Abstract
Isoforms of RUSH interact with a RING-finger binding protein (RFBP), which is a splice variant of the Type IV P-Type ATPase, ATP11B. Spice arrays and RT-PCR showed that although most splice variants in RUSH and ATP11B are conserved in human and rabbit, the RFBP isoform is specific to rabbit. Interactions between the discontinuous PVITHC – HAKCPL sequence in the RING-domain of RUSH and the KVIRLIKIS sequence in the catalytic loop of RFBP were first identified with pull-down assays. Fine mapping involved probing CLIPS-constrained RING peptides with GST-tagged KVIRLIKIS. When the companion site in RFBP was fine mapped by replacement analysis with MBP-tagged RING, a 4-fold increase in binding was noted for the KVIRLDKIS mutant. Direct comparison of splicing events in the RUSH and ATP11B genes between human and rabbit shows high structural stability in these protein interactions sites, which are 100% conserved in all mammalian orthologs.
Keywords: RING-finger, RUSH, SMARCA3/HLTF, ATP11B, alternative splicing, CLIPS-technology
1. Introduction
The RUSH (Rabbit, Uteroglobin promoter-binding, SWI/SNF-related, Helicase) acronym recognizes two alternatively spliced transcription factors: a full length, progesterone-dependent, α isoform, and a truncated, estrogen-dependent, β isoform in rabbit endometrium (1). The amino acid sequence for RUSH-1α is 95% similar and 91% identical to the human ortholog HLTF (helicase-like transcription factor) formerly known as SMARCA3 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 3).
RUSH-1α (1005 amino acids) and HLTF (1009 amino acids) contain seven consecutive motifs (I, Ia, II-VI) that are characteristic of ATPases and DNA helicases, and a RING-type zinc finger interposed between motifs III and IV. The insertion of a 57-bp exon that contains an in frame stop codon produces RUSH-1β. This truncated isoform (836 amino acids) is identical to the alpha isoform through the RING-finger motif and for 33 amino acids there after. Then because of an exon inclusion event the protein extends for 5 unique amino acids and stops.
The RING (really interesting new gene) motif or C3HC4-type zinc finger of RUSH is a protein interaction domain known to bind the transcription factors Egr-1 and c-Rel (2). When the RING-domain of RUSH was used to screen an expression library from a rabbit uterine epithelial cell line (3), an atypical Type-IV P-type ATPase (4) was identified. This so-called RING-finger binding protein (RFBP) was devoid of the region containing transmembrane domain 4. All working models of membrane topology for P-type ATPases are characterized by an even number of highly conserved transmembrane passes and eight signature motifs designated A–H. ATP11B has four transmembrane passes in the NH2-terminus and six in the C-terminus. A small, strongly hydrophilic loop (motifs A–C) is positioned between the second and third NH2-terminal transmembrane passes, and a larger loop (motifs D–H) which contains the aspartate phosphorylation motif and ATP-binding domain is positioned between the fourth NH2-terminal transmembrane pass and the first C-terminal transmembrane pass. However, RFBP has an odd number of transmembrane passes. Region D that contains transmembrane domain 4, corresponding to exon 12 in the human, is missing.
This structural alteration placed the major catalytic loop of the enzyme on the opposite side of the membrane from the small hydrophilic loop. Western analysis with affinity-purified, antipeptide (IgG) antibodies against amino acids (663–678) in the major catalytic loop of RFBP (3) was used to identify a 128-kDa immunoreactive protein in nuclear fractions from endometrium. In vitro protein-protein interactions were verified by GST pull-down assays and immunoblotting. Immunoelectron microscopy showed RFBP associates with the inner nuclear envelope. The same immunolabeling pattern was identified when the outer nuclear membrane was selectively removed with Triton X-100. Halleck et al (5) used a PCR strategy to show that both full-length ATPIIB and RFBP are expressed in a cDNA library prepared from rabbit leukocytes. The independent confirmation of the existence of RFBP, a putative ATP11B splice variant, prompted questions about alternative splicing patterns relevant to the functional importance of conserved domains. Alternative splicing is a powerful mechanism for macromolecular complexity because splice variants can display different signaling activities, enzymatic properties and subcellular locations. As a result, RFBP expression was evaluated with custom designed splice arrays. Six different oligonucleotide probes, so-called exon, junction and event probes, were used to detect and quantify splice variants in rabbit and human uterus. Expression levels were confirmed by competitive RT-PCR.
In vitro protein-protein interactions between RUSH and RFBP were verified by pull-down assays in which recombinant RING peptide bound to native RFBP in a dose-dependent manner. Next, pull-down assays were used to identify the subregions of each protein that participated in the binding interactions. Fine mapping the RING domain was achieved with peptide-microarrays (n=450 peptides). This included overlapping 15-mers covering the RING domain (n=50 peptides), and CLIPS™ technology (Chemical LInkage of Peptides onto Scaffolds), an integrated technology that combines chemical linkage of linear (n=400) peptides to a synthetic scaffold with conformational fixation of the peptide (6, 7). Fine mapping the companion-binding site in RFBP included testing all possible single-position variants (n=300 peptides) in 15-mers covering relevant sequence in core region G of the major catalytic loop. The possibility that splicing alters the binding domains was evaluated.
2. Material and methods
2.1 Reagents and Tools
Staff at Midland Certified Reagent Company (Midland, TX) synthesized custom oligonucleotides for cloning and primers for PCR reactions. The pMAL-c2E vector and E. coli TB1 cells were purchased from New England Biolabs (Beverly, MA). The pGEX-2TK vector was purchased from GE Healthcare (Piscataway, NJ). BRL(DE3)pLysS cells were purchased from Novagen a division of EMD Chemicals Inc (Gibbstown, NJ). NuSieve® GTG®-agarose gel was purchased from Cambrex Bioscience Rockland, Inc. (Rockland, ME). QIAEX II was purchased from Qiagen, Inc. (Valencia, CA). Human uterine poly (A+) RNA and the BD Advantage-GC 2 PCR kit were purchased from BD Biosciences Clontech (Palo Alto, CA). Nucleotrap was purchased from Macherey-Nagel (Bethlehem, PA). Invitrogen (Carlsbad, CA) was the commercial source of SuperScript III First-Strand synthesis System for RT-PCR and the TA Cloning Kits that contain the pCRII-TOPO vector. TaKaRa ExTaq enzyme and 10X LA PCR buffer were purchased from Takara Bio Inc USA (Madison, WI). The QuikChange Site-Directed Mutagenesis Kit that contains Pfu Turbo DNA polymerase, 10X reaction buffer, and DpnI endonuclease was purchased from Stratagene (La Jolla, CA). The RNA 6000 Nano LabChip kit was purchased from Agilent Technologies (Santa Clara, CA). Staff at ExonHit Therapeutics Inc. (Gaithersburg, MD) synthesized the custom DNA microarrays, performed hybridizations and data analyses. Staff at Pepscan Therapeutics BV (Lelystad, The Netherlands) prepared custom peptide-microarrays on polypropylene supports (6, 7), performed ELISA and data analyses. We collaborated with staff scientists at these companies because of their technical expertise and facilities.
