Abstract
Mutations in the dystrophin gene cause Duchenne muscular dystrophy and result in the loss of dystrophin and the entire dystrophin–glycoprotein complex (DGC) from the sarcolemma. We show that sarcospan (SSPN), a unique tetraspanin-like component of the DGC, ameliorates muscular dystrophy in dystrophin-deficient mdx mice. SSPN stabilizes the sarcolemma by increasing levels of the utrophin–glycoprotein complex (UGC) at the extrasynaptic membrane to compensate for the loss of dystrophin. Utrophin is normally restricted to the neuromuscular junction, where it replaces dystrophin to form a functionally analogous complex. SSPN directly interacts with the UGC and functions to stabilize utrophin protein without increasing utrophin transcription. These findings reveal the importance of protein stability in the prevention of muscular dystrophy and may impact the future design of therapeutics for muscular dystrophies.
Introduction
The dystrophin–glycoprotein complex (DGC) is composed of integral and peripheral membrane proteins that span the plasma membrane and connect the extracellular matrix with the intracellular actin cytoskeleton (Campbell and Kahl, 1989; Ervasti et al., 1990, 1991; Yoshida and Ozawa, 1990; Ervasti and Campbell, 1991, 1993). In skeletal muscle, the DGC provides mechanical stability to the sarcolemma during contraction (Petrof et al., 1993). Mutations in the dystrophin gene are responsible for X-linked Duchenne muscular dystrophy (DMD), which is characterized by progressive wasting of skeletal muscles eventually resulting in cardiac and respiratory failure (for review see Durbeej and Campbell, 2002). In DMD patients, loss of dystrophin results in the absence of the entire DGC complex, leading to severe membrane damage and muscle degeneration (for review see Durbeej and Campbell, 2002). mdx mice, which are an established model for DMD, possess a genetic mutation in exon 23 of the murine dystrophin gene, resulting in loss of dystrophin protein. As a result, the entire DGC is also absent from the sarcolemma, likely because of rapid protein degradation in the absence of a fully assembled complex. Muscles from mdx mice are pathologically similar to DMD patients and display marked membrane disruption as a result of sarcolemmal instability. Akt signaling is hyperactivated in muscles from DMD patients and mdx mice (Peter and Crosbie, 2006), suggesting that the DGC may also play a role in cellular signaling in addition to its role in mechanical stability of the sarcolemma (Judge et al., 2006).
The transmembrane proteins of the DGC serve as important anchorages for the peripheral membrane DGC components. These integral membrane proteins include sarcospan (SSPN), the sarcoglycans (SGs; α-, β-, γ-, and δ-SG), and β-dystroglycan (DG; for review see Michele and Campbell, 2003). The SGs and β-DG are single-pass transmembrane glycoproteins. Dystrophin, an actin-binding protein, is localized adjacent to the sarcolemma by attachment to the intracellular N terminus of β-DG (for review see Michele and Campbell, 2003). On the extracellular face of the membrane, β-DG interacts with α-DG to form a receptor for ligands in the extracellular matrix (Ervasti and Campbell, 1993). The SGs form a tight subcomplex with SSPN (Crosbie et al., 1999; Miller et al., 2007). Together, the SG–SSPN subcomplex functions to anchor α-DG attachment to the sarcolemma (Holt and Campbell, 1998). As a whole, the DGC provides a physical linkage across the sarcolemma between the extracellular matrix and the intracellular actin cytoskeleton protecting the membrane from contraction-induced damage (for review see Barresi and Campbell, 2006).
It is well established that stable interactions among the integral membrane proteins are critical for DGC function and prevention of muscular dystrophy (for review see Durbeej and Campbell, 2002). Despite their importance, the factors that determine the structural integrity of the DGC are not well understood. The observation that SSPN possesses some sequence homology to the tetraspanin superfamily of proteins raises the possibility that SSPN may serve an important role in mediating and stabilizing protein interactions within the DGC (Crosbie et al., 1997, 1998, 1999). The tetraspanins each possess four transmembrane domains and function to cluster and organize transmembrane protein complexes, thereby controlling a wide range of cellular functions (for reviews see Hemler, 2003; Levy and Shoham, 2005). Using a site-directed mutagenesis approach, we have demonstrated that SSPN exhibits the structural characteristics that define the tetraspanin superfamily of proteins (Miller et al., 2007).
