577–590. doi: 10.1083/jcb.140.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels JJ, Payne GS. A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 1998;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]