2.2 Animal Treatments
All studies were conducted according to the NIH Guidelines for the Care and Use of Laboratory Animals, as reviewed and approved by the Animal Care and Use Committee at Texas Tech University Health Sciences Center. Adult female New Zealand white rabbits (six months of age) were housed for three weeks prior to experimentation.
2.3 RNA isolation
Three rabbits were treated with progesterone (3 mg/kg/24 h) for 4 days and killed 24 h after the last injection. Ethanol-corn oil (50:50) was used as the vehicle. Total RNA was isolated from the combined endometrium of two of these rabbits (Rabbit1) by the cold precipitation method of Han et al. [8]. Total RNA was isolated from the endometrium of the third rabbit (Rabbit 2) and from two preparations of human Ishikawa cells (Human1 and Human2) with TriReagent according to the manufacturers instructions. The integrity of each RNA sample was confirmed by electrophoretic fractionation through formaldehyde containing agarose (1.2%) gels and ethidium bromide staining. Staff at ExonHit Therapeutics verified total RNA quality and quantity using the RNA 6000 Nano LabChip kit with the Agilent Bioanalyzer 2100.
Poly (A+) RNA was isolated from total (rabbit) RNA with the Nucleo Trap Acid Purification Kit as previously described (9). RNA concentrations were determined spectrophotometrically (A260). The A260/280 ratio for each sample was > 1.8.
2.4 Splice arrays
Interspecies microarray analyses were used to profile alternative splicing events in 32 pairs of human and rabbit genes based on conservation at genome and transcript levels in the uterus. A total of 245 putative events were evaluated with 3438 probes in 4 custom 11K SpliceArrays on the Agilent platform. Putative splice events were individually profiled with nine oligonucleotide probes, i.e. 44-mers that contained 24 nucleotides of human gene-specific sequence. Probe sequences were derived from a Blat (10) alignment of all publicly available RefSeq, mRNA and qualified spliced EST sequences. ESTs were qualified according to the strength of a match to the genome with more than 1 exon in common with the selected reference sequence. Following alignment, a proprietary algorithm was used to perform a difference comparison between the reference sequence for a given locus and all available overlapping sequences present in the databases. Data were extracted and generated for each event. As a result, the two possible molecular species from each splice event, i.e. a long isoform product of exon retention or a short isoform product of exon exclusion, were detected with nine probes, two directed against specific exons, four directed against exon-exon junctions, and three directed against exon-intron junctions in human genes.
For the hybridization assays, total RNA (5µg/reaction) from rabbit endometrium and human Ishikawa cells was reverse transcribed into first strand cDNA using random primers and labeled post synthesis with either Cy3 or Cy5 for dye swap hybridizations. The cDNA products were purified and equal amounts of labeled material were used for the hybridization experiments. All incubations were performed in an ozone-free atmosphere to prevent any degradation of signal. The array layout for the project was as follows:
After hybridization, staff at ExonHit Therapeutics scanned the arrays using Agilent’s Microarray Scanner. The images were analyzed with Feature Extraction software (v. 9.9.3), using all default settings. Data files, which contained processed signals (background subtracted, dye normalized, floored values) were important into Partek® Genomics Suite (Partek GS) and analyzed at ExonHit Therapeutics. Partek GS integrates advanced statistics and interactive visualization to extract data patterns from Affymetrix GeneChips (http://www.partek.com). Data were quantile normalized across arrays, and an ANOVA analysis was performed to select significant probes.
2.5 RT-PCR and sequencing
Poly (A+) RNA samples (500 ng) were reverse transcribed with the SuperScript II First-Strand Synthesis system for RT-PCR. Reaction products were amplified with gene-specific primers and the BD Advantage-GC 2 PCR kit for the amplification of GC-rich templates. PCR primer pairs that target sequences in exons 11 and 13, and flank exon 12, were derived from the GenBank RFBP sequence (AF236061) for rabbit as previously described by us (4) and used by Halleck et al. (5). Comparable human probes were derived from GenBank ATP11B sequence (NT 005612.15) with a BLASTN algorithm for pairwise DNA-DNA sequence alignment (11). Hot start PCR amplifications were performed as follows: 30 s at 94°C, followed by 5 cycles of 94°C, 5 s; 63.5 °C, 90 s; 5 cycles of 94°C, 5 s; 64°C, 90 s; 5 cycles of 94°C, 5 s; 64.5 °C, 90 s; 20 cycles of 94°C, 5 s; 65°C, 90 s; and a final extension for 10 min at 68°C. Samples were rapidly cooled to 4°C. PCR products were subcloned and sequenced.
2.6 Synthesis of MBP-RING fusion proteins
The RING motif (nucleotides 2433–2579) was amplified from 50 ng of a 1030-bp partial RUSH cDNA clone as previously described (1). The forward primer [5′-GCG GTA CCG GAA GAA TGT GCT-3′] had a unique Kpn I site (bold) and the reverse primer [5′-GCA AGC TTG CTC ATC CAT GTA T-3′ had a unique Hind III site (bold). Hot start PCR amplifications were performed as follows: 30 s at 94 °C, followed by 5 cycles of 94 °C, 5 s; 50 °C, 120 s; 5 cycles of 94 °C, 5 s; 53 °C, 120 s; 5 cycles of 94 °C, 5 s; 56 °C, 120 s; 5 cycles of 94 °C, 5s; 59 °C, 120s; 20 cycles of 94 °C, 5 s; 62 °C, 120 s; and a final extension of 10 minutes at 68 °C. Samples were rapidly cooled to 4 °C. A single 169-bp PCR product was cloned into pCRII, then excised with Kpn I and Hind III, and directionally subcloned into the polylinker of the pMAL-c2E vector. This construct was designated RING1.
