Abstract
Studies over the past 50 years revealing the molecular events that promote normal T lymphocyte cycle competence and progression led to a detailed understanding of how cytokines function to regulate normal hematopoietic cell proliferation. During that same period, the molecular and genetic changes introduced by the Philadelphia chromosome in chronic myelogenous leukemia were unraveled, and these have led to an understanding of how mutations that constitutively activate normal cytokine signaling pathways can cause unregulated cell proliferation and malignant transformation. Based on the paradigm established by these data, it is inescapable that going forward, investigators will operate under the hypothesis that transformation of additional cells and tissues will have a similar pathogenesis.
Introduction
Current understanding of leukemogenesis evolved over almost 50 years, involving research contributions from many different scientific disciplines. Indeed, 50 years ago, a complete lack of knowledge of the molecular mechanisms involved in the regulation of the maturation, growth, and differentiation of normal hematopoietic cells meant that an understanding of exactly what might be responsible for leukemia was simply unapproachable. At that time, as leukemic cells had the microscopic morphology of immature progenitors, most hematologists subscribed to the notion that the underlying fundamental problem in leukemia was one of arrested maturation rather than loss of normal control of cell growth. However, we have now come to realize that cells constituting metazoan organisms are regulated from without, via cytokine molecules that direct their proliferative behavior. Cytokines, and their receptors, signaling pathways, and transcriptional activators, were first shown to function as the mediators of cell-cycle expression in T lymphocytes, which became a model system for the study of normal hematopoietic cell proliferation.
Independent studies that were performed in parallel over the past 50 years have made it apparent that leukemias result from mutations in genes that encode key molecules that usurp the normal strict cytokine/receptor-dependent digital control of the decision of hematopoietic cells to undergo proliferative expansion. Of the various kinds of leukemia that are recognized by their clinical course (i.e., acute or chronic) and cellular morphology (i.e., myeloid or lymphoid), our understanding of the pathogenesis of chronic myelogenous leukemia (CML) is now the most complete and is thus the focus of this Review.
At this juncture, it is germane to chronicle the crucial discoveries that have led to our present understanding of the signals controlling the growth of both normal hematopoietic cells and CML cells (see A chronology of leukemogenesis), to reveal not only what we now know, but how and why we came to our present knowledge, as well as who was responsible. This is a story of how science progresses, via contributions from fields with no apparent initial connections. Despite the long time interval involved, the data that accumulated establish the paradigm that the mutational usurpation of normal cell growth regulation underlies malignant transformation of many, if not all, other cells and tissues. It therefore follows that revealing the cytokine molecules and their receptors that deliver key signals to promote cell-cycle progression in each type of cell and tissue will make possible the development of effective therapies for other cancers.
The 1960s: proliferating lymphocytes, abnormal chromosomes, 3T3 cell cycles, and viral and cellular proto-oncogenes
The early years: proliferating lymphocytes and mitogenic factors.
Prior to 1960, lymphocytes were described in textbooks as uninteresting, terminally differentiated cells that were thought to be incapable of proliferating. No one really understood their function, let alone that antigens and cytokines are involved in their exquisite growth control and that they are the primary cells responsible for immunity. The beginnings of the molecular dissection of normal lymphocyte growth regulation are attributable to the work of a single young scientist, Peter Nowell of the University of Pennsylvania. In 1960, Nowell discovered that lymphocytes are actually able to proliferate (1). Using the kidney bean extract phytohemagglutinin (PHA) to agglutinate red blood cells, so as to separate them from the plasma (which contained white blood cells), Nowell inadvertently left the white blood cells in the incubator for several days and found that all the cells had become large and resembled lymphocyte leukemic blast cells, with many cells undergoing mitosis (1). Nowell’s serendipitous discovery that lymphocytes can proliferate in response to mitogenic lectins, and the subsequent observations that a similar phenomenon occurs in response to antigens (2–4), led to an explosion of enthusiasm for the study of lymphocyte cultures and created an entirely new field, that of cellular immunology, which dominated immunology for the next two decades.
Only a few years passed after Nowell’s publication before two independent groups simultaneously reported that the medium from cultures of alloantigen-stimulated lymphocytes contained a mitogenic activity (5, 6). This mitogenic activity was christened blastogenic factor and was subsequently found in medium conditioned by PHA-stimulated lymphocytes, as well as medium conditioned by peripheral blood leukocytes activated by soluble protein antigen (medium conditioned by any form of lymphocyte activation will be referred to hereafter as lymphocyte-conditioned medium). Over the next ten years, many mitogenic activities were reported in medium conditioned by stimulated leukocyte cultures. However, the molecular nature of these mitogenic activities remained obscure.
The Philadelphia chromosome and CML.
Also in 1960, together with David Hungerford, Nowell made the novel observation that cells from patients with CML contained a small abnormal chromosome that was absent in the chromosomes found in PHA-stimulated normal lymphocytes (7). This abnormal chromosome was named the Philadelphia (Ph) chromosome, after the city in which it was first observed. In this regard, it is noteworthy that chromosome abnormalities pathognomonic for other recognizable types of leukemia, such as acute myeloid leukemia (AML), other chronic myeloproliferative disorders, and most acute and chronic lymphoid leukemias, were not readily demonstrable at the time. However, the Ph chromosome was later to provide the genetic key to begin to unravel what went wrong with myeloid cells to cause CML.
It is noteworthy that more than a decade elapsed before Janet Rowley, using techniques that were new at the time to stain chromosomes, revealed that the Ph chromosome abnormality is generated by reciprocal translocation, whereby the tip of the long arm of chromosome 22 is replaced by the tip of the long arm of chromosome 9 (8). This translocation phenomenon proved ultimately to be very important in the evolving understanding of CML leukemogenesis.
3T3 cells and cell cycles.
Also in the 1960s, investigators interested in the control of cell growth established cultures of murine embryonic fibroblasts (9, 10). When these cells were passed in serum-containing medium at 3,000 cells every 3 days (a protocol that gave rise to their name, 3T3 cells), they assumed a phenotype of “normal” adult fibroblasts, in that when grown to confluence, they would become contact inhibited and their growth would cease. It was realized subsequently that the cells ceased proliferating because they had consumed the cytokines and/or growth factors in the serum. However, as the growth stimulus was provided by serum, the dissection of the critical molecules responsible for stimulating cell-cycle progression was unapproachable. Even so, 3T3 cells became established as the cell culture system of choice for investigators interested in understanding normal cell growth as well as malignant transformation.
Cancer viruses and cellular proto-oncogenes.
It has now been 100 years since cell-free filtrates were first demonstrated to cause leukemia (11) and a rapidly fatal sarcoma in chickens (12). But it was in the 1950s and 1960s that leukemogenesis C-type RNA tumor viruses were described in mice (13–16) and cats (17). Then, in 1970, another new murine retrovirus was reported to cause lymphosarcomas together with a marked increase in the number of polymorphonuclear leukocytes in the blood (18). This virus, termed Abelson lymphosarcoma virus, was destined to play a major role in the understanding of CML leukemogenesis.
To explain how RNA viruses could cause cancer in a cell that was then passed on to all of the cellular progeny, Howard Temin proposed the “DNA provirus hypothesis” in 1964 (19). This hypothesis went against the “central dogma” that the flow of information goes only from DNA to RNA to protein. This led him to devise experiments that revealed RNA virus–directed DNA synthesis and eventually to the discovery of the RNA-dependent DNA polymerase that came to be known as reverse transcriptase (20). Similar experiments were reported simultaneously by David Baltimore (21). Soon thereafter, Hildesaburo Hanafusa and coworkers (22) provided convincing evidence that uninfected normal avian cells contain DNA complimentary to the 70S RNA of avian tumor viruses.
All of these findings led Temin to refine his hypothesis to the “protovirus hypothesis” (23). To explain how RNA viruses could cause tumors, he speculated that the capacity of the retroviruses to redirect the flow of genetic information from RNA to DNA and then back to RNA allowed the virus to “pick up” bits of normal cellular DNA and incorporate them into the viral genome. Thus, he proposed that normal cellular genes involved in fundamental cellular processes, such as proliferation and differentiation, were the cause of cancer. These genes are now called cellular proto-oncogenes.