As a first test of SSPN function, we generated SSPN transgenic (SSPN transgene [Tg]) mice with moderate (10-fold) levels of SSPN protein overexpression in skeletal muscle (Peter et al., 2007). Forced elevation of SSPN caused a concomitant increase in DGC protein expression but did not disrupt localization of the complex to the sarcolemma. We found that overexpression of exogenous SSPN dramatically reduced endogenous SSPN to levels that were barely detectable, suggesting that SSPN expression is tightly regulated. 10-fold elevation of SSPN disrupted normal interactions within the SG–SSPN subcomplex, which, in turn, weakened α-DG attachment to the sarcolemma (Peter et al., 2007). As a result, assembly of the extracellular matrix was disrupted, giving rise to severe congenital muscular dystrophy in mice with moderate levels of SSPN overexpression (Peter et al., 2007). Furthermore, membrane instability was not detected in 10-fold SSPN-Tg mice, demonstrating that pathogenetic mechanisms resulting from SSPN overexpression are distinct from dystrophin deficiency. Despite our exhaustive efforts, we were never able to isolate free, unassociated SSPN in 10-fold SSPN-Tg muscle, which strongly supports our conclusion that SSPN's toxicity is directly related to its association with other molecules within the sarcolemma.
SSPN-Tg mice with low levels of SSPN overexpression (two- to threefold) were also characterized (Peter et al., 2007). In contrast to SSPN-Tg mice with 10-fold levels of overexpression, SSPN-Tg mice with low levels of SSPN overexpression did not exhibit detectable signs of muscle pathology. DGC levels were mildly up-regulated in these mice, but interactions with the extracellular matrix were not adversely affected. Endogenous SSPN was not affected by this low level of exogenous SSPN expression. Collectively, these findings lead us to conclude that SSPN levels control DGC structure and function. Furthermore, this work leads to the hypothesis that SSPN may function to orchestrate assembly and stability of the DGC.
Results and discussion
Our previous work has shown that SSPN functions as a tetraspanin to coordinate protein interactions within the DGC (Peter et al., 2007), which led us to rationalize that SSPN expression in dystrophin-deficient muscle might anchor the transmembrane components of the DGC within the sarcolemma. To test this hypothesis, we engineered transgenic mice to overexpress SSPN on the dystrophin-deficient mdx background (SSPN-Tg:mdx). The mdx phenotype is inherited as an X-linked recessive trait, which stems from a premature stop codon in the dystrophin gene, leading to complete absence of the dystrophin protein. The DGC is absent from the sarcolemma of mdx muscle likely because of premature degradation of the protein components in the absence of a fully assembled complex (Ohlendieck and Campbell, 1991). For these experiments, we chose to use low (two- to threefold) SSPN-transgenic (SSPN-Tg) mice, lines 31.6 and 29.1, which displayed normal muscle morphology (Peter et al., 2007). SSPN-Tg mice were engineered to carry the human SSPN (hSSPN) gene under control of the human skeletal muscle α-actin promoter, which limited SSPN protein expression to skeletal muscles (Peter et al., 2007). Although human and mouse SSPN proteins are >90% identical at the amino acid level, antibodies specific to human and mouse SSPN permitted us to distinguish between exogenous (human) and endogenous (mouse) SSPN. SSPN-Tg males were crossed with mdx females to generate dystrophin-deficient mice carrying the SSPN Tg (SSPN-Tg:mdx).