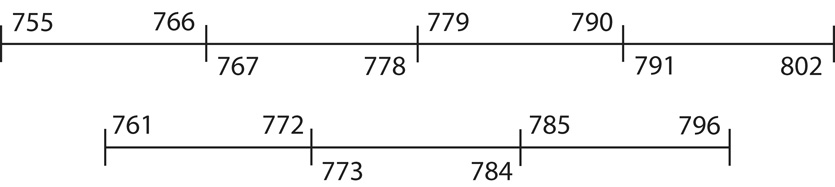
The RING-finger region of RUSH was subdivided into 7 peptides according to the strategy in Figure 1. Sequences encoding these peptides were synthesized as oligonucleotides with Kpn1 and Hind III overhangs. Each coding strand was synthesized as a 41-mer and each complementary strand was synthesized as a 49-mer. Reciprocal pairs of oligonucleotides were annealed and directionally cloned into the Kpn1 – Hind III restriction sites in the polylinker region of the pMAL-c2E vector. The eighth and final pair of oligonucleotides, which encodes the binding site (PVITHC-HAKCPL) was similarly synthesized and cloned. Thus full-length RING and all RING-fragments were inserted down-stream from the malE gene, which encodes maltose-binding protein (MBP), resulting in the expression of a series of MBP-RING fusion proteins.
Figure 1.
Strategy for subdividing the RING-domain of RUSH/HLTF. The RING-finger plus 2 amino acids at each end equals a 48 amino acid region (amino acids 755–802) that was divided into four non-overlapping peptides of 12 amino acids (36 nucleotides) each. Three additional double-stranded oligonucleotides were designed to encode peptides that span the ends of the original four peptides in case the boundaries of those peptides interrupted the sequence(s) required for binding. Sufficient spacer nucleotides were introduced at the 5′-end of each RING-fragment to ensure that the desired reading frame was preserved.
2.7 Synthesis of GST-RFBP fusion proteins
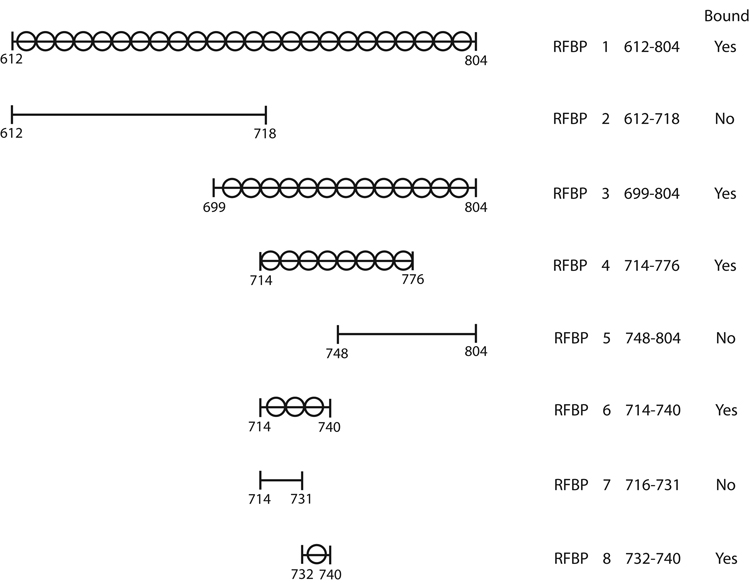
The major catalytic loop (nucleotides 1835–2413) was amplified (Figure 2) from the original 1584-bp RFBP clone (4). The forward primer [5′-GCG GAT CCG ACA GAC TGC AA-3′] had a unique BamH I site (bold) and the reverse primer [5′-GCG AAT TCC TCT AAT ATA ATA-3′] had a unique EcoR I site (bold). Hot start PCR amplifications were performed as follows: 30 s at 94 °C, followed by 5 cycles of 94 °C, 5 s; 43 °C, 120 s; 5 cycles of 94 °C, 5 s; 47 °C, 120 s; 5 cycles of 94 °C, 5 s; 51 °C, 120 s; 5 cycles of 94 °C, 5s; 55 °C, 120s; 20 cycles of 94 °C, 5 s; 59 °C, 120 s; and a final extension of 10 minutes at 68 °C. Samples were rapidly cooled to 4 °C. A single 594-bp PCR product was cloned into pCRII, then excised with BamH 1 and EcoR I, and directionally subcloned into the polylinker region of the pGEX-2TK vector. This construct was designated RFBP1. All subdivisions of RFBP1 were PCR amplified using these same reaction conditions with location-specific primer pairs and directionally subcloned into pGEX-2TK.
Figure 2.
Strategy for subdividing a portion of the conformationally flexible loop of RFBP. Amino acids 612–804 were subdivided into overlapping, unequal regions by a process of elimination based on empirical evidence that confirmed either positive or negative binding. The search converged to reveal that amino acids 732–740 are capable of binding MBP-RING1.
RFBP1 was subdivided into two 338-bp overlapping segments (Figure 2). RFBP2 was synthesized with the same forward primer as used in the synthesis of RFBP1 and a new reverse primer [5′-GCG AAT TCC TGA ACA ATT CCT-3′]. RFBP3 was synthesized with a new forward primer [5′-GCG GAT CCA GCC TCT CTC TT-3′] and the reverse primer used in the RFBP1 synthesis reaction. RFBP4 (203-bp) and RFBP5 (188-bp) were synthesized from RFBP3. RFBP4 was synthesized with new forward [5′-GCG GAT CCA GGA ATT GTT CA-3′] and reverse [5′-GCG AAT TCC AGC CTG TCT CG-3′] primers. RFBP5 was synthesized with a new forward primer [5′-GCG GAT CCG GCT GTT GGG AT-3′] and the reverse primer used in the synthesis of RFBP1. RFBP6 (94-bp) and RFBP7 (67-bp) were synthesized from RFBP4 using the same forward primer as for RFBP4 and new reverse primers for RFBP6 [5′-GCG AAT TCC GGA GAT TTT TAT CAA TCT-3′] and RFBP7 [5′-GCG AAT TCC CGC TTT CTG CA-3′].