The 1970s: the G1 phase of the cell cycle, long-term T lymphocyte growth, and viral and cellular proto-oncogenes
Competence and progression in the G1 phase of the cell cycle in 3T3 cells.
In 1974, Arthur Pardee published a paper that was destined to become a classic in the cell-cycle field (24). Through a series of experimental approaches, he showed that 3T3 cells induced by serum to proliferate in vitro underwent a serum-dependent interval early in the G1 phase of the cell cycle, but subsequently the cells became independent of serum at a point late in G1, just before DNA replication. The point between the serum-dependent and serum-independent transition he termed the restriction point, which soon became abbreviated to the R-point.
A few years later, Jack Pledger, Charles Stiles, and coworkers separated the G1 phase of the cell cycle into two distinct subphases: a serum-dependent phase responsible for initiating the cell cycle that is now known as the G0-G1 transition but that they termed competence; and cell-cycle progression to DNA synthesis and replication, a phase now known as the G1-S transition (25). They showed that competence was dependent upon the addition of an extract of platelets, now known to be PDGF, whereas G1 progression to S phase was dependent upon a factor(s) that was contained in platelet-poor plasma, now known to be EGF. At the time, the molecular nature of these cytokines and/or growth factors remained a mystery because it was difficult to separate the molecules in serum. However, the distinction of competence and progression as separate functional subphases of G1 became important once it was possible to define the various molecules involved in these cellular processes.
Long-term T cell growth and monoclonal functional T lymphocytes.
Just as blastogenic factor was found to promote the proliferation of lymphocytes, in 1974, lymphocytes stimulated with PHA were found to release hematopoietic colony stimulating activity (CSA) into lymphocyte-conditioned medium (26). This finding prompted attempts to use lymphocyte-conditioned medium to grow leukemia cells from bone marrow samples of patients with AML. This effort failed, but the lymphocyte-conditioned medium supported the long-term culture of T lymphocytes (27). However, because bone marrow had been used, it was not entirely clear whether the cultured cells were immature T lymphocyte precursors or mature T lymphocytes.
These results prompted attempts to establish antigen-specific T lymphocyte lines in long-term culture. However, according to immunologic dogma, the growth stimulus for T lymphocytes resided entirely with antigen, not a soluble mitogenic factor. Despite the dogma, the first antigen-specific cytolytic T lymphocyte lines (CTLLs) were created in the very first experiments testing the use of lymphocyte-conditioned medium to support the growth of antigen-selected cytolytic T lymphocytes (28). From these CTLLs, the first monoclonal cytolytic T lymphocytes were generated (29), something that revolutionized the study of T lymphocytes and subsequently proved as important for immunology as mAbs.
Mutations, viral oncogenes, cellular proto-oncogenes, and protein tyrosine kinases.
In 1976, Nowell published a prescient hypothesis for the clonal evolution of tumor cell populations (30). Using CML as an example, he postulated that the initiating event of the transition of a normal cell to a neoplastic cell had to involve escape from normal control of cell growth, thereby providing the cell with a selective growth advantage over the normal cell from which it was derived. Today, this would be termed the founding mutation. He further postulated that evolution to the accelerated phase of CML, also called blast crisis, would depend upon additional acquired mutations followed by selection of the most malignant sublines (30).
However, a mutational etiology of cancer was superseded by a group led by Michael Bishop and Harold Varmus, who presented the first evidence that the transforming gene of Rous sarcoma virus (v-src) had a normal cellular counterpart (c-Src), thereby providing support for Temin’s proto-oncogene hypothesis (31). Subsequently, c-Abl, homologous to the transforming gene, v-abl, of Abelson lymphosarcoma virus, was defined (32).
Also, soon thereafter, both the c-SRC and c-ABL proteins were found to have tyrosine-specific kinase activity (33, 34). However, many proteins in the cell were phosphorylated by these protein tyrosine kinases (PTKs), and it was not clear which of the substrates, if any, were associated with the malignant transforming properties of these viral oncogenes and cellular proto-oncogenes.
The 1980s: the first interleukin molecules and their receptors, T lymphocyte cycle competence and progression, mutations in cellular proto-oncogenes, and cyclins
The first interleukins.
The ability to grow normal T lymphocyte clones indefinitely in lymphocyte-conditioned medium allowed the creation of a rapid (24 hours) quantitative bioassay for T cell growth factor (TCGF) activity in the medium (35). This assay proved instrumental in a series of biochemical experiments that defined the characteristics of the TCGF molecule for the first time (36). Then, the generation of an mAb reactive with TCGF allowed the first purification of milligram quantities of a homogeneous single protein molecule with TCGF activity (37). Preparation of purified radiolabeled TCGF led to the identification of the cell-surface receptor for TCGF (38), which was the first cytokine receptor to be discovered, and the identification of the first mAb specific for a cytokine receptor (39).
All of these advances ushered in a new era, that of molecular immunology, which began in the 1980s and is still ongoing. For the first time it was possible to distinguish between lymphocyte-activating factor (LAF), a macrophage product mitogenic for lymphocytes (40), and TCGF, a T lymphocyte mitogenic product (41). Experiments revealed that LAF promoted the proliferation of lymphocytes by augmenting T lymphocyte production of TCGF and that TCGF was the true T lymphocyte mitogen in lymphocyte-conditioned medium (42, 43). Thus, because LAF was found to be functioning before TCGF, it was renamed IL-1, and TCGF was renamed IL-2, in anticipation that there might be more cytokine molecules yet to be discovered (43). The term interleukin was coined to indicate that the molecules were functioning to carry messages between leukocytes.
In this regard, it is important to recognize that the lymphocyte-conditioned medium described originally as containing blastogenic factor must have contained IL-1, IL-2, and CSA, as well as many other interleukins that still remained undiscovered. Thus, it was necessary to purify each individual component and to test each one for the various activities described. It is noteworthy that the molecular characterization of IL-1 lagged behind that of IL-2, primarily because the bioassay for IL-1, which used thymocytes, was more cumbersome and time consuming (3 days) than the 24-hour IL-2 bioassay, which used cloned T lymphocytes. It is probable that blastogenic factor and IL-2 are synonymous, but as the blastogenic factor molecule(s) were never characterized, it is impossible to know now.
T lymphocyte cycle competence and progression, a new model to study cell proliferation.
Armed with these new molecular reagents and the understanding that IL-2, rather than antigen, functions to promote T lymphocyte cycle progression, for the first time, the critical molecular determinants responsible for cell-cycle progression could be determined at the single-cell level (44). With the newly available flow cytometer, Doreen Cantrell demonstrated convincingly that each T lymphocyte proliferates as a result of a critical number of intermolecular interactions between IL-2 and its receptor (IL-2R) at the cell surface and that the cell seems capable of counting these molecular interactions, so that it only commits to the irrevocable decision to replicate its DNA when it has received the requisite number of receptor “hits” (45, 46). This provided a new model system for cell growth and, for the first time, a molecular explanation of the R-point. This understanding led to the speculation that loss of this exquisite cytokine/receptor control of the decision to divide could lead to autonomous growth, i.e., cancer (47, 48).
