It is a great honor and privilege to stand before you today to deliver the 2008 American Society of Clinical Investigation Presidential Address. I have chosen a controversial topic — and one that I know intimately — that challenges the standard operating procedures of our profession at many levels. While no one will object to the goal of applying basic biomedical research insights to clinical medicine, we can certainly debate the methods by which we encourage this process. Translational medicine has rapidly emerged as the rallying cry for many stakeholders in biomedical research, most notably the Congress and the public, who rightfully want to hold us accountable for their investment of taxpayer dollars into our enterprise. Major funding sources have channeled significant portions of their budgets into new translational research programs, many of which prescribe specific projects and even link delivery of resources to completion of defined milestones. Is this the best way to ensure bench-to-bedside science? Can translational research be directed from headquarters?
Before I begin that discussion, I want to reflect on the history of ASCI on our 100th anniversary, as there are many lessons in the past. As the story goes, ASCI was founded in 1908 by 8 young physicians, all in their early 30s (1). They had been regular attendees at the annual Association of American Physicians meeting, but were frustrated with the conventionalism of the old guard and eager to incorporate more science into medicine. In response they created the ASCI, a new society of young physician-scientists with a greater focus on the interface between science and medicine. Membership was restricted, as today, to those in the early stages of their careers. The phrase “Young Turks” comes from the coincident Young Turk Revolution against Sultan Abdul Hamid II of the Ottoman empire.
I should disclose that I majored in history as an undergraduate at Princeton and was taught to dig deep into primary source material. In the same way that we all view early scientific data from our labs with a skeptical eye, I looked into this fairy tale of renegade youth a bit more closely. It turns out that the true instigator for the formation of ASCI was not so young. This was Dr. Samuel Meltzer (Figure 1), a New York City clinician in his mid-50s!
Figure 1. Samuel Meltzer, first ASCI president.

Dr. Meltzer was born in Russia and trained in Berlin, where he was exposed to science-based medicine in the European tradition. For unclear reasons, after this high-level training, he established a clinical practice in Harlem, New York, rather than pursue a medical research career in Germany. Then, at the age of 53, he was selected to head the first Department of Physiology and Pharmacology at the newly established Rockefeller Institute. This seems a remarkable turn of events for a New York City clinician. But in his spare time on nights and weekends, Dr. Meltzer had been conducting experiments in physiology at Columbia — riding his horse over from Harlem after seeing his last patients — and publishing his results in the medical literature. A sampling of his publications can be found on PubMed (Figure 2).
Figure 2. Selected publications by Samuel Meltzer.
Dr. Meltzer clearly took his suboptimal experience in blending a science-medicine career to heart in his new position at Rockefeller. He became a very strong, nationally visible advocate for proper physician-scientist training. It was he who instigated the first 8 Young Turks to establish the ASCI. In appreciation of his role as their mentor, the Young Turks selected him as their first president.
Dr. Meltzer delivered the first ASCI Presidential Address, entitled “The science of clinical medicine: what it ought to be and the men to uphold it,” one year later. In it, he laid out the fundamental principles of obtaining proper laboratory-based scientific training and of maintaining a focus on clinical science rather than clinical practice.
There can be little doubt that over the past century we have met Dr. Meltzer’s goals. The evidence is obvious: the proliferation of state-sponsored medical schools across the US, the establishment of scientific standards of excellence through the Flexner report, the endowment of many leading private medical institutions to enable the development of pure medical science unfettered by the demands of clinical practice, and, of course, the establishment of combined MD-PhD programs at medical schools throughout the country.
But with these successes, significant challenges remain. I list only a few here. These include unpredictable swings in research funding that discourage young investigators from following in our footsteps, the misalignment of the goals of revenue-driven hospitals with the academic medical science mission, the growing distractions of administrative work, and so on. These and related issues have been eloquently covered in prior Presidential Addresses (2, 3).
When I was a house officer at the University of California, San Francisco, my attending, physician-scientist Joel Ernst, handed me a copy of Joe Goldstein’s 1986 ASCI Presidential speech where he reported several cases of PAIDS (Paralyzed Academic Investigator Disease Syndrome). Having just reviewed nominations for ASCI membership, Joe had seen far too many applications from investigators who were inadequately trained to move important clinically relevant observations forward. He offered this prescription: basic science training and technical courage (4).
In 1988 I began my postdoctoral studies in Owen Witte’s laboratory at UCLA, working on the BCR-ABL translocation found in chronic myeloid leukemia. Having made progress studying the signaling properties of this fusion tyrosine kinase oncogene and mapping the requirements for leukemic transformation, I began my independent career on the 80/20 (80 percent lab, 20 percent clinical) physician-scientist track familiar to us all. We enter this track with the idea that these 2 parts of our lives are connected, but we know this rarely happens. In fact, we are often warned against trying to do both because clinical time becomes a service obligation and a distraction from our primary focus. I was fortunate to have them converge.