RFBP8, i.e. the construct that encodes the binding site (KVIRLIKIS) was obtained from oligonucleotides (27-mers) with BamH 1 and EcoR I overhangs. Reciprocal pairs of oligonucleotides were annealed and directionally cloned into the BamH 1-EcoR I restriction sites in the polylinker region of the pGEX-2TK vector. The I→D change at position 6 of KVIRLIKIS in RFBP8 to produce the RFBP mutant (RFBP8m) KVIRLDKIS was achieved with the QuikChange site-directed mutagenesis kit (12). Thus all RFBP-fragments were inserted down-stream from the Glutathione S-Transferase (GST) region resulting in the expression of a series of GST-RFBP fusion proteins.
2.8 Assays previously described by us
Hot start PCR reactions were performed (4) such that each DNA sample was amplified in a 50 µl PCR reaction mix containing LA PCR buffer (1X), TaKaRa ExTaq DNA polymerase (2.5U/50 µl), TaqStart antibody (0.55 µg/50 µl), dNTPs (0.2 mM each) and primers (0.2 mM each). PCR products were fractionated by electrophoresis on 2.5% NuSieve® GTG®-agarose gels, extracted with QIAEX II according to the manufacturer’s instructions, and ligated to pCRII-TOPO. Insert orientation was confirmed by sequencing in both directions by the dideoxy chain termination method.
TB1 host cells were transformed with either the pMAL-c2E vector (negative control) or the pMAL-c2E-RING constructs. BLR(DE3)pLysS host cells were transformed with either pGEX-2TK (negative control) or the pGEX-2TK-RFBP constructs. All clones were screened for isopropylthiogalactoside (IPTG)-induced fusion protein expression (4). MBP-RING fusion proteins were affinity purified based on MBP binding to amylose resin. GST-RFBP fusion proteins were affinity purified based on binding to glutathione sepharose. Purified proteins were used in pull-down assays and Western analysis (4), and ELISA. Rabbit anti-MBP (1:10,000) and rabbit anti-GST (1:40,000) were used for Western analysis. The secondary antibody was horseradish peroxidase-conjugated donkey anti-Rabbit IgG (1:5,000). Signals on Western blots were detected by chemiluminescence with Renaissance reagents. Relative intensities of signals were quantified (13) with a computer-assisted image analysis system (Bio-Image Visage 2000).
2.9 Peptide-microarrays
Using peptides to identify binding sites required an empirical approach in which 450 peptides covering the complete RING domain were synthesized in an attempt to reconstruct both linear and discontinuous binding sites. First, linear 15-mer overlapping peptides (50 total) derived from LIKKMKLILSSGSDEECAICLDSLTVPVITHCAHVFCKPCICQCIQNEQPHAKCPLCRNDIH GDNLLECPP were synthesized directly onto polypropylene plates (6). Second, CLIPS™ technology was used to synthesize all positional variants of the peptide PVITHC(StBu)AC(StBu)VFAKPAIAVAIQNEQPHAKC(StBu)PLC(StBu)R in which the four cysteines [C (StBu)] were covalently coupled to a synthetic scaffold with four connection points to mimic the zinc bridge. Complex 30-mer constructs (400 peptides) were derived from ligations of 15-mers such that PVITHC(StBu)AC(StBu)VFAKPAIC was ligated to all positional variants of AIQNEQPHAKC(StBu)PLC(StBu)R, and vice versa. Protein binding interactions were determined in ELISA. Arrays were probed with affinity purified GST-RFBP8 (1:1,000) and horseradish peroxidase-conjugated swine anti-Rabbit IgG (1:1,000). Signals were assessed using ABTS substrate.
Replacement analysis was used to test all single positional variants of the peptide QKAKVIRLIKISPEK containing the KVIRLIKIS binding site from the conformationally flexible loop of ATP11B. Overlapping 15-mer peptides (50) derived from LIKKMKLILSSGSDEECAICLDSLTVPVITHCAHVFCKPCICQCIQNEQPHAKCPLCRNDIH GDNLLECPP were synthesized directly onto polypropylene plates (6). Protein binding interactions were determined in ELISA. Arrays were probed with affinity purified MBP-RING1 (1:1,000) and horseradish peroxidase-conjugated swine anti-Rabbit IgG (1:1,000). Signals were assessed using ABTS substrate.
3. Results
3.1 Analysis of splice arrays
Partek GS analysis of the interspecies microarray data files showed a significant (P < 0.001) difference between human and rabbit for 279 of 3438 probe-specific events. Two of the 20 most significant events (Table 1) occurred in locus 23200, i.e. the ATP11B gene. Probe sets that monitored all possible splicing events across this gene confirmed exon 27 is preferentially retained and exon 5 is preferentially skipped. The expression patterns were the same for human and rabbit but isoform expression was up regulated in rabbits (Table 1).
Table 1. Top 20 Evidence Events.
Top scoring alternative splicing events. Candidates on this list were selected from the evidenced event (1175) probes based on descending pValue (significance at p < 0.001). Important events in the pair-wise comparison of human vs. rabbit are shown (GFoldChange).