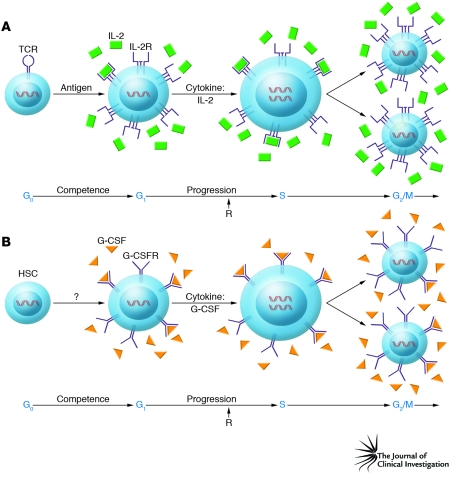
In another important advance, human antigen-specific cytolytic T lymphocyte clones were created by Ellis Reinherz and colleagues and used to generate murine mAbs that led to the first description of the TCR, its signaling component (CD3), and the MHC restriction molecules CD4 and CD8 (49). These unique cellular and molecular reagents then allowed experiments in which it was established that the TCR stimulates mature resting T lymphocytes to pass through the G0-G1 transition (i.e., competence), which at the molecular level was manifest by the production of IL-2 and the expression of IL-2R. By comparison, IL-2R triggering induced progression through the G1 phase of the cell cycle and initiated the G1-S transition (50). Thus, the TCR signal was found to have made the cells competent to receive the IL-2 signals driving cell-cycle progression (Figure 1).
Figure 1. Cell-cycle competence and progression.
(A) T lymphocytes are normally in the resting, or G0, phase of the cell cycle. Signals from the TCR promote the transition from G0 to G1, which renders the cells competent to progress further through the cell cycle by stimulating the expression of the genes encoding IL-2 and the subunits of IL-2R. Subsequently, the IL-2–IL-2R interaction is responsible for blastic transformation and G1 progression, which moves the cell through the G1 phase of the cell cycle to the point that the cells no longer require signals from the IL-2–IL-2R interaction (i.e., past the R-point [R]) to enter S phase and subsequently undergo mitosis. (B) HSCs, like resting mature T lymphocytes, are also normally in G0. Unlike the T lymphocyte lineage, little is known regarding the cytokines and receptors responsible for stimulating the HSCs to leave G0 and enter the cell cycle. Even so, once this G0-G1 transition occurs and an HSC has given rise to a committed progenitor cell, such as a committed granulocyte precursor, the cytokines responsible for G1 progression are known, and this is exemplified in the figure by G-CSF acting on a committed granulocyte precursor to promote the progression from the G1 to the S, G2, and M phases.
Viral and cellular proto-oncogenes, the Ph chromosome, and BCR-ABL.
As more and more viral oncogenes and cellular proto-oncogenes were reported in the early 1980s, Robert Weinberg and Geoffrey Cooper independently reported that cellular DNA from both animal and human tumor cells could induce the formation of transformed foci in normal 3T3 cells (51–54). Thus, it was speculated that oncogenesis could involve dominant genetic mechanisms resulting in the activation of transforming cellular proto-oncogenes (55, 56).
A significant advance in CML research occurred in the early 1980s, when it was first reported that the 3′ end of the human c-ABL proto-oncogene mapped within the region of chromosome 9 that is found on the Ph chromosome (57, 58). Subsequently, the region of chromosome 22 that is disrupted by the translocation that generates the Ph chromosome was found to contain a novel gene, which was termed the breakpoint cluster region (BCR) gene. Further, the 3′ end of the BCR gene was lost during the translocation (58). The fused RNA BCR-ABL transcript found in CML cells encodes a 210-kDa BCR-ABL protein that has constitutive PTK activity, unlike the normal c-ABL protein (145 kDa), which is usually inactive. Thus, for the first time, an oncogene discovered in a retrovirus and capable of inducing cancer in animals was found to play a role in genetically altered human cancer. However, there still remained a large knowledge gap between these findings and the mechanism whereby the expression of these genes caused malignant transformation. To fill in this gap, developments in the cell-cycle and cytokine fields were necessary.
The cyclins, intracellular recipients of cytokine signals.
During the 1980s, the cyclin proteins were discovered, first in dividing fertilized sea urchin (59) and clam (60) oocytes and then in yeast (61). Of particular importance for our discussion are the discoveries of the mammalian G1 cyclin D proteins by several laboratories in 1991. It is noteworthy that cyclin D1 was uncovered by cloning genes induced by murine M-CSF, a cytokine mitogenic for macrophages (62). Of additional significance, cyclin D1 was also independently identified at the breakpoint of a human chromosome 11 inversion in parathyroid adenomas, where its coding sequences were juxtaposed to the parathyroid hormone promoter (63). The cyclin D1 locus was also found to be deregulated in cancer by translocation (64), retroviral insertion (65), and gene amplification (66), implying that cyclin D1 could act as a proto-oncogene.
Subsequently, using synchronized normal human T lymphocytes, the sequential IL-2 induction of cyclin D2 and then cyclin D3 mRNA and protein during mid- to late G1 was demonstrated by Julia Turner in 1993 (67). Simultaneously, work by many others uncovered how the cyclin D proteins and the cyclin-dependent kinases move the cell through G1 beyond the R-point into S phase and then cytokinesis (reviewed in ref. 68). Despite this insight into the transcriptional changes underlying the cytokine/receptor dependency of a T lymphocyte for its progression through the R-point from G1 to S phase, the molecular events actually triggered by the cytokine receptors that induce expression of the cyclin D proteins remained obscure.
Hematopoietic cytokine–dependent cell lines and v-abl transformation.
The new model system for assessing cell growth at the molecular and single-cell level provided by the ability to grow cloned T lymphocytes in suspension in the presence of IL-2 (44) led to similar experiments in all other cells of the hematopoietic system during the 1980s. In this regard, the work of Donald Metcalf and Nicholas Nicola is noteworthy (69, 70). However, the hematopoietic cytokine field lagged behind the interleukin field because although the colony assay, the traditional assay for identification of the hematopoietic cytokines, was quantitative, it was cumbersome. A week was required for macroscopic colonies to become visible, compared with 24 hours for the IL-2 bioassay. It was not possible to replicate the IL-2 work until cell lines responsive to individual hematopoietic cytokines were generated.
Thus, after hematopoietic cytokine–dependent cell lines had been created, experiments were performed with Abelson lymphosarcoma virus using cell lines that were dependent on GM-CSF for growth. It was readily shown that infection by the virus conferred cytokine independence (71). Moreover, similar experiments showed that IL-3–dependent mast cell lines could be made to grow independently upon infection with Abelson lymphosarcoma virus (72), and IL-2–dependent T lymphocyte lines behaved similarly after infection with the same virus (73). In each of these cases, the mechanism(s) responsible for cytokine-independent proliferation in vitro and tumorigenicity in vivo could not be ascribed to an autocrine mechanism due to virus-promoted persistent cytokine production and action. Accordingly, attention turned to the intracellular signaling pathways normally activated by these cytokines.
The 1990s: signaling molecules, their inhibitors, and persistent signaling of cell-cycle progression
The JAKs and STATs.
After several interleukins and hematopoietic cytokines and their receptors had been identified and sequenced, it was realized that they constituted a whole new superfamily of ligands and receptor molecules (74). However, based upon their sequences, none of the receptors had PTK domains, or any other enzymatic domain, that could be construed to initiate signaling to the cell interior. Thus, it was fortunate that a new family of PTKs was discovered by investigators searching for genes containing DNA sequences homologous to those encoded by known PTKs (75–77). The Janus kinases (JAKs) were named after the Roman god Janus, because analysis of their DNA sequence predicted that they contained 2 PTK domains arranged in opposite orientation. By 1994, the four members of the JAK family of cytoplasmic and/or membrane-associated kinases, JAK1, JAK2, JAK3, and Tyk2, had been discovered (75–79). Of interest, JAK1 and JAK3 were found to be phosphorylated upon IL-2 signaling (Figure 2), whereas Tyk2 and JAK2 were not (78, 80).
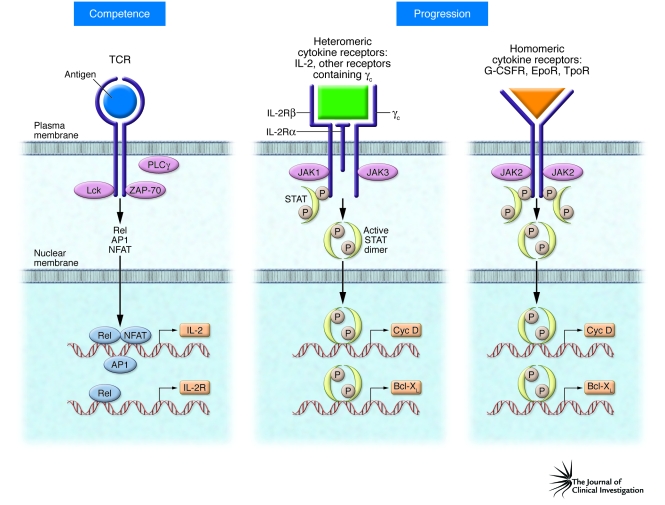
Figure 2. Structure-function relationships of the receptors controlling T lymphocyte and myeloid cell cycle competence and progression.