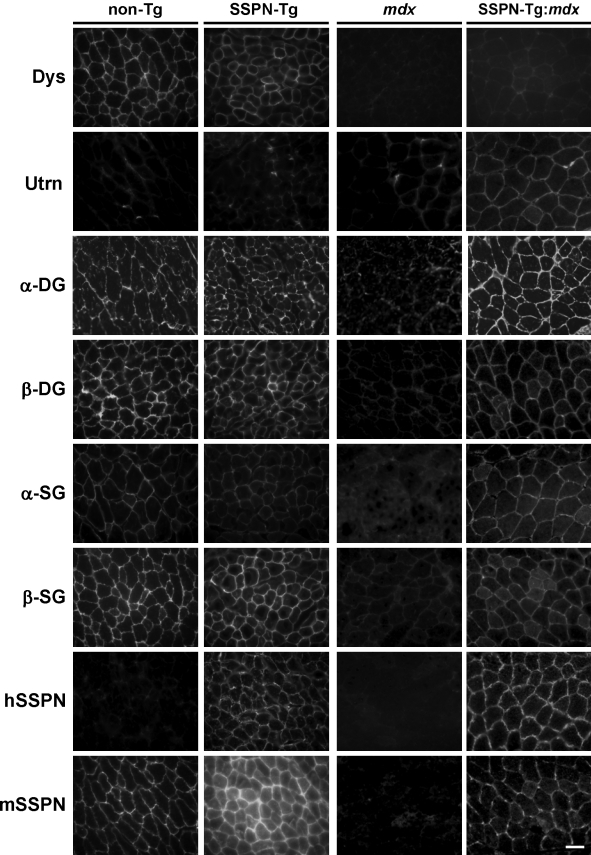
We first investigated whether SSPN overexpression affected membrane association of DGC components in dystrophin-deficient muscle. Indirect immunofluorescence was performed on transverse cryosections of SSPN-Tg:mdx quadriceps muscle to determine the subcellular localization of the other DGC components. In addition to the core components of the DGC, we also analyzed the localization of utrophin, a protein closely related to dystrophin. At the postsynaptic region of the neuromuscular junction in wild-type and mdx muscle, utrophin replaces dystrophin to form an otherwise identical utrophin–glycoprotein complex (UGC; Matsumura et al., 1992). Analysis of wild-type non-Tg and SSPN-Tg muscle served as positive controls, and mdx muscle was used as a negative control. As shown in Fig. 1, DGC expression was robust in positive controls (wild-type non-Tg and SSPN-Tg) but was not detected in negative control samples (mdx and SSPN-Tg:mdx). As expected, staining for exogenous SSPN (hSSPN) was only detected in SSPN-Tg muscle, and utrophin expression was restricted to the neuromuscular junction in wild-type non-Tg and SSPN-Tg muscles (Fig. 1). In agreement with previous results (Matsumura et al., 1992), we found that utrophin expression was slightly up-regulated in mdx muscle at the extrasynaptic sarcolemma. Surprisingly, we found that overexpression of SSPN in mdx muscle broadened the localization of utrophin protein so that it was no longer restricted to the neuromuscular junction but was present throughout the sarcolemma (Fig. 1). Grady et al. (2000) have demonstrated that expression and localization of the UGC is important for proper synaptic formation and clustering of acetylcholine receptors at the postsynaptic membrane. Introduction of utrophin in dystrophin-deficient muscle broadens localization of the UGC to the extrasynaptic sarcolemma, where it ameliorates muscular dystrophy (Tinsley et al., 1996, 1998). The observation that utrophin is localized to the extrasynaptic sarcolemma in SSPN-Tg:mdx muscle raises the possibility that utrophin displays elevated protein levels leading to the observed increased sarcolemmal localization and possible functional replacement of the DGC. We also found that SSPN overexpression in mdx muscle increased α-/β-DG as well as α-/β-SG levels at the sarcolemma (Fig. 1). These data suggest that SSPN has restored expression of a complete UGC at the sarcolemma in mdx muscle, where it may functionally compensate for the loss of dystrophin and its associated proteins.
Figure 1.
SSPN stabilizes the UGC at the sarcolemma. Transverse cryosections of quadriceps muscle from non-Tg (wild type), SSPN-Tg (wild type), mdx, and SSPN-Tg:mdx mice were stained with the antibodies to dystrophin (Dys), utrophin (Utrn), DGs (α- and β-DG), mouse SSPN (mSSPN), transgenic hSSPN, and SGs (α- and β-SG). Protein staining was visualized by indirect immunofluorescence. Bar, 100 μm.