| Event Tested | Outcome | Locus | Gene Name | GFold Change Human vs Rabbit | |
|---|---|---|---|---|---|
| Human | Rabbit | ||||
| Novel Exon in 5’ UTR | ◓ | ◓ | 2625 | GATA 3 | 23.50 |
| Skipped exon 1 | ● | ● | 2625 | GATA 3 | 24.97 |
| Skipped exon 3 | ● | ● | 2625 | GATA 3 | 22.39 |
| Alternative splice donor for exon 9 | ○ | ◓ | 4772 | NFATC1 | −5.47 |
| Skipped exon 7 | ○ | ○ | 4773 | NFATC2 | −6.13 |
| Skipped exon 6 | ● | ● | 4775 | NFATC3 | −10.02 |
| Alternative splice donor for exon 10 | ○ | ○ | 4800 | NFYA | −2.58 |
| Skipped exon 4 | ○ | ● | 5079 | PAX5 | −45.75 |
| Novel exon between exons 3–4 | ○ | ● | 5079 | PAX5 | −39.45 |
| Skipped exon 4 | ● | ● | 5079 | PAX5 | −46.18 |
| Skipped exon 3 | ○ | ○ | 5079 | PAX5 | −37.86 |
| Novel exon between exon 2–3 | ○ | ○ | 5915 | RARB | −11.90 |
| Intron retained between exons 25–26 | ◓ | ◓ | 6596 | SMARCA3 | −3.57 |
| Skipped exon 25 | ○ | ○ | 6596 | SMARCA3 | −3.54 |
| Skipped exon 7 | ● | ● | 6670 | SP3 | −5.24 |
| Novel exon at exon 7 | ◓ | ◓ | 6670 | SP3 | −3.77 |
| Skipped exon 18 | ◓ | ◓ | 6776 | STAT5A | −3.05 |
| Skipped exon 6 | ● | ◓ | 10488 | CREB3 | −5.65 |
| Skipped exon 27 | ○ | ○ | 23200 | ATP1IB | −4.35 |
| Skipped exon 5 | ◓ | ◓ | 23200 | ATP1IB | −2.85 |
○ - Event did not occur
◓ - Event did occur
● - Both isoforms present
No other unexpected splicing events were detected for the human or rabbit ATP11B gene with the splice arrays. Expression data for the probes monitoring the exon 12 skip event in this gene indicated that the reference gene (long isoform) was expressed at a relatively low level and the variant (RFBP, short isoform) was not expressed in either rabbit or human. Since the variant was cloned from a rabbit library with the RING-domain of RUSH as the probe, we assumed that the probes on this array did not hybridize to the rabbit sequences because of mismatches. In fact, sequence divergence became apparent when we designed primer pairs for the PCR amplification of products from RT reactions.
3.2 RT-PCR analysis of exon 12 in ATP11B
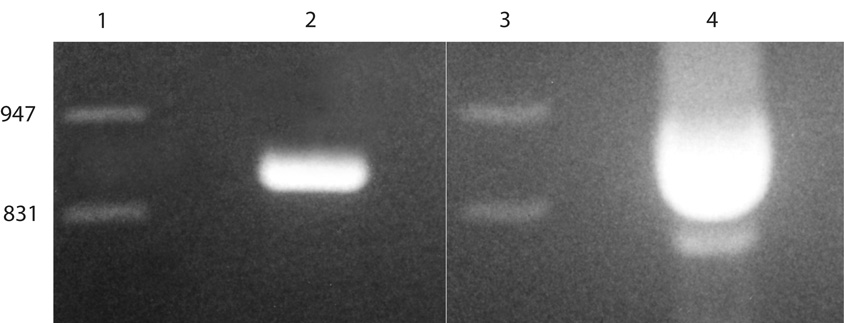
Analysis of RT-PCR reaction products by agarose gel electrophoresis (2.5% NuSieve) and ethidium bromide fluorescence allowed the visualization (Figure 3) of a single 939-bp reaction product from human uterus. Sequence analysis confirmed this population of amplicons contains exon 12. Similar analysis of reaction products from competitive RT-PCR allowed the visualization of two reaction products from rabbit endometrium. Sequence analysis showed the larger band of 939-bp amplicons contains exon 12. Sequence analysis of the smaller band of 741-bp amplicons confirmed the exon-skip event. The presence of two ATP11B isoforms in rabbit endometrium prompted careful analysis of the region that binds the RING domain of RUSH.
Figure 3.
RT-PCR confirms the ATP11B gene is alternatively spliced in rabbit endometrium. Lanes 1 and 3, Lambda DNA/EcoR I + Hind III markers (bp). Lane 2, the single. 939-bp product from Ishikawa cells. Lane 4, two populations of ATP11B amplicons from a single competitive PCR reaction, i.e. the 939-bp, full-length product whose sequence matches that of the human, and the 741-bp, abbreviated (RFBP) product resulting from the exon-skip event.
3.3 Pull-down assays
The original RFBP cDNA clone that bound the RUSH RING-finger contained the C-terminal 496 amino acids of the protein (4). This region was reduced to 194 amino acids that comprise much of the large loop and eliminated the transmembrane domains. Subregions were isolated by PCR and directionally subcloned into the pGEX-2TK expression vector. GST-fusion protein was screened with a pull-down assay to determine which peptide sequences associated with the MBP-RING1 fusion protein. Amino acids 612–804 were subdivided into overlapping, unequal regions by a process of elimination based on empirical evidence that confirmed either positive or negative binding (Figure 2). The search converged to reveal a small segment (K*-V-I-R*-L-I-K*-I-S) of Core Region G (amino acids 732–740) that is capable of binding MBP-RING1.
3.4 Characterization of the binding site in RFBP
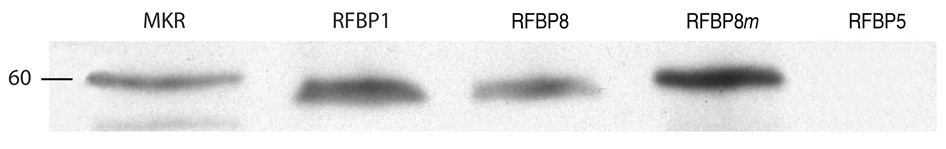
Replacement analysis of the QKAKVIRLIKISPEK peptide revealed binding (values of 250–600), albeit low, to MBP-RING1. In these peptide arrays changing one residue did not eliminate binding. Replacement analysis also showed that the charged residues (*) that comprise an amphipathic alpha helix are required for binding. Changing the residue at position 737 from I, which is hydrophobic, to D, which is charged, improved binding (value of 2423). This was confirmed with pull-down assays (Figure 4) in which binding of MBP-RING1 to RFBP8m (KVIRLDKIS) was greater than binding to RFBP8 (KVIRLIKIS).
Figure 4.
Western analysis confirms enhanced binding of KVIRLDKIS to MBP-RING1. Left lane, molecular size marker (60 kDa). Binding of MBP-RING1 to RFBP1 (second lane) and RFBP8 (KVIRLIKIS, third lane) is comparable. Binding of MBP-RING1 to RFBP8m (KVIRLDKIS, fourth lane) is increased 2–4-fold. Negligible binding of MBP-RING1 to RFBP5 (fifth lane) is provided as a negative result (please see Figure 2). All binding comparisons were determined by computer-assisted image analysis as described.