Many of you are familiar with the story of imatinib (Gleevec) in chronic myeloid leukemia. I want to share a brief vignette of this story to illustrate how our laboratory and clinical lives can be linked in this era of molecular medicine and offer an example of the kind of translational research our society should champion. In the winter of 1999, Brian Druker and I witnessed dramatic drops in the blood counts of 6 CML patients we treated with imatinib in the phase I study. This single result was the culmination of a series of advances dating back to the first description of the Philadelphia chromosome by Peter Nowell (5); the recognition of the reciprocal translocation of chromosomes 9 and 22 by Janet Rowley (6); the molecular identification of the BCR-ABL fusion tyrosine kinase by Owen Witte, Eli Canaani, Nora Heistercamp, and John Groffen (7–9); and the demonstration that BCR-ABL causes CML in mice by George Daley and David Baltimore (10). In parallel, advances in combinatorial chemistry and in robotic high-throughput screening allowed the synthesis of large chemical libraries containing millions of compounds that could be quickly screened against different protein targets. Imatinib emerged as an optimized hit from the screening program at Ciba-Geigy Pharmaceuticals, directed by Alex Matter and Nick Lyden, against the platelet-derived growth factor receptor. Only later was it appreciated that imatinib inhibits ABL. Brian Druker showed that imatinib was effective in preclinical models of CML, and, together with Novartis, we moved it into the clinic, where it succeeded beyond our wildest dreams (11).
What you may not recall is that less than a year after we saw these dramatic responses in CML, we began to see patients lose their response to imatinib, initially those in blast crisis. Since these relapses were occurring while patients were still taking the drug, we reasoned that either BCR-ABL–independent subclones had arisen or that the drug was no longer inhibiting BCR-ABL. After ruling out trivial explanations such as enhanced drug metabolism, we ran the definitive experiment — assessing BCR-ABL activity in tumor cells at relapse by measuring phosphorylation of a downstream substrate CRKL. In all cases, kinase activity was restored! Furthermore, this biochemical resistance to kinase inhibition was cell autonomous because we could not inhibit BCR-ABL when the cells were exposed to imatinib ex vivo. We made a short list of the possible explanations and, in very short order, discovered a novel amino acid substitution in the BCR-ABL kinase domain in the resistance subclones. Reconstitution studies proved that this mutation conferred drug resistance in biochemical and cell growth assays — and we were done (12).
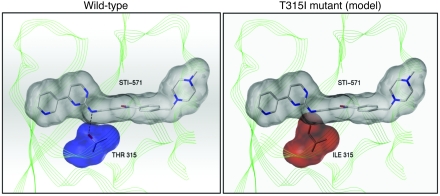
But the story gets better because John Kuriyan, a structural biologist interested in very basic questions about kinase regulation, had just published the cocrystal structure of ABL bound to imatinib (13). The threonine residue that we had identified in patients as the site of the point mutation formed a hydrogen bond with imatinib at the base of the ATP-binding pocket. John and I did not know each other, and likely would never have crossed paths had it not been for this remarkable convergence. Within 20 minutes of an introductory phone call during which I outlined our findings, John sent me a PowerPoint slide by e-mail — which clearly delineates a model of steric hindrance caused by the T315I substitution (Figure 3). This threonine residue, which is conserved across many kinases, is now called the gatekeeper residue and forms a hydrogen bond with many of the ATP-competitive kinase inhibitors now in clinical development. And, as one might predict, analogous gatekeeper residue mutations are associated with resistance to other kinase inhibitors, such as erlotinib, in EGFR-driven lung cancer.
Figure 3. Model of imatinib binding to ABL kinase domain.
The 2 panels depict wild-type Abl in complex with STI-571 (imatinib; left) and predicted structure of STI-571 binding pocket of T315I mutant Abl in complex with STI-571 (right). In the molecular structures representing STI-571 and Abl residue 315, nitrogen atoms are shown in blue, and oxygen atoms are shown in red. A hydrogen bond between Thr315 (left) and STI-571 is shown by the dashed line. Van der Waals interactions are depicted in gray for STI-571 (both panels), in blue for wild-type Abl residue Thr315 (left), and in red for mutant Abl residue Ile315 (right). The polypeptide backbone of the Abl kinase domain is represented in green. Reprinted with permission from Science.