To determine how SSPN expression affects UGC protein levels, skeletal muscle lysates from 6-wk-old mdx and SSPN-Tg:mdx mice were analyzed by immunoblotting with antibodies to each of the UGC components. To confirm expression of the Tg, immunoblots were probed with antibodies specific to hSSPN. Exogenous SSPN was abundant in SSPN-Tg:mdx muscle samples, demonstrating that stable SSPN protein was produced from the Tg (Fig. 2 A). Levels of α- and β-DG as well as α-, β-, and γ-SG were dramatically increased in SSPN-Tg:mdx mice relative to mdx controls (Fig. 2 A). DG and SG protein expression in SSPN-Tg:mdx muscle was identical to age-matched non-Tg (wild type) muscle (Fig. 2 A and not depicted). As expected, dystrophin protein was not detected in samples from either mdx or SSPN-Tg:mdx mice. Utrophin protein levels were elevated in SSPN-Tg:mdx muscle relative to mdx muscle (Fig. 2 A). To investigate whether SSPN increased utrophin expression by enhancing transcription, we performed quantitative RT-PCR analysis. Utrophin mRNA levels were similar in SSPN-Tg:mdx and mdx muscle (Fig. 2 B). This finding demonstrates that utrophin up-regulation by SSPN does not involve transcriptional regulation. Furthermore, these data suggest that SSPN maintains utrophin protein at the sarcolemma by providing a tetraspanin scaffold, which stabilizes and promotes protein interactions.
Figure 2.
SSPN regulates UGC levels by interacting directly with this complex. (A) Skeletal muscle from mdx and SSPN-Tg:mdx tissue was solubilized in modified radioimmunoprecipitation assay buffer, and 60-μg protein samples were resolved by SDS-PAGE and immunoblotted using antibodies against dystrophin (Dys), utrophin (Utrn), DGs (α- and β-DG), exogenous SSPN (hSSPN), and SGs (α-, β-, and γ-SG). As expected, exogenous hSSPN was only detected in SSPN-Tg:mdx muscle. Expression levels of DGs and SGs were dramatically elevated in SSPN-Tg:mdx muscle samples. Utrophin, a homologue of dystrophin, is up-regulated in SSPN-Tg:mdx tissue. Equal loading of mdx and SSPN-Tg:mdx lysates was confirmed by Coomassie blue (CB) staining. Molecular masses of individual proteins are indicated in parentheses. (B) Quantitative RT-PCR analysis was used to determine the level of utrophin mRNA in both SSPN-Tg:mdx and mdx tissue. Data are normalized to GAPDH controls and are represented relative to control values (100%). Utrophin mRNA levels were not statistically different (standard t test; P = 0.258) between SSPN-Tg:mdx and mdx controls, suggesting that SSPN-Tg expression did not affect utrophin transcription. Values represent mean ± SEM (error bars). (C) SSPN is a core component of the UGC in SSPN-Tg:mdx mice. Purified UGC proteins were separated by ultracentrifugation through 5–30% sucrose gradients. Fraction 1 represents the lightest region of the gradient. Fractions were analyzed by immunoblotting with antibodies to utrophin (Utrn), DGs (α- and β-DG), SGs (α-, and γ-SG), and exogenous SSPN (hSSPN) as indicated. SSPN and utrophin are enriched in fraction 4, demonstrating that SSPN is a core component of the UGC. The molecular mass of each component is identical to that indicated in A.
We established that SSPN is a core component of the DGC using rigorous biochemical methods (Crosbie et al., 1997, 1998). In normal skeletal muscle, SSPN is enriched at the postsynaptic region of the neuromuscular junction in a utrophin-dependent manner (Crosbie et al., 1999). Our findings that SSPN expression alters utrophin localization and, conversely, that SSPN enrichment at the neuromuscular junction is dependent on utrophin suggest that SSPN functions to regulate utrophin through direct association with the UGC. To biochemically determine whether SSPN is a core component of the UGC, we isolated this complex from skeletal muscle of SSPN-Tg:mdx mice using lectin affinity chromatography followed by ultracentrifugation on 5–30% sucrose gradients. Only proteins that are tightly associated will be maintained as a complex and migrate together during ultracentrifugation (Crosbie et al., 1997, 1998). Fractions were collected and analyzed by immunoblotting with antibodies to components of the UGC. We found that SSPN protein is highly enriched in fraction 4, which contains utrophin and utrophin-associated proteins (Fig. 2 C). We conclude that SSPN regulates utrophin levels in a mechanism that involves direct association of SSPN with the UGC.