The TCR activates cytoplasmic kinases (including Lck, ZAP-70, and PLCγ), which signal via intermediates to induce the activation of members of three distinct families of transcription factors, Rel, activator protein–1 (AP1), and nuclear factor of activated T cells (NFAT). These transcription factors then coordinate the expression of the genes encoding IL-2 and the subunits of IL-2R, thereby rendering the cell competent to progress through the cell cycle. Heteromeric cytokine receptors, here represented by IL-2R, are composed of two or three distinct transmembrane chains. Binding of the cytokine to the external receptor chain domains brings the cytoplasmic domains into close enough proximity for their respective receptor-associated JAK molecules to initiate signaling. Subsequently, the activated STAT dimers translocate to the nucleus and initiate transcription of genes encoding proteins such as cyclin D2 (Cyc D) to promote progression beyond the R-point and also genes encoding cell-survival proteins, such as Bcl-XL. By comparison, homomeric receptors already have the same bound JAK molecules in close proximity, and ligand binding readily initiates signaling by promoting transphosphorylation of JAK2, the receptor chains, and eventually STAT5. G-CSFR, G-CSF receptor; EpoR, erythropoietin receptor; TpoR, thrombopoietin receptor.
Even before these developments, work on the mechanisms by which IFNs signal to modulate gene transcription had begun (81, 82). Initially, James Darnell’s group isolated two new proteins associated with IFN activity, and because they were shown to act as both signal transducers and activators of transcription, they were given the names signal transducer and activator of transcription 1 (STAT1) and STAT2 (83–86). Soon thereafter, Darnell’s group isolated STAT3 and STAT4 (87, 88). STAT5 was discovered to be activated by prolactin in mammary tissue of lactating sheep (89) and subsequently found to exist as two almost identical (95%) isomers, which were designated as STAT5a and STAT5b (90, 91). STAT6 was identified as being induced by IL-4 (92).
As the mechanism by which STATs mediate transcriptional activation was being resolved, work in the laboratories of George Stark and Ian Kerr on cell mutants incapable of an IFN response showed that the JAKs activated the STATs (refs. 93, 94; reviewed in ref. 95). James Ihle’s group then quickly demonstrated that JAK2 is phosphorylated following the binding of erythropoietin (Epo) (96) and IL-3 to their respective receptors (97), while Yoshiyuki Niho’s group reported that JAK2 is phosphorylated by G-CSF binding to its receptor (98).
These experiments provided the impetus to investigate whether JAK/STAT pathways are involved in transferring the IL-2/IL-2R signal to the nucleus. Carol Beadling, working in Cantrell’s group, found that TCR triggering did not lead to JAK/STAT activation, but IL-2 was found to activate JAK1 and JAK3, which then promoted the tyrosine phosphorylation of two STAT proteins (80), subsequently identified as STAT5a and STAT5b (Figure 2) (99, 100).
PTK inhibitors.
Unlike the interleukin and hematopoietic cytokine receptors, the intracellular domain of EGFR functions as a tyrosine kinase and mediates autophosphorylation upon EGF binding. As early as 1986, a natural product (erbstatin) that inhibited the autophosphorylation of EGFR at concentrations in the low micromolar range was isolated from an actinobacterium (101). Subsequently, using the structure of erbstatin as a base, Alexander Levitzki and coworkers synthesized a series of compounds that inhibited both EGFR autophosphorylation and EGF-dependent cell proliferation at low micromolar concentrations (100). In addition, this group reported several inhibitors of the BCR-ABL kinase, all with IC50 in the low micromolar range (102). They speculated that the mechanism of inhibition involved interference with the interaction between the kinase and its substrate, rather than interference with ATP binding to the kinase, which is required for phosphorylation of the substrate (103).
With this as a background, the report of small molecular inhibitors of PTKs by Brian Druker and colleagues in 1996 was incendiary for the whole cancer field (104). Working at about the same time as Levitzki’s group, investigators at Ciba-Geigy (now Novartis) searching for molecules that could inhibit the function of PTKs by blocking the ATP-binding site, identified a lead compound by screening a large chemical library. Subsequently, a series of compounds were synthesized, and one compound in particular blocked in vitro substrate phosphorylation by BCR-ABL with an IC50 of 25 nM and inhibited cellular BCR-ABL PTK activity with an IC50 of 250 nM. This compound (STI571) is now called imatinib (Gleevec). Druker’s group showed that this compound inhibits BCR-ABL–expressing cells from proliferating in vitro and forming tumors in vivo without inhibiting normal granulocyte colony formation. In the phase I clinical trials of this compound, when doses of 300 mg were reached, 53 of 54 patients with CML achieved a complete hematological response (105).
Persistent STAT activation and malignant transformation.
Accordingly, it was established both in vitro and in vivo that inhibition of BCR-ABL PTK activity via Gleevec effectively lysed BCR-ABL–transformed CML cells. However, it still remained to be discerned precisely how BCR-ABL transforms cells so that it could be understood how Gleevec works.
Soon after the discovery of the STATs, Richard Jove showed that persistently phosphorylated/activated STAT3 was required for SRC-mediated transformation of 3T3 cells, and this was subsequently confirmed by Jacqueline Bromberg and Darnell (106–110). Simultaneously, transformation of pre-B cells by v-Abl was found to result in persistent phosphorylation and activation of STAT5 by Paul Rothman’s group (111). Then, in 1997, Oliver Bernard and coworkers (112) and Peter Marynen’s group (113) independently reported that a second chromosomal translocation involving JAK2 and TEL, a gene encoding a member of the ETS transcription factor family, was associated with several types of leukemia. The TEL-JAK2 fusion protein contains the catalytic domain of JAK2 and the TEL-specific helix-loop-helix dimerization domain. Accordingly, this dimeric fusion protein has constitutive PTK activity and does not require a cytokine receptor to bring two JAK2 PTK domains together to serve as docking sites to activate STAT5 (114).
A new millennium and the first molecular understanding of leukemogenesis
Over the course of several years, many reports accumulated supporting the notion that the BCR-ABL gene product mediates malignant transformation of HSCs by usurping the normal regulatory control of cytokine/receptor/JAK signals to persistently activate STAT5 directly, thereby leading to autonomous cell-cycle progression and cell survival via cyclin D2 and cyclin D3, and cell survival via Bcl-X expression (115–117). Moreover, convincing evidence from Tomasz Skorski’s group indicated that BCR-ABL activates the SRC family kinase hematopoietic cell kinase (Hck), which in turn activates STAT5 via phosphorylation of Tyr699 (118). Thus, the weight of the data indicated that transformation occurred via the persistent activation of STAT5.
However, experiments with mice in which the genes encoding both STAT5a and STAT5b were deleted (Stat5a–/–Stat5b–/– mice) failed to support this contention, in that these mice were still susceptible to v-abl transformation (119). Accordingly, the role of the STAT proteins in oncogenic transformation remained unclear, leading some to question whether persistent STAT activity itself is the cause or the result of the transforming process (120). Subsequently, it was realized that the original Stat5a–/–Stat5b–/– mice still express N-terminally truncated STAT5 protein, as only the most N-terminal exon was deleted, leaving the rest of the coding regions still in-frame. Therefore, Veronika Sexl, who had performed the original Abelson lymphosarcoma virus transformation experiments while in Ihle’s group, repeated the experiments with mice in which the entire Stat5a/b locus had been deleted. When inoculated with Abelson lymphosarcoma virus, these mice do not succumb to lymphosarcomas (121). Accordingly, this report closed the loop, and for the first time, the picture of the molecular pathways leading to malignant transformation of a human cancer (i.e., CML) became evident.