We initially wondered if drug discovery efforts could be refocused on compounds that might retain activity against this imatinib-resistant mutant. But this approach quickly became untenable as we and other groups began to enumerate over 50 different amino acid substitutions associated with imatinib resistance. Fortunately, I had maintained contact with John Kuriyan. As we mapped the growing list of mutations to the structures solved in John’s lab, a theme emerged. Most of these mutations share the common property of destabilizing the inactive, closed conformation of ABL that is optimal for binding to imatinib (14). And it raised the enchanting notion that one might overcome these resistance mutations with an inhibitor that binds the active, open conformation. That inhibitor, dasatinib, surfaced serendipitously from a SRC kinase inhibitor project at Bristol Myers Squibb. Dasatinib is now approved by the FDA for treatment of imatinib-resistant CML. As predicted from the structural modeling studies, dasatinib was active against all but one of the imatinib-resistant mutants in preclinical models and in patients (15, 16).
Today CML patients are treated sequentially, first with imatinib, and, if they relapse, with dasatinib or a third ABL inhibitor, nilotinib, that has also recently been approved. Some of you may be wondering whether sequential treatment might select different types of mutants, as was seen with antivirals in HIV, where combination therapy is required to overcome resistance. Indeed, last year we reported in the Journal of Clinical Investigation that an increasingly complex spectrum of mutations arise in CML patients who relapse after sequential therapy, including compound mutations that would have never emerged had we used the drugs in combination (17).
I realize this vignette may seem self serving, but I hope this example, as well as many of those we heard at the meeting today, makes the point that basic science, when properly blended with clinical observation and molecularly based human investigation, is what I call translational research. The impact can be enormous, and fast.
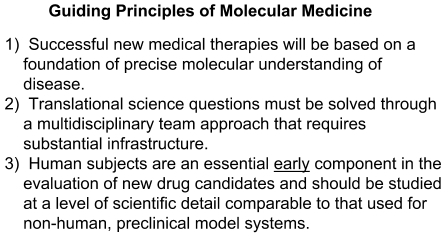
The focus on translational research today, whether we agree or not, is a reality. I believe this focus is justified, but only if done right, and I believe that we, as physician-scientists, are uniquely qualified to address it. I do not have all the answers but will share some thoughts based on these 3 principles (Figure 4). One: Successful new medical therapies will be based on a foundation of precise molecular understanding of disease. Two: Translational science questions can only be solved through a multidisciplinary team approach that requires substantial infrastructure. Three: Human subjects are an essential early component in the evaluation of new drug candidates and should be studied at a level of scientific detail comparable to that used for nonhuman preclinical model systems.
Figure 4. Guiding principles of translational medicine.
We are all aware of efforts at institutions across the country to build up translational research — often without precisely specifying what translational research is. In my opinion, all of these programs are controversial, and none are assured of success. Most of you are familiar with federal initiatives such as the institutional Clinical and Translational Science Awards (CTSA grants), which have been awarded to 24 institutions over the past 2 years, and the Specialized Program of Research Excellence (SPORE grants), awarded for organ-specific translational cancer research through the NCI. We are also witnessing growing efforts by disease-focused philanthropic foundations to steer their funding toward translational research projects and even play an active role in directing the research.
I am very concerned about how these programs are unfolding. The successes that I have been a part of largely bubbled up from traditional investigator-initiated research programs directed by well-trained physician-scientists, and enabled by truly collaborative partnerships with the pharmaceutical sector. These projects evolved naturally, and quickly, when the goals of the various partners in the collaboration were aligned and, most importantly, when the scientific rationale was compelling. I worry that successful translational research cannot be prescribed through overly detailed grant mechanisms with long checklists of required components. I suspect many of us have seen examples of poorly conceived science forced into clinical experiments solely to qualify for translational funding. As Rick Lifton showed us today in his lecture, the reviewers of these large program grants are often not qualified to judge the science.
I am also concerned about the impact of the unavoidable shift of funds away from basic science research. Imagine if John Kuriyan had not been funded to study basic structural biology of kinases. I want more translational research just as much as every disease advocate and Congressional leader, but it must be guided by and conducted with the same scientific rigor that we require of basic science. Rather than prescribe specific translational projects, I favor spending resources on the infrastructure needed to conduct the kind of molecularly focused human studies that I described. In the context of cancer, I am talking about the ability to annotate the molecular status of the tumor by cataloguing relevant somatic alterations in tumor DNA. We need better tools to quantify the engagement of a drug with its target to measure dose response using biochemical parameters rather than toxicity parameters. Finally, we need to serially track the impact of a treatment intervention on distinct molecular subsets in the tumor.