To investigate the functional consequences of increased UGC levels in dystrophin-deficient muscle, we examined SSPN-Tg:mdx muscle for dystrophic pathology. mdx pathology is characterized by progressive muscle degeneration, compensatory hypertrophy, and necrosis of damaged muscle fibers. Histological analysis was performed by staining transverse cryosections of quadriceps muscle with hematoxylin and eosin (H&E). Images of whole quadriceps muscle taken from mdx mice revealed numerous patches of necrosis, which were not present in SSPN-Tg:mdx muscle (Fig. 3 A). Central nucleation, a marker of myofiber regeneration, was quantitated by analyzing H&E-stained quadriceps muscle at higher magnification (Fig. 3 B). mdx muscles at 6 wk of age display elevated levels (60%) of regenerated myofibers with central nuclei (Fig. 4 A). We found a dramatic (40%) reduction in central nucleation in muscle from SSPN-Tg:mdx mice (Fig. 4 A). Although the level of centrally nucleated fibers in SSPN-Tg:mdx mice remained slightly elevated compared with non-Tg controls, this is likely explained by fiber to fiber variation in promoter activation (unpublished data). In addition, expression of SSPN in mdx mice was associated with a reduction in the number of smaller, atrophic muscle fibers (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200808027/DC1). This reduction of small, newly regenerating fibers provides an additional measure of muscle pathology and is similar to the results reported for microdystrophin (Gregorevic et al., 2006).
Figure 3.
SSPN-Tg:mdx mice display near normal muscle pathology. (A) Transverse cryosections of whole quadriceps muscle from age-matched mdx and SSPN-Tg:mdx mice were stained with H&E to visualize muscle pathology. Many necrotic patches are visible in mdx quadriceps. Necrotic fibers were never observed in SSPN-Tg:mdx quadriceps. Boxed regions indicated on H&E staining of quadriceps muscle from mdx and SSPN-Tg:mdx mice are shown at higher magnifications in B. Note the presence of numerous myofibers with central nucleation as well as the variation in fiber size evident in mdx tissue. Muscle from SSPN-Tg:mdx muscle displays reduced central nucleation and improvements in fiber size variation. Bar, 100 μm.
Figure 4.
SSPN improves muscle degeneration and sarcolemmal stability. (A) Central nucleation (percent of total fibers) was quantified for quadriceps isolated from mdx and SSPN-Tg:mdx (n = 4). SSPN-Tg:mdx mice display a threefold reduction in central nuclei compared with mdx age-matched controls. Each value represents mean ± SEM (error bars) of the total quadriceps analyzed (*, P = 6.0 × 10−4). (B) To examine infiltration of blood serum proteins into damaged muscle fibers, mdx and SSPN-Tg:mdx mice were intraperitoneally injected with Evans blue dye, a marker for membrane instability. mdx quadriceps displays many Evans blue dye–positive fibers (visualized by red fluorescence), which is a marker for membrane damage. Evans blue dye was not detected in muscle from SSPN-Tg:mdx mice, demonstrating that SSPN expression restored membrane stability in dystrophin-deficient muscle. Bar, 20 μm.
Loss of the DGC from the sarcolemma causes membrane instability, which is the primary defect of dystrophin-deficient muscular dystrophy (Weller et al., 1990; Menke and Jockusch, 1991; Petrof et al., 1993; Straub et al., 1997). Unrepaired tears in the sarcolemma allow proteins that are normally restricted to the blood serum to freely diffuse across the sarcolemma and accumulate in the sarcoplasm. To further test the functionality of the UGC in SSPN-Tg:mdx mice, we performed a tracer assay with fluorescent Evans blue dye that binds albumin in blood serum (Straub et al., 1997). mdx mice displayed severe sarcolemmal fragility marked by Evans blue dye accumulation in numerous muscle fibers (Fig. 4 B). In contrast, we never observed Evans blue–positive fibers in SSPN-Tg:mdx tissue (Fig. 4 B).