The importance of the competence/progression paradigm and cytokine receptor structure in leukemogenesis
A selective growth advantage must occur in a single mutated cell, thereby allowing its progeny to undergo proliferative expansion, which ultimately leads to accumulation and dissemination of leukemia cells throughout the body. By definition, a mutation that leads to a block in maturation without a growth advantage cannot lead to accumulation and dissemination. These distinct outcomes are axiomatic, given that the early stages of the cell cycle are separated into two molecularly distinct decision points necessary for proliferative clonal expansion, i.e., the transition from G0-G1, when the cell becomes competent to receive the progression signals that move the cell from G1 to the S phase of the cell cycle.
In this regard, T lymphocytes are normally in the G0 phase of the cell cycle and require activation via the TCR to enter the G1 phase of the cell cycle and become competent to progress further through the cell cycle by virtue of expression of IL-2 and IL-2R (Figure 1A). As shown in Figure 1B, HSCs are analogous to resting naive mature T lymphocytes, in that they also remain in G0 until called upon to undergo cell division and give rise to committed precursors. Moreover, committed hematopoietic precursors are analogous to antigen-activated competent T lymphocytes, with the lineage-specific cytokines such as G-CSF and their receptors serving as the progression factors, promoting the transition from G1 to S, then beyond to complete the G2 and M phases of the cell cycle. This separation between competence and progression accounts for the observations that T lymphocytic leukemias have not been found to result from mutations leading to constitutive activation of the TCR signaling complex or signaling pathways — this receptor system promotes only cell-cycle competence and not progression (44). This difference in the TCR versus the signaling pathways activated by T lymphocyte cytokine receptors was recently underscored by a report showing that the TCR is unnecessary for STAT5-mediated transformation of CD8+ T lymphocytes (122). Furthermore, most of the myeloid hematopoietic malignancies have now been traced to persistent activation of STAT5, the transcription factor responsible for signaling cell-cycle progression of the normal committed precursors.
The similarities between resting T lymphocytes and HSCs, both of which are in G0 and require signals to leave G0 and enter G1, have obvious implications for the cancer stem cell hypothesis, which holds that only some cancer cells have the capacity for self renewal, while most of the progeny cells cannot continue to proliferate indefinitely (123). It follows, by analogy to T lymphocytes, that efforts should now be made to grow HSCs in proliferative culture and to identify the cytokines responsible for activating cell-cycle competence versus cell-cycle progression.
The molecular differences between competence and progression are now known for T lymphocytes (Figure 2), where the antigen receptors activate kinases, transcription factors, and genes that are distinct from the kinases, transcription factors, and genes activated by the cytokine receptors. By comparison, little is known about the cytokines, receptors, and intracellular signaling molecules that regulate the self-renewal decisions of HSCs.
As also shown in Figure 2, the signaling components of the heteromeric T lymphocyte cytokine receptors, exemplified by IL-2R, are made up of at least two distinct signaling chains, and each receptor chain is bound by distinct JAKs, e.g., JAK1 is associated with the β-chain of IL-2R and JAK3 is associated with the common γ-chain (γc) component of IL-2R. A similar arrangement exists for all other receptors containing γc, including the receptors for several T lymphocyte cytokines (IL-4, IL-7, IL-9, IL-13, IL-15, and IL-21) as well the receptor for IL-3, which is expressed by mast cells, and the receptor for GM-CSF, which is expressed by the myeloid cell precursors of granulocytes and macrophages.
The differing dependence of heteromeric receptors on several distinct JAKs and homodimeric receptors on only one JAK has important implications when considering that activating mutations occur in genes that encode JAK proteins. For example, by comparison with the heteromeric T lymphocyte cytokine receptors, the receptors for Epo, G-CSF, and thrombopoietin (Tpo) are homodimers bound by two JAK2 molecules. Thus, if a random JAK2 mutation occurs in one of the myeloid committed precursor cells that expresses one of the cell-specific homodimeric cytokine receptors, it is readily understandable how leukemia can arise more easily in these cells than in the T lymphocyte compartment (Figure 2) (124). Thus, a random mutation causing a persistent activation of a single JAK protein in T cells, which express heteromeric receptors and at least two JAKs, is far less likely to cause leukemia. However, mutations involving other kinases, such as BCR-ABL (125) and TEL-JAK2 (114), can transform cells that do not express homodimeric cytokine receptors, as we have seen for IL-2–, IL-3–, and GM-CSF–dependent cells, thereby bypassing the cytokine receptor as a necessary docking site for STAT activation (126). It is noteworthy that BCR-ABL activates STAT5 indirectly via the SRC family kinase Hck, while the downstream STAT5-activated genes transform the cells. Accordingly, these oncogenic PTKs can transform hematopoietic precursor cells, either HSCs or committed progenitors, that do not express homodimeric cytokine receptors, as long as they express SRC family kinases and STAT molecules (124, 126).
Conclusions
Now that the pathogenesis of CML is understood at the molecular level, it is possible to identify the critical contributions that clearly stand out as furthering the progress to the present knowledge base. Thus, the discoveries of both normal T lymphocyte blastic transformation and the Ph chromosome associated with CML were seminal. It is now clear that most human leukemias are caused not by oncogenic retroviruses, but rather by random mutations exemplified by Ph, the first chromosomal abnormality identified. However, the study of oncogenic retroviruses that cause leukemia in animals clearly pointed the way to genes that ultimately proved to be some of the most commonly mutated cellular proto-oncogenes. Although JAK and STAT genes were not identified as cellular proto-oncogenes usurped by oncogenic retroviruses, many genes that were identified by studying retroviruses proved to encode key molecules in the cytokine/receptor/JAK/STAT/cyclin D pathway, which regulates normal hematopoietic cell proliferation.
The generation of cloned IL-2–dependent T lymphocyte lines pointed the way for the creation of additional hematopoietic cytokine–dependent cell lines that could be used for transformation studies using Abelson virus, which first demonstrated that autonomous, cytokine-independent cell growth (malignant transformation) could occur by circumventing the normal cytokine control of cell proliferation. These experiments were key, because they led to the discovery of the genes and molecules inside the cell that carry the cytokine/receptor signals from the membrane to the nucleus and to the genes that regulate cell-cycle progression, such as the D cyclins.
The discovery of a small molecule inhibitor of the PTK activity of BCR-ABL, and the consequential inhibition of the downstream persistent STAT5/cyclin D promotion of cell-cycle progression, was key in pointing the way to new, relatively nontoxic therapies for leukemias and other cancers. However, understanding the molecular regulation of normal T lymphocyte and hematopoietic cell proliferation, and the role played by cytokines, their receptors, and the signaling pathways, transcription factors, and key cell-cycle genes that they regulate, was clearly necessary to piece together the puzzle of the cellular proto-oncogenes into a coherent picture of their role in oncogenesis and provide the mechanistic understanding as to why the PTK inhibitors are effective therapeutics for the treatment of individuals with CML.
The molecular understanding of leukemogenesis took almost five decades and the combined efforts of many scientists from many different disciplines. At this juncture, it is important to emphasize that our understanding of additional cancers can now be based upon the hypothesis that abnormalities arising in genes encoding the proteins involved in cytokine signaling pathways that regulate the normal growth of other, nonhematopoietic cells and tissues may well be responsible for their malignant transformation. Accordingly, we propose this as the next obvious working hypothesis for the cancer field.
The lessons learned from the past 50 years point to the establishment of cloned cytokine-dependent normal cell lines as a key step in identifying the molecules responsible for normal cell growth control. For the future, based on the CML story, we can look forward to the development of effective agents for the treatment of many cancers based upon a sound knowledge of the molecular pathogenesis of each of the different mutants. In this regard, it is important to emphasize that although mutations may arise in many different upstream genes, eventually they must converge onto the final common mitogenic pathway to cause malignant transformation, exemplified by the cytokine/receptor/JAK/STAT/cyclin D genes and gene products in hematopoietic cells. Moreover, when effective agents are available, it will be important to detect the first mutations and to treat early, before multiple mutations accumulate. The lesson from CML is that autonomous growth stimulated by the BCR-ABL mutation occurs several years before additional mutations occur and become selected for more virulent malignancy.