A total of 104 blast hits were found with a computer search of sequence databases with the BLASTP 2.2.17 program (14) with KVIRLIKIS as the query. Importantly, this sequence is 100% identical in ATP11B proteins from a variety of mammals including humans, long-tailed macaque, rats, mice, dogs, horses and cattle. In contrast, when the search was performed with KVIRLDKIS as the query, the D substitution was highly conserved in protists.
3.5 Characterization of the binding site in the RING-finger
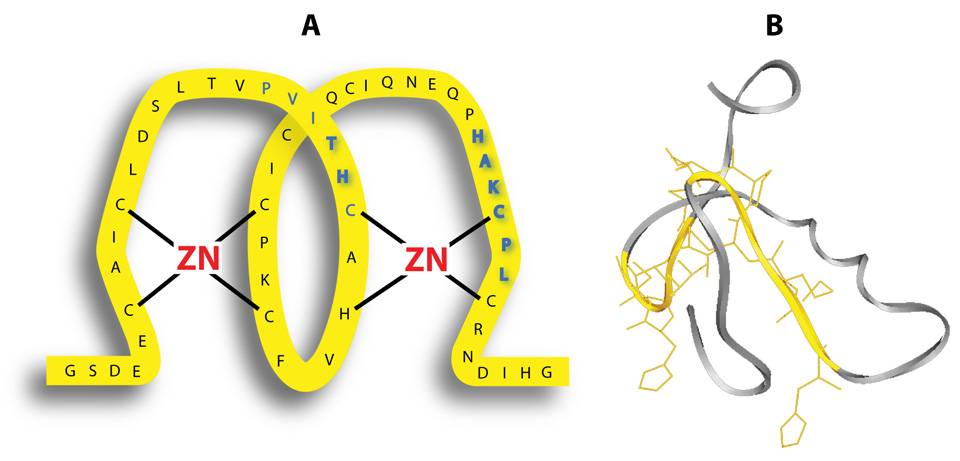
A computer search with the BLASTP 2.2.17 program (14) showed the RING-domain of RUSH is 100% identical in the alpha and beta isoforms in the rabbit, and 96% identical to the human ortholog, HLTF. To identify what was likely to be a highly conserved binding domain, the RING-finger of RUSH was subdivided (Figure 1) into 7 regions that were used in pull-down assays. These assays provided strong evidence for a discontinuous binding site (Figure 5A; bold blue) when they revealed the PVITHC (amino acids 767–772) and HAKCPL (amino acids 791–796) residues are required for binding.
Figure 5.
The RING-domain in RUSH is the protein interaction site. Panel A. Schematic representation of the cross-brace zinc ligation in the RING domain of RUSH shows the binding site (PVITHC – HAKCPL) in blue with an emphasis (bold blue) on the essential binding site (TH – HAKCPL). Panel B. The ribbon diagram generated with the ExPASy Proteomics Server of the Swiss Institute of Bioinformatics (SIB) emphasizes the relationship of the discontinuous binding site (yellow) to the central helix (gray).
Replacement analysis in which overlapping 15-mers covering amino acids LIKKMKLILSSGSDEECAICLDSLTVPVITHCAHVFCKPCICQCIQNEQPHAKCPLCRNDIH GDNLLECPP were probed with GST-tagged KVIRLIKIS failed to show binding. Peptides containing only one of the two regions of the putative binding site were unrecognized in the array format. However, validation of the binding site in RING was obtained by probing scaffolded RING peptides (30-mers) with GST-tagged KVIRLIKIS. The RING peptide PVITHC(StBu)AC(StBu)VFAKPAIAVAIQNEQPHAKC (StBu)PLC(StBu)R was tested such that PVITHC(StBu)AC(StBu)VFAKPAIC was ligated to all positional variants of AIQNEQPHAK C(StBu)PLC(StBu)R, and vice versa. Cysteines [C(StBu)] were covalently coupled to the scaffolds via four connection points to mimic zinc bridges. Only 2 of the 400 different peptide combinations exhibited significant binding (values > 100) after correction for background. Peptide QAIQNEQPHAKAPLC – AIALDSLTVPVITHZ (value 730) contains the binding regions in the correct orientation such that the link between the C and Z in the synthetic peptide is a mimic of a zinc-bridge in the native peptide. The second peptide CIQNEQPHAKAPLAR-THAAHVFAKPAIAQZ (value 688) where the C is coupled to Z, showed the amino acid sequence HAKAPL – TH is essential for binding. The substitution of any one amino acid produced a complete loss of binding activity (Table 2), and no binding was observed for either half-site alone. Thus all of the other non-binding combinations were in fact controls showing that this particular discontinuous peptide contains the essential binding sequence.
Table 2. Fine mapping with CLIPS-constrained peptides. Light gray boxes indicate charged residues. Values are corrected for background.
Nine single positional changes that eliminated RING-binding. The complex molecular architecture of the discontinuous binding site was maintained via fixation of linear peptides. Thus changes in binding are due to changes in individual amino acids not changes in the molecular scaffold. Candidates on this list were selected from fine mapping of CLIPS-constrained RING peptides.
| Consensus sequence | Value | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | V | I | T | H | C | ACVFAKPAIC | + | AIQNEQP | H | A | K | C | P | L | CR | |
| Changes that eliminated binding | ||||||||||||||||
| M | + | 3 | ||||||||||||||
| R | + | 2 | ||||||||||||||
| D | + | 1 | ||||||||||||||
| T | + | 0 | ||||||||||||||
| + | Q | −4 | ||||||||||||||
| S | + | −16 | ||||||||||||||
| + | P | −23 | ||||||||||||||
| K | + | −46 | ||||||||||||||
| P | + | −61 | ||||||||||||||
A total of 103 blast hits were found with a computer search of sequence databases with the BLASTP 2.2.17 program (14) at the National Center for Biotechnology Information and the PVITHC-HAKCPL binding site as query. Importantly, the components of the discontinuous binding site are 100% conserved in all HLTF orthologs.