In this study, we provide biochemical evidence that SSPN ameliorates dystrophic pathology in mdx mice by stabilizing the UGC at the sarcolemma through direct interaction with this complex. We demonstrate that SSPN causes redistribution of the UGC to the extrasynaptic sarcolemma, where it functionally replaces the DGC. The primary defect in dystrophin-deficient mice is the loss of sarcolemma integrity, and we demonstrate that SSPN provides stability to the sarcolemma. These data suggest that the actin-UGC–extracellular matrix connection is fully and functionally restored. Future studies will be performed to determine whether SSPN overexpression improves muscle strength in mdx mice.
Tinsley et al. (1996, 1998) have shown that increased expression of utrophin in mdx mice rescues muscular dystrophy by stabilizing the UGC at the sarcolemma in a dose-dependent manner. Complete reduction of mdx pathology to normal wild-type levels (as determined by central nucleation and muscle strength measurements) occurred when utrophin was overexpressed at twice the levels of mdx mice (note that endogenous utrophin levels are already up-regulated in mdx muscle compared with wild-type controls). Their data nicely show that pathology was directly correlated with the level of utrophin overexpression. Lower levels of utrophin expression reduced the pathology, but not completely back to normal levels. Based on these findings, we speculate that increasing the level of SSPN overexpression would have an even greater impact on improving mdx pathology.
Overexpression of DGC proteins, signaling molecules, and compensatory molecules can improve the pathology of muscle fibers in the mdx mouse. Utrophin, dystrophin, integrin, neuronal nitric oxide synthase, and cytotoxic T cell N-acetylgalactosamine transferase have been shown to improve muscular dystrophy. Most of these efforts have been focused on the use of dystrophin and utrophin to restore normal muscle function in mdx mice. Elegant work from Chamberlain's group has established that full-length (DelloRusso et al., 2002) and internally truncated forms of dystrophin are highly effective at ameliorating mdx pathology in addition to increasing muscle strength (for review see Odom et al., 2007). SSPN is unique and offers numerous advantages over current therapeutic strategies. The SSPN cDNA is under 1 kb, which is well within the range for easy packaging into adeno-associated viral vectors for delivery, circumventing the necessity to generate recombinant forms of SSPN. Because SSPN is expressed in a variety of nonmuscle tissues, even in dystrophin deficiency, increasing the expression of SSPN in skeletal muscle should not pose an immune threat. One therapeutic angle for the treatment of DMD is to identify small molecules that up-regulate utrophin protein expression. Our findings now establish that SSPN regulates utrophin protein levels and localization within the myofiber.
The results in this study were unexpected because, for nearly 10 yr, the precise function of SSPN has remained elusive. Our experiments show that SSPN is a critical mediator of protein interactions within UGC and raise the possibility that SSPN contributes to the assembly and targeting of the UGC. This is impressive considering that SSPN, the smallest member of the complex, must orchestrate proper structural arrangement of over seven large peripheral and integral membrane proteins. These properties are unique to SSPN and suggest that gene therapeutic strategies targeting SSPN expression may be beneficial for the treatment of DMD.
Materials and methods
Generation of SSPN-Tg:mdx mice
Transgenic constructs were engineered with the human skeletal actin promoter and the VP1 intron upstream of hSSPN as described previously (Crawford et al., 2000; Spencer et al., 2002; Peter et al., 2007). SSPN transgenic males, line 29.1, were bred with mdx females (The Jackson Laboratory) to produce male SSPN-Tg:mdx mice. Female SSPN-Tg (wild type) and non-Tg (wild type) mice as well as male non-Tg:mdx littermates were used as controls. Mice were analyzed at 6 wk of age. Transgenic mice used for this study expressed an approximately twofold increase in SSPN protein expression in skeletal muscle compared with non-Tg (wild type) mice (Peter et al., 2007). All mice were maintained in the Life Sciences Vivarium, and all procedures were carried out in accordance with guidelines set by the University of California, Los Angeles Institutional Animal Care and Use Committee.