Acknowledgments
The authors thank Carol Beadling, Jacqueline Bromberg, Doreen Cantrell, and James Darnell for helpful criticism and advice on the manuscript.
Footnotes
Nonstandard abbreviations used: BCR, breakpoint cluster region; CML, chronic myelogenous leukemia; IL-2R, IL-2 receptor; LAF, lymphocyte-activating factor; Ph, Philadelphia; PHA, phytohemagglutinin; PTK, protein tyrosine kinase; TCGF, T cell growth factor.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:3564–3573 (2008). doi:10.1172/JCI35819.
References
- 1.Nowell P.C. Phytohemagglutinin: an initiator of mitosis in cultures of normal human leukocytes. Cancer Res. 1960;20:462–468. [PubMed] [Google Scholar]
- 2.Hirschhorn K., Bach F., Kolodny R., Firschein I., Hashem N. Immune response and mitosis of human peripheral blood lymphocytes in vitro. Science. 1963;142:1185–1187. doi: 10.1126/science.142.3596.1185. [DOI] [PubMed] [Google Scholar]
- 3.Bach F., Hirschhorn K. lymphocyte interaction: a potential histocompatibility test in vitro. Science. 1964;143:813–814. doi: 10.1126/science.143.3608.813. [DOI] [PubMed] [Google Scholar]
- 4.Bain B., Lowenstein L. Genetic studies on the mixed leukocyte reaction. Science. 1964;145:1315–1316. doi: 10.1126/science.145.3638.1315. [DOI] [PubMed] [Google Scholar]
- 5.Kasakura S., Lowenstein L. A factor stimulating DNA synthesis derived from the medium of leukocyte cultures. Nature. 1965;208:794–795. doi: 10.1038/208794a0. [DOI] [PubMed] [Google Scholar]
- 6.Gordon J., MacLean L.D. A lymphocyte-stimulating factor produced in vitro. Nature. 1965;208:795–796. doi: 10.1038/208795a0. [DOI] [PubMed] [Google Scholar]
- 7.Nowell P., Hungerford D. Chromosome studies on normal and leukemic human leukocytes. J. Natl. Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- 8.Rowley J. A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 9.Todaro G., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established cell lines. . J. Cell. Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todaro G., Lazar G., Green H. The initiation of cell division in a contact-inhibited mammalian cell line. J. Cell. Physiol. 1965;66:325–333. doi: 10.1002/jcp.1030660310. [DOI] [PubMed] [Google Scholar]
- 11.Ellerman V., Bang O. Experimentelle leukamie bei huhnern. Zentralbl Bakteriol. [Orig.]. 1908;46:595–609. [Google Scholar]
- 12.Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J. Exp. Med. 1911;132:397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross L. “Spontaneous” leukemia developing in C3H mice following inoculation, in infancy, with AK-leukemic extracts, or AK-embryos. Proc. Soc. Exp. Biol. Med. 1951;76:27–32. [PubMed] [Google Scholar]
- 14.Friend C. Cell-free transmission in adult swiss mice of a disease having the character of a leukemia. J. Exp. Med. 1957;105:307–318. doi: 10.1084/jem.105.4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moloney J. Biological studies on a lymphoid-leukemia virus extracted from sarcoma 37. I. Origin and introductory investigations. J. Natl. Cancer Inst. 1960;24:933–951. [PubMed] [Google Scholar]
- 16.Rauscher F. A virus-induced disease of mice characterized by erythrocytopoiesis and lymphoid leukemia. J. Natl. Cancer Inst. 1962;29:515–532. [PubMed] [Google Scholar]
- 17.Hardy W., et al. Feline leukemia virus: occurrence of viral antigen in the tissues of cats with lymphosarcoma and other diseases. Science. 1969;166:1019–1021. doi: 10.1126/science.166.3908.1019. [DOI] [PubMed] [Google Scholar]
- 18.Abelson H., Rabstein L. Lymphosarcoma: virus-induced thymic independent disease in mice. Cancer Res. 1970;30:2213–2222. [PubMed] [Google Scholar]
- 19.Temin H. Nature of the provirus of Rous sarcoma. Natl. Cancer. Inst. Monogr. 1964;17:557–570. [Google Scholar]
- 20.Temin H., Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. . 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 21.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumor viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 22.Rosenthal P., Robinson H., Robinson W., Hanafusa T., Hanafusa H. DNA in uninfected and virus-infected cells complimentary to avian tumor virus RNA. Proc. Natl. Acad. Sci. U. S. A. 1971;68:2336–2340. doi: 10.1073/pnas.68.10.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Temin H. The protovirus hypothesis: speculations on the significance of RNA-directed DNA synthesis for normal development and for carcinogenesis. J. Natl. Cancer Inst. 1971;46:3–7. [PubMed] [Google Scholar]
- 24.Pardee A. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 1974;71:1286–1292. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pledger W., Stiles C., Antoniades H., Scher C. Induction of DNA synthesis in BALB/c 3T3 cells by serum components: reevaluation of the commitment process. Proc. Natl. Acad. Sci. U. S. A. 1977;74:4481–4485. doi: 10.1073/pnas.74.10.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cline M., Golde D. Production of colony-stimulating activity by human lymphocytes. Nature. 1974;248:703–704. doi: 10.1038/248703a0. [DOI] [PubMed] [Google Scholar]
- 27.Morgan D.A., Ruscetti F.W., Gallo R. Selective in vitro growth of T lymphocytes from normal human bone marrows. Science. 1976;193:1007–1008. doi: 10.1126/science.181845. [DOI] [PubMed] [Google Scholar]
- 28.Gillis S., Smith K.A. Long term culture of tumour-specific cytotoxic T cells. Nature. 1977;268:154–156. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- 29.Baker P.E., Gillis S., Smith K.A. Monoclonal cytolytic T-cell lines. J. Exp. Med. 1979;149:273–278. doi: 10.1084/jem.149.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowell P.C. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 31.Stehelin D., Varmus H.E., Bishop J.M., Vogt P.K. DNA related to the transforming gene(s) of avian sarcoma viruses present in normal avian DNA. Nature. 1976;260:170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 32.Goff S., Gilboa E., Witte O., Baltimore D. Structure of the Abelson murine leukemia virus genome and the homologous cellular gene: studies with cloned viral DNA. Cell. 1980;22:777–785. doi: 10.1016/0092-8674(80)90554-1. [DOI] [PubMed] [Google Scholar]
- 33.Collett M.S., Erikson R.L. Protein kinase activity associated with the avian sarcoma virus src gene product. Proc. Natl. Acad. Sci. U. S. A. 1978;75:2021–2024. doi: 10.1073/pnas.75.4.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponticelli A., Whitlock C., Rosenberg N., Witte O. In vivo tyrosine phosphorylations of the Abelson virus transforming protein are absent in its normal cellular homologue. Cell. 1982;29:953–960. doi: 10.1016/0092-8674(82)90458-5. [DOI] [PubMed] [Google Scholar]
- 35.Gillis S., Ferm M.M., Ou W., Smith K.A. T cell growth factor: parameters of production and a quantitative microassay for activity. . J. Immunol. 1978;120:2027–2032. [PubMed] [Google Scholar]
- 36.Robb R.J., Smith K.A. Heterogeneity of human T-cell growth factor(s) due to variable glycosylation. Mol. Immunol. 1981;18:1087–1094. doi: 10.1016/0161-5890(81)90024-9. [DOI] [PubMed] [Google Scholar]
- 37.Smith K.A., Favata M.F., Oroszlan S. Production and characterization of monoclonal antibodies to human interleukin 2: strategy and tactics. J. Immunol. 1983;131:1808–1815. [PubMed] [Google Scholar]
- 38.Robb R.J., Munck A., Smith K.A. T cell growth factor receptors: quantitation, specificity, and biological relevance. J. Exp. Med. 1981;154:1455–1474. doi: 10.1084/jem.154.5.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leonard W.J., et al. A monoclonal antibody that appears to recognize the receptor for human T-cell growth factor; partial characterization of the receptor. Nature. 1982;300:267–269. doi: 10.1038/300267a0. [DOI] [PubMed] [Google Scholar]