4. Discussion
RUSH is structurally complex. It is a SWI/SNF-family member because it has seven consecutive motifs that are characteristic of ATPases and DNA helicases. Human HLTF binds to the SPH repeats of the SV40 enhancer and to the HIV-1 promoter (15). It displays strong ATPase activity. It is a common target for methylation and epigenetic gene silencing in colon cancer (16), and a candidate colon cancer suppressor gene. Rabbit RUSH-1α mediates the ability of prolactin to augment progesterone-dependent transcriptional activation of the uteroglobin gene through direct binding to the proximal promoter (13). In its own promoter, DNA-bound RUSH interacts physically with liganded Egr-1 via a DNA-looping mechanism, to mediate repression by c-Rel (2). RUSH is also grouped with hundreds of unrelated RING-finger proteins based on the common possession of a C3HC4 type RING with the consensus sequence C-X2-Cys-X9–39-C-X1–3-H-X2–3-C-X2-C-X4–48-C-X2-C in which X is any amino acid.
Zinc-finger proteins are the most abundant protein superfamily in the mammalian genome (17). They have an above average rate of alternative splicing that may have evolved in a modular fashion (18). If domains are encoded by individual exons, the out come from any single exon exclusion/retention event could be a functionally different isoform with an altered subcellular address. One example is the RIKEN transcript C330026E23Rik, which encodes a protein with a C-terminal C2H2-type finger and an N-terminal KRAB-repressor domain. The two isoforms that encode the C2H2 finger but lack the KRAB domain may compete with full-length wild-type protein to relieve repression (17).
Zinc finger domains are usually encoded by a single exon (17). However, the RING-domain of RUSH and the discontinuous-binding site (Figure 5A), are encoded by two exons. The PVITHC component of the site from exon 20 is located between C2–C3 where the spacing is 9–39 amino acids. The HAKCPL component of the site from exon 21 is located between C5–6 where the spacing is 4–48 amino acids. Thus the two modules of this protein interaction site are located in regions of the RING that have the greatest potential for structural and therefore functional diversity (19). Moreover, a single exon exclusion event would eliminate half of the binding site. Based on the experimental outcomes with the CLIPS™ technology an exon exclusion event would eliminate one-half of the site and all of the binding potential. However, Partek GS analysis of the interspecies microarray data files showed that two of the 20 most significant events (Table 1) occurred in this gene (locus 6596), but not in exons 20 and 21. This direct comparison of splicing events in the RUSH gene in rabbits with the HLTF gene in humans shows there is high structural stability in the zinc-binding domains and in the protein interaction site. This conclusion was extended to all other species with a computer search of sequence databases. All non-redundant GenBank CDS translations + PDB + SwissProt + PIR + PRF excluding environmental samples from WGS project were aligned with the BLASTP 2.2.17 program (14) at the National Center for Biotechnology Information and the PVITHC-HAKCPL binding site as query. This search showed that all of the components of the binding site are 100% conserved in all HLTF orthologs regardless of the diversity in the intervening sequence. In comparison, the RING domain is only 45% identical to the BRCA1 gene product. Only the PV-T-C----CPL components of the binding site intact. Based on the binding analysis with the CLIPS™ technology, this site is predicted to be nonfunctional.
According to the ribbon diagram (Figure 5B), the two components of the binding site (yellow) are located on either side of the well-defined central helix (gray) that is composed of two turns. However, the BLASTP 2.2.17 analysis (14) shows this region is only conserved at the level of the zinc binding ligands in the BRCA1 gene. All but two of the ten remaining amino acids have been substituted. Generally, RING proteins share very little sequence homology outside of the consensus RING. The BLASTP alignment shows that the RING motif in RUSH/HLTF is very different from the RING motif in BRCA1. This comparison is important because mutations in the RING domain of the BRCA1 gene predispose individuals to breast and ovarian cancer (20).
More is known about the effects of alternative splicing on the functional diversity of plasma membrane calcium-ATPases (21) than Type IV P-type ATPases, The loss or malfunction of specific calcium pump isoforms is associated with heart failure, ataxia, and deafness (22). This current study confirms that alternative splicing is responsible for species differences in the expression of ATP11B. The splice array data support the conclusion that RFPB results from an exon deletion event that is unique to rabbit compared with human. The function of this species-specific alternatively spliced product must be further examined as it is generally accepted that non-conserved alternative splicing events are less likely to generate functional proteins than the conversed events (23). However, in a search for evolutionarily conserved exon skip events Yeo et al (24) found that only one out of every 18 events in the human EST database was conserved in mouse. Collectively these studies support an initiative to identify and characterize exons whose exclusion or inclusion is subject to evolutionary conservation.
In humans and rabbits the catalytic loop of the ATP11B isoforms, which contains consensus sequences for a phosphorylation site and an ATP binding site, is encoded by exons 12–22. The highly conserved phosphorylation site (DKTGTLT) in core region E is located in exon 13. The highly conserved ATP binding site is located in exon 19. In contrast, the KVIRLIKIS binding site straddles two exons. The K in position 732 is the last amino acid in exon 20. The V in position 733 is the first amino acid in exon 21. However, based on the splice arrays, there is no evidence in either human or rabbit that the binding site is altered by an exon skip event. This direct comparison of splicing events in the ATP11B gene between human and rabbit shows there is high structural stability in the protein interaction site.
This conclusion was extended to other species with a computer search of non-redundant sequence databases with the BLASTP 2.2.17 program (14) with KVIRLIKIS as the query. Importantly, this sequence is 100% identical in ATP11B proteins from a variety of mammals. In contrast, when the search was performed with KVIRLDKIS as the query, the D substitution was highly conserved in protists. This finding supports the speculation that evolution in the ATP11B protein has occurred by point mutation, and not in a modular fashion.
Cross-species splicing studies are the only way to identify functionally important alternative splicing events. The easiest comparisons are between human and mouse lineages because arrays are available. Humans and mice diverged 96–110 million years ago, enough time for mutations to accumulate and functional sequences to be conserved because of selection pressure. More comparative genomic studies will be possible because of the NIH-funded initiative to expand the current genome coverage of mammals by sequencing the whole-genome of 24 additional mammals including rabbit. For now, this is the first demonstration that functional domains in RUSH and ATP11B isoforms are highly conserved in humans and rabbits which diverged 90–110 million years ago (25, 26).