The goals are within our grasp with the technologies available to us today, but we often compromise our clinical trial design due to lack of infrastructure. This infrastructure includes molecular pathology, molecular imaging, and bioinformatic integration of molecular and clinical datasets. It may be possible to solve some of these problems with core facilities — but these core facilities, focused on human research, need to be different from traditional models. These cores need to be directed by highly trained scientists — perhaps physician-scientists — who can work in a team science model and conduct original, innovative work in addition to service work. I have no illusions; this is a very difficult challenge at multiple levels — requiring resources, space, creating new academic promotion tracks, incentives, and so on. But we are the most qualified individuals in the biomedical research enterprise to address them.
At Memorial Sloan-Kettering Cancer Center, Harold Varmus, Bob Wittes, and Tom Kelly decided to address some of these issues by creating a new program with laboratory-based translational oncology as its mission. Our goals are to assemble an outstanding group of laboratory-trained physician-scientists across clinical disciplines — each of whom has a passionate commitment to human disease. These investigators become integral members of the clinical disease-management teams responsible for delivery of clinical care and work together with their full-time clinical colleagues on the development of protocols. We are also establishing infrastructure to make precise, innovative molecular measurements on tissues obtained from the patients treated on these trials — at the level we would expect in our own laboratories — but centralized to enable larger throughput and quality control. This is but one pilot experiment currently underway at one institution. I hope we have a continuing dialogue about others.
Acknowledgments
A number of people have made my 3 years on the ASCI Council truly special. First, I thank my immediate predecessors, Eric Fearon and Barb Weber, for their leadership, and my successors, Nancy Andrews and Jon Epstein, for their wisdom and advice. Second, I thank my ASCI Council colleagues — David Altshuler, Lynda Chin, Hal Deitz, Theresa Guise, Jake Liang, Jon Licht, Beth McNally, and Larry Turka — for their camaraderie and support. Third, I thank Judy Swain, Elliott Kieff, and Denny Ausiello for their hard work on the planning committee for this 100th anniversary meeting. Fourth, I thank my wife, Susan, and my 2 children, Sophie and Sam, for supporting my passion for this exciting career. Fifth, I give special thanks to John Hawley and to Karen Guth, who have worked tirelessly behind the scenes to administrate our ASCI programs. Lastly, I congratulate you all on a remarkable first 100 years of success and urge us to embrace our new challenges.
Footnotes
This article is adapted from a presentation at the ASCI/AAP Joint Meeting, April 26, 2008, in Chicago, Illinois, USA.
Address correspondence to: Charles L. Sawyers, Howard Hughes Medical Institute, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, New York 10065, USA. Phone: (646) 888-2138; Fax: (646) 888-2595; E-mail: sawyersc@mskcc.org.
Citation for this article: J. Clin. Invest. 118:3798–3801 (2008). doi:10.1172/JCI37557.
References
- 1.Brainard E.R. History of the American Society for Clinical Investigation, 1909–1959. J. Clin. Invest. 1959;38:1784–1864. doi: 10.1172/JCI103958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber B.L. In the palace of the sultan: 2007 American Society for Clinical Investigation Presidential Address. J. Clin. Invest. 2007;117:1727–1731. doi: 10.1172/JCI32575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginsburg D. The history and evolution of the ASCI: deja vu all over again. American Society for Clinical Investigation. J. Clin. Invest. 2002;110:S1–S4. [PubMed] [Google Scholar]
- 4.Goldstein J.L. On the origin and prevention of PAIDS (Paralyzed Academic Investigator’s Disease Syndrome). J. Clin. Invest. 1986;78:848–854. doi: 10.1172/JCI112652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowell P.C., Hungerford D.A. Chromosome studies in human leukemia. II. Chronic granulocytic leukemia. J. Natl. Cancer Inst. 1961;27:1013–1035. [PubMed] [Google Scholar]
- 6.Rowley J.D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 7.Konopka J.B., Watanabe S.M., Singer J.W., Collins S.J., Witte O.N. Cell lines and clinical isolates derived from Ph1-positive chronic myelogenous leukemia patients express c-abl proteins with a common structural alteration. Proc. Natl. Acad. Sci. U. S. A. 1985;82:1810–1814. doi: 10.1073/pnas.82.6.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heisterkamp N., et al. Localization of the c-ab1 oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature. 1983;306:239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 9.Shtivelman E., Lifshitz B., Gale R.P., Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315:550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 10.Daley G.Q., Van Etten R.A., Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 11.Druker B.J., et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 12.Gorre M.E., et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 13.Schindler T., et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. . Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 14.Shah N.P., et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 15.Shah N.P., et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 16.Talpaz M., et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 17.Shah N.P., et al. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J. Clin. Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]