- 40.Gery I., Gershon R.K., Waksman B. Potentiation of the T-lymphocyte response to mitogens. J. Exp. Med. 1972;136:128–142. doi: 10.1084/jem.136.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith K.A. T-cell growth factor. Immunol. Rev. 1980;51:337–357. doi: 10.1111/j.1600-065X.1980.tb00327.x. [DOI] [PubMed] [Google Scholar]
- 42.Smith K.A., Gilbride K.J., Favata M.F. Lymphocyte activating factor promotes T-cell growth factor production by cloned murine lymphoma cells. Nature. 1980;287:853–855. doi: 10.1038/287853a0. [DOI] [PubMed] [Google Scholar]
- 43.Smith K.A., Lachman L.B., Oppenheim J.J., Favata M.F. The functional relationship of the interleukins. J. Exp. Med. 1980;151:1551–1556. doi: 10.1084/jem.151.6.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantrell D.A., Smith K.A. The interleukin-2 T-cell system: a new cell growth model. Science. 1984;224:1312–1316. doi: 10.1126/science.6427923. [DOI] [PubMed] [Google Scholar]
- 45.Smith K.A. Interleukin-2: inception, impact, and implications. Science. 1988;240:1169–1176. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- 46.Smith K.A. The interleukin 2 receptor. Annu. Rev. Cell Biol. 1989;5:397–425. doi: 10.1146/annurev.cb.05.110189.002145. [DOI] [PubMed] [Google Scholar]
- 47.Smith K.A. Cell growth signal transduction is quantal. Ann. N. Y. Acad. Sci. 1995;766:263–271. doi: 10.1111/j.1749-6632.1995.tb26674.x. [DOI] [PubMed] [Google Scholar]
- 48. Smith, K. 1997. Why do cells count? InNonlinear cooperative phenomena in biological systems. L. Matsson, editor. World Scientific Publishing Co. Pte. Ltd. Singapore, Singapore. 13–19. [Google Scholar]
- 49.Meuer S.C., et al. Clonotypic structures involved in antigen-specific human T cell function. Relationship to the T3 molecular complex. J. Exp. Med. 1983;157:705–719. doi: 10.1084/jem.157.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meuer S.C., et al. Triggering the T3-Ti antigen-receptor complex results in clonal T cell proliferation through an interleukin 2-dependent autocrine pathway. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1509–1513. doi: 10.1073/pnas.81.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shih C., Shilo B.-Z., Goldfarb M.P., Dannenberg A., Weinberg R.A. Passage of phenotypes of chemically transformed cells via transfection of DNA and chromatin. Proc. Natl. Acad. Sci. U. S. A. 1979;76:5714–5718. doi: 10.1073/pnas.76.11.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cooper G., Okenquist S., Silverman L. Transforming activity of DNA of chemically transformed and normal cells. Nature. 1980;284:418–421. doi: 10.1038/284418a0. [DOI] [PubMed] [Google Scholar]
- 53.Krontiris T.G., Cooper G.M. Transforming activity of human tumor DNAs. Proc. Natl. Acad. Sci. U. S. A. 1981;78:1181–1184. doi: 10.1073/pnas.78.2.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shih C., Padhy L., Murray M., Weinberg R. Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature. 1981;290:261–264. doi: 10.1038/290261a0. [DOI] [PubMed] [Google Scholar]
- 55.Cooper G. Cellular transforming genes. Science. 1982;217:801–806. doi: 10.1126/science.6285471. [DOI] [PubMed] [Google Scholar]
- 56.Weinberg R. Fewer and fewer oncogenes. Cell. 1982;30:3–4. doi: 10.1016/0092-8674(82)90003-4. [DOI] [PubMed] [Google Scholar]
- 57.Heisterkamp N., et al. Localization of the c-abl oncogene adjacent to a translocation break point in chronic myelocytic leukemia. . Nature. 1983;306:239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 58.Groffen J., et al. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 59.Evans T., Rosenthal E., Youngbloom J., Distel D., Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 60.Rosenthal E., Tansey T., Ruderman J. Sequence-specific adenylations accompany changes in the translation of maternal messenger RNA after fertilization of Spinsula oocytes. . J. Mol. Biol. 1983;166:309–327. doi: 10.1016/S0022-2836(83)80087-4. [DOI] [PubMed] [Google Scholar]
- 61.Solomon M., Booher R., Kirschner M., Beach D. Cyclin in fission yeast. Cell. 1988;54:738–740. doi: 10.1016/S0092-8674(88)90933-6. [DOI] [PubMed] [Google Scholar]
- 62.Matsushime H., Roussel M., Ashmun R., Sherr C. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 63.Motokura T., Bloom T., Kim H., Juppner H., Ruderman J. A novel cyclin encoded by a bcl1-linked candidate oncogene. . Nature. 1991;350:512–515. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- 64.Withers D., et al. Characterization of a candidate bcl-1 gene. . Mol. Cell. Biol. 1991;11:4846–4853. doi: 10.1128/mcb.11.10.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lammie G., et al. Proviral insertions near cyclin D1 in mouse lymphomas: a parallel for BCL-1 translocations in human B-cell neoplasms. Oncogene. . 1992;7:2381–2387. [PubMed] [Google Scholar]
- 66.Lammie G., et al. D11S287, a putative oncogene on chromosome q13, is amplified and expressed in squamous cell and mammary carcinomas and linked to BCL-1. Oncogene. . 1991;6:439–444. [PubMed] [Google Scholar]
- 67.Turner J. IL2-dependent induction of G1 cyclins in primary T cells is not blocked by rapamycin or cyclosporin A. Int. Immunol. 1993;5:1199–1209. doi: 10.1093/intimm/5.10.1199. [DOI] [PubMed] [Google Scholar]
- 68.Nevins J., Leone G., DeGregori J., Jakoi L. Role of the Rb/E2F pathway in cell growth control. J. Cell. Physiol. 1997;173:233–236. doi: 10.1002/(SICI)1097-4652(199711)173:2<233::AID-JCP27>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 69.Nicola N., Metcalf D., Matsumoto M., Johnson G. Purification of a factor inducing differentiation in murine myelomonocytic leukemia cells: identification as granulocyte colony stimulating factor. J. Biol. Chem. 1983;258:9017–9023. [PubMed] [Google Scholar]
- 70.Nicola N., Metcalf D. Binding of the differentiation-inducer, granulocyte-colony-stimulating factor, to responsive, but not unresponsive leukemic cell lines. Proc. Natl. Acad. Sci. U. S. A. 1984;81:3765–3769. doi: 10.1073/pnas.81.12.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cook W., Metcalf D., Nicola N., Burgess A., Walker F. Malignant transformation of a growth factor-dependent myeloid cell line by Abelson virus without evidence of an autocrine mechanism. Cell. 1985;41:677–683. doi: 10.1016/S0092-8674(85)80048-9. [DOI] [PubMed] [Google Scholar]
- 72.Pierce J., et al. Neoplastic transformation of mast cells by Abelson-MuLV: abrogation of IL3-dependence by a nonautocrine mechanism. Cell. 1985;41:685–693. doi: 10.1016/S0092-8674(85)80049-0. [DOI] [PubMed] [Google Scholar]
- 73.Cook W., Fazekas de St. Groth B., Miller J., MacDonald H., Gabathuler R. Abelson virus transformation of an interleukin-2-dependent antigen-specific T-cell line. Mol. Cell. Biol. 1987;7:2631–2635. doi: 10.1128/mcb.7.7.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bazan J.F. Structural design and molecular evolution of a cytokine receptor superfamily. Proc. Natl. Acad. Sci. U. S. A. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilks A. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc. Natl. Acad. Sci. U. S. A. 1989;86:1603–1607. doi: 10.1073/pnas.86.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilks A., et al. Two Novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol. Cell. Biol. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Firmbach-Kraft I., Byers M., Shows T., Dalla-Favera R., Krolewski J. tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. . Oncogene. 1990;5:1329–1336. [PubMed] [Google Scholar]
- 78.Johnston J., et al. Phosphorylation and activation of the JAK-3 Janus kinase in response to interleukin-2. Nature. 1994;370:151–153. doi: 10.1038/370151a0. [DOI] [PubMed] [Google Scholar]
- 79.Witthuhn B., et al. Involvement of the JAK-3 Janus kinase in signaling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature. 1994;370:153–157. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]