Immunofluorescence
Quadriceps from female SSPN-Tg (wild type), female non-Tg (wild type), male non-Tg:mdx, and SSPN-Tg:mdx littermates were dissected from 6-wk-old mice. Muscles were covered in 10.2% polyvinyl alcohol/4.3% polyethylene glycol and rapidly frozen in liquid nitrogen–cooled isopentane. Mounted muscle was stored at −80°C until analyzed. 8-μm transverse sections were prepared using a cryostat (CM 3050S; Leica) and stored on positively charged glass slides (VWR). Sections were stored at −80°C for future analysis. Sections were acclimated to RT for 15 min and blocked using 3% BSA diluted in PBS for 1 h at RT. Sections were then incubated at 4°C for 18 h with antibodies to the following proteins (antibody dilutions are indicated): dystrophin (MANDYS1, 1:10; Developmental Studies Hybridoma Bank), utrophin (NCL-DRP2, 1:5; Vector Laboratories), α-DG (IIH6C-4, 1:100; Upstate Signaling), β-DG (MANDAG2, 1:50; Developmental Studies Hybridoma Bank), α-SG (VP-A105, 1:100; Vector Laboratories), β-SG (VP-B206, 1:50; Vector Laboratories), hSSPN (affinity-purified rabbit 15, 1:25), and mouse SSPN (affinity-purified rabbit 18, 1:25). Polyclonal antibodies to endogenous (mouse) and exogenous (human) SSPN have been described previously (Peter et al., 2007). Primary antibodies were detected by FITC-conjugated anti–rabbit and anti–mouse (Jackson ImmunoResearch Laboratories and Millipore) secondary antibodies diluted at 1:500. Secondary antibodies were incubated for 1 h at RT. To preserve the fluorescence signal, sections were mounted in Vectashield (Vector Laboratories). To determine the level of nonspecific staining, secondary antibodies alone were incubated with sections. Mounted sections were visualized using a fluorescent microscope (Axioplan 2; Carl Zeiss, Inc.) equipped with a Plan Neofluar 40× NA 1.3 oil differential interference contrast objective, and images were captured using Axiovision 3.0 software (Carl Zeiss, Inc.).
Evans blue tracer assay
To establish sarcolemmal integrity, mice were injected with 50 μl of Evans blue dye (10 mg/ml in 10 mM of sterile phosphate buffer and 150 mM NaCl, pH 7.4) per 10 g of body weight as described previously (Straub et al., 1997). Peritoneal cavity injection was performed on 6-wk-old SSPN-Tg:mdx and non-Tg:mdx littermates. 24 h after injection, quadricepses were excised and mounted as described in the Immunofluorescence section. 8-μm transverse sections were briefly fixed in ice-cold acetone, washed in PBS (10 mM phosphate buffer and 150 mM NaCl, pH 7.4), and mounted with Vectashield. Evans blue–positive myofibers were observed using an Axioplan 2 fluorescent microscope and a Plan Neofluar 40× NA 1.3 oil differential interference contrast objective.
Histology
H&E staining was used for visualization of fibrosis, central nuclei, and fiber diameter. 8-μm transverse quadriceps sections were left at RT for 15 min before beginning the staining procedure. Sections were incubated with hematoxylin for 3 min, washed with water for 1 min, incubated with eosin for 3 min, and dehydrated in solutions of 70, 80, 90, and 100% ethanol. Sections were then incubated in xylene for a total of 6 min. Sections were left at RT briefly to dry before mounting with Permount. All supplies for the H&E staining were purchased from Thermo Fisher Scientific.
To measure the fiber area and count centrally nucleated fibers, digitized images were captured under identical conditions using an Axioplan 2 fluorescent microscope and Axiovision 3.0 software (Carl Zeiss, Inc.). The percentage of centrally nucleated fibers and fiber areas were assessed for four SSPN-Tg:mdx and four non-Tg:mdx mice. Data for central nucleation are represented as a percentage of the total fibers counted. Fiber areas and central nucleation percentages were averaged, and the standard error is represented. Sigma Plot software (Systat Software, Inc.) was used to perform statistical analysis. Entire quadriceps images were attained using a microscope (AxioImager M1, Carl Zeiss, Inc.) and Axiovision Rel. 4.5 software (Carl Zeiss, Inc.).
Protein preparation
Skeletal muscle was snap frozen in liquid nitrogen and ground to a fine powder with a mortar and pestle. Pulverized muscle was added to ice-cold modified radioimmunoprecipitation assay lysis buffer with phosphatase inhibitors (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 5 mM N-ethylmaleimide, 50 mM sodium fluoride, 2 mM β-glycerophosphate, 1 mM sodium orthovanadate, 100 nM okadaic acid, 5 nM microcystin-l-arginine, and 20 mM Tris-HCl, pH 7.6) and protease inhibitors (0.6 μg/ml pepstatin A, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.75 mM benzamidine, and 0.1 mM PMSF). Homogenates were rocked at 4°C for 1 h. After clarification by centrifugation at 15,000 g for 15 min, tissue lysates were stored at −80°C.