Acknowledgements
We thank Larry Starr, Medical Photography, TTUHSC, for artwork; and the NIH (HD29457) for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayward-Lester A, Hewetson A, Beale EG, Oefner PJ, Doris PA, Chilton BS. Cloning, characterization and steroid-dependent posttranscriptional processing of RUSH-1α and β two uteroglobin promoter-binding proteins. Mol. Endocrinol. 1996;10:1335–1349. doi: 10.1210/mend.10.11.8923460. [DOI] [PubMed] [Google Scholar]
- 2.Hewetson A, Chilton BS. Progesterone-dependent DNA looping between RUSH/SMARCA3 and Egr-1 mediates repression by c-Rel. Mol. Endocrinol. 2008 doi: 10.1210/me.2007-0432. (Epub ahead of print Jan 3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li WI, Chen CL, Chou JY. Characterization of a temperature-sensitive β-endorphin-secreting transformed endometrial cell line. Endocrinology. 1989;125:2862–2867. doi: 10.1210/endo-125-6-2862. [DOI] [PubMed] [Google Scholar]
- 4.Mansharamani M, Hewetson A, Chilton BS. Cloning and characterization of an atypical type IV P-type ATPase that binds to the RING motif of RUSH transcription factors. J. Biol. Chem. 2001;276:3641–3649. doi: 10.1074/jbc.M004231200. [DOI] [PubMed] [Google Scholar]
- 5.Halleck MS, Schlegel RA, Williamson PL. Reanalysis of ATP11B, a type IC P-type ATPase. J. Biol. Chem. 2002;277:9736–9740. doi: 10.1074/jbc.M200240200. [DOI] [PubMed] [Google Scholar]
- 6.Timmerman P, Beld J, Puikj WC, Meloen RH. Rapid and quantitative cyclization of multiple peptide loops onto synthetic scaffolds for structural mimicry of protein surfaces. ChemBioChem. 2005;6:821–824. doi: 10.1002/cbic.200400374. [DOI] [PubMed] [Google Scholar]
- 7.Timmerman P, Wouter CP, Meloen RH. Functional reconstruction and synthetic mimicry of a conformational epitope using CLIPS technology. J. Mol. Recognit. 2007;20:283–299. doi: 10.1002/jmr.846. [DOI] [PubMed] [Google Scholar]
- 8.Han JH, Stratowa C, Rutter WJ. Isolation of full-length putative rat lysophospholipase cDNA using improved methods for mRNA isolation and cDNA cloning. Biochemistry. 1987;26:1617–1625. doi: 10.1021/bi00380a020. [DOI] [PubMed] [Google Scholar]
- 9.Hewetson A, Moore SL, Chilton BS. Prolactin signals through RUSH/SMARCA3 in the absence of a physical association with Stat5a. Biol. Reprod. 2004;71:1907–1912. doi: 10.1095/biolreprod.104.031435. [DOI] [PubMed] [Google Scholar]
- 10.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Hewetson A, Chilton BS. An Sp1-NF-Y/progesterone receptor DNA binding-dependent mechanism regulates progesterone-induced transcriptional activation of the rabbit RUSH/SMARCA3 gene. J. Biol. Chem. 2003;278:40177–40185. doi: 10.1074/jbc.M303921200. [DOI] [PubMed] [Google Scholar]
- 13.Hewetson A, Hendrix EC, Mansharamani M, Lee VH, Chilton BS. Identification of the RUSH consensus-binding site by cyclic amplification and selection of targets: demonstration that RUSH mediates the ability of prolactin to augment progesterone-dependent gene expression. Mol. Endocrinol. 2002;16:2101–2112. doi: 10.1210/me.2002-0064. [DOI] [PubMed] [Google Scholar]
- 14.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheridan PL, Schorpp M, Voz ML, Jones KA. Cloning of an SNF2/SWI2-related protein that binds specifically to the SPH motifs of the SV40 enhancer and to the HIV-1 promoter. J. Biol. Chem. 1995;270:4575–4587. doi: 10.1074/jbc.270.9.4575. [DOI] [PubMed] [Google Scholar]
- 16.Moinova HR, Chen WD, Shen L, Smiraglia D, Olechnowicz J, Ravi L, Kasturi L, Myeroff L, Plass C, Parsons R, Minna J, Wilson JK, Green SB, Issa JP, Markowitz SD. HLTF gene silencing in human colon cancer. Proc. Natl. Acad. Sci. USA. 2002;99:4562–4567. doi: 10.1073/pnas.062459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravasi T, Huber T, Zavolan M, Forrest A, Gaasterland T, Grimmond S, Hume DA RIKEN GER Group, GSL Members. Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome. Genome Res. 2003;6B:1430–1442. doi: 10.1101/gr.949803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgenstern B, Atchley WR. Evolution of bHLH transcription factors: modular evolution by domain shuffling? Mol. Biol. Evol. 1999;12:1654–1663. doi: 10.1093/oxfordjournals.molbev.a026079. [DOI] [PubMed] [Google Scholar]
- 19.Saurin AJ, Borden KLB, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem. Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 20.Brzovic PS, Rajagopal P, Hoyt DW, King M-C, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 21.Stauffer TP, Hilkifer H, Carafoli E, Strehler EE. Quantitative analysis of alternative splicing options of human plasma membrane calcium pump genes. J. Biol. Chem. 1994;268:25993–26003. [PubMed] [Google Scholar]
- 22.Strehler EE, Treiman M. Calcium pumps of plasma membrane and cell interior. Curr. Mol. Med. 2004;4:323–335. doi: 10.2174/1566524043360735. [DOI] [PubMed] [Google Scholar]
- 23.Kan Z, Garrett-Engele PW, Johnson JM, Castle JC. Evolutionarily conserved and diverged alternative splicing events show different expression and functional profiles. Nucleic Acids Res. 2005;33:5659–5666. doi: 10.1093/nar/gki834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 26.Su C, Nei M. Fifty-million-year-old polymorphism at an immunoglobulin variable region gene locus in the rabbit evolutionary lineage. Proc. Natl. Acad. Sci. 1999;96:9710–9715. doi: 10.1073/pnas.96.17.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]