- 80.Beadling C., et al. Activation of JAK kinases and STAT proteins by interleukin-2 and interferon alpha, but not the T cell antigen receptor, in human T lymphocytes. EMBO J. 1994;13:5605–5615. doi: 10.1002/j.1460-2075.1994.tb06898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Larner A., et al. Transcriptional induction of two genes in human cells by beta interferon. Proc. Natl. Acad. Sci. U. S. A. 1984;81:6733–6737. doi: 10.1073/pnas.81.21.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Larner A., Chaudhuri A., Darnell J.E., Jr. Transcriptional induction by interferon. . J. Biol. Chem. 1986;261:453–459. [PubMed] [Google Scholar]
- 83.Schindler C., Shuai K., Prezioso V., Darnell J. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 84.Fu X., Kessler D.S., Veals S.A., Levy D.E., Darnell J.E., Jr. ISGF3, the transcriptional activator induced by interferon {alpha}, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schindler C., Fu X., Improta T., Aebersold R., Darnell J.E., Jr. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84- kDa ISGF-3 proteins that are activated by interferon {alpha}. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fu X., Schindler C., Improta T., Aebersold R., Darnell J.E., Jr. The proteins of ISGF-3, the interferon {alpha}-induced transcriptional activator, define a gene family involved in signal transduction. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhong Z., Wen Z., Darnell J. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 88.Zhong Z., Wen Z., Darnell J. Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc. Natl. Acad. Sci. U. S. A. 1994;91:4806–4810. doi: 10.1073/pnas.91.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wakao H., Gouilleux F., Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu X., Robinson G.W., Gouilleux F., Groner B., Hennighausen L. Cloning and expression of stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mui A., Wakao H., O’Farrell A.-M., Harada N., Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologues. EMBO J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hou J., et al. An interleukin-4-induced transcription factor: IL4 Stat. Science. 1994;265:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 93.Pelligrini S., John J., Shearer M., Kerr I., Stark G. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Velazquez L., Fellous M., Stark G., Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-L. [DOI] [PubMed] [Google Scholar]
- 95.Darnell J., Kerr I., Stark G. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 96.Witthuhn B., et al. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-L. [DOI] [PubMed] [Google Scholar]
- 97.Silvennoinen O., et al. Structure of the murine Jak2 protein-tyrosine kinase and its role in interleukin 3 signal transduction. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8429–8433. doi: 10.1073/pnas.90.18.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shimoda K., et al. G-CSF induces tyrosine phosphorylation of the JAK2 protein in the human myeloid G-CSF responsive and proliferative cells, but not in mature neutrophils. Biochem. Biophys. Res. Commun. 1994;203:922–928. doi: 10.1006/bbrc.1994.2270. [DOI] [PubMed] [Google Scholar]
- 99.Hou J., Schindler U., Henzel W., Wong S., McKnight S. Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity. 1995;2:321–329. doi: 10.1016/1074-7613(95)90140-X. [DOI] [PubMed] [Google Scholar]
- 100.Lin J.-X., Mietz J., Modi W.S., John S., Leonard W.J. Cloning of human Stat5B. J. Biol. Chem. 1996;271:10738–10744. doi: 10.1074/jbc.271.18.10738. [DOI] [PubMed] [Google Scholar]
- 101.Yaish P., Gazit A., Gilon C., Levitzki A. Blocking of EGF-dependent cell proliferation by EGF receptor kinase inhibitors. Science. 1988;242:933–935. doi: 10.1126/science.3263702. [DOI] [PubMed] [Google Scholar]
- 102.Anafi M., Gazit A., Zehavi A., Ben-Neriah Y., Levitzki A. Tyrphostin-induced inhibition of p210bcr-abl tyrosine kinase activity induces K562 to differentiate. . Blood. 1993;82:3524–3529. [PubMed] [Google Scholar]
- 103.Levitzki A., Gazit A. Tyrosine kinase inhibition: an approach to drug development. Science. 1995;267:1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 104.Druker B., et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 105.Druker B., et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 106.Yu C.-L., et al. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the src oncoprotein. . Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 107.Bromberg J.F., Horvath C.M., Besser D., Lathem W.W., Darnell J.E., Jr. Stat3 activation is required for cellular transformation by v-src. Mol. Cell. Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Turkson J., et al. Stat3 activation by src induces specific gene regulation and is required for cell transformation. Mol. Cell. Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bromberg J., et al. Stat3 as an oncogene. . Cell. 1999;98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 110.Bromberg J. Stat proteins and oncogenesis. J. Clin. Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Danial N., Pernis A., Rothman P. Jak-STAT signaling induced by the v-abl oncogene. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 112.Lacronique V., et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 113.Peeters P., et al. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90:2535–2540. [PubMed] [Google Scholar]
- 114.Lacronique V., et al. Transforming properties of chimeric TEL-JAK proteins in Ba/F3 cells. Blood. 2000;95:2076–2083. [PubMed] [Google Scholar]
- 115.Horita M., et al. Blockade of the Bcr-Abl kinase activity induces apoptosis of chronic myelogenous leukemia cells by suppressing signal transducer and activator of transcription 5-dependent expression of Bcl-XL. . J. Exp. Med. 2000;191:977–984. doi: 10.1084/jem.191.6.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gesbert F., Griffin J. Bcr-Abl activates transcription of the Bcl-X gene through STAT5. . Blood. 2000;96:2269–2276. [PubMed] [Google Scholar]
- 117.Xie S., et al. Involvement of Jak2 tyrosine phosphorylation in Bcr-Abl transformation. Oncogene. 2001;20:6188–6195. doi: 10.1038/sj.onc.1204834. [DOI] [PubMed] [Google Scholar]
- 118.Klejman A., et al. The Src family kinase Hck couples BCR/ABL to STAT5 activation in myeloid leukemia cells. EMBO J. 2002;21:5766–5774. doi: 10.1093/emboj/cdf562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sexl V., et al. Stat5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl- and bcr/abl-induced transformation are independent of Stat5. Blood. 2000;96:2277–2283. [PubMed] [Google Scholar]
- 120.Benekli M., Baer M., Bauman H., Wetzler M. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101:2940–2954. doi: 10.1182/blood-2002-04-1204. [DOI] [PubMed] [Google Scholar]
- 121.Hoelbl A., et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–4906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bessette K., et al. A Stat5b transgene is capable of inducing CD8+ lymphoblastic lymphoma in the absence of normal TCR/MHC signaling. Blood. 2008;111:344–350. doi: 10.1182/blood-2007-04-084707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Clarke M., et al. Cancer stem cells — Perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 124.Lu X., et al. Expression of a homodimeric type I cytokine receptor is required for JAKV617F-mediated transformation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18962–18967. doi: 10.1073/pnas.0509714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Daley G., Baltimore D. Transformation of an interleukin 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210BCR-ABL protein. Proc. Natl. Acad. Sci. U. S. A. 1988;85:9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wong S., et al. IL-3 signaling is dispensable for BCR-ABL-induced myeloproliferative disease. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11630–11635. doi: 10.1073/pnas.2035020100. [DOI] [PMC free article] [PubMed] [Google Scholar]