Immunoblot analysis
Protein concentrations were determined using the DC Protein Assay (Bio-Rad Laboratories). Equal concentrations (60 μg) of protein samples were resolved by 5, 10, or 15% SDS-PAGE and transferred to nitrocellulose membranes (Millipore) for subsequent immunoblotting. Primary antibodies against dystrophin (MANDYS1, 1:10), utrophin (NCL-DRP2, 1:5), α-DG (IIH6C-4, 1:1,000), β-DG (MANDAG2, 1:250), α-SG (VP-A105, 1:100), β-SG (VP-B206, 1:100), γ-SG (VP-G803, 1:100; Vector Laboratories), and hSSPN (affinity-purified rabbit 15, 1:500) were incubated at RT while shaking for 18 h. Horseradish peroxidase–conjugated anti–rabbit IgG (GE Healthcare) and anti–mouse IgG (GE Healthcare) and IgM (Upstate Signaling) secondary antibodies were used at 1:3,000 dilutions. All immunoblots were developed using enhanced chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Thermo Fisher Scientific).
Purification of the UGC
0.1 g/ml of SSPN-Tg:mdx total skeletal muscle protein samples was prepared using standard DGC solubilization buffer (50 mM Tris-HCl, pH 7.8, 500 mM NaCl, and 1% digitonin) and protease inhibitors (0.6 μg/ml pepstatin A, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.75 mM benzamidine, 0.1 mM PMSF, 5 μM calpain I, and 5 μM calpeptin; Crosbie et al., 1998). Homogenates were rocked at 4°C for 1 h. After clarification by centrifugation at 15,000 g for 20 min, lysate was then rotated overnight at 4°C with 1 ml agarose-bound succinylated WGA (AL-1023; Vector Laboratories). Succinylated WGA agarose was washed by rotating samples at 4°C four times for 10 min with wash buffer (50 mM Tris-HCl, pH 7.8, 500 mM NaCl, and 0.1% digitonin), including protease inhibitors (0.75 mM benzamidine, 0.1 mM PMSF, 5 μM calpain I, and 5 μM calpeptin) to remove unbound proteins. Bound proteins were eluted with 0.3 M N-acetylglucosamine (Sigma-Aldrich) and concentrated using columns (Centricon Ultracel; Millipore) by centrifugation at 4,000 g for 20 min. Protein concentration was determined with the DC Protein Assay. A 200-μg sample was layered on top of a 5–30% sucrose gradient prepared by adding 6 ml of a 5% sucrose solution into an open top centrifuge tube (Polyclear; Seton) using a 14-gauge Hamilton syringe and underlaying with an equal volume of a 30% sucrose solution. Gradients were mixed using the Gradient IP station (Biocomp). Sucrose gradients were centrifuged at 35,000 g in an ultracentrifuge (Optima L-90K; Beckman Coulter) for 12 h at 4°C using an SW41 rotor (Beckman Coulter). 12 1-ml fractions were collected, and 80 μl of each fraction was resolved by 8% isocratic or 4–20% gradient SDS-PAGE and transferred to nitrocellulose membranes for subsequent immunoblotting. All immunoblotting was performed as described in the previous section with the exception of utrophin (MANCHO3, 1:200; Developmental Studies Hybridoma Bank), which was incubated at 4°C.
RNA extraction and quantitative PCR analysis
RNA was extracted from snap-frozen total skeletal muscle from male non-Tg:mdx and SSPN-Tg:mdx littermates using a RiboPure kit (Ambion) as per the manufacturer's instructions. Purified RNA was quantitated using a spectrophotometer (DU 640; Beckman Coulter) as described in the RiboPure instruction manual. Equal RNA concentrations (2.3 μg) were reverse transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems) as described by the manufacturer. Reverse transcription was performed on a Mastercycler Gradient (Eppendorf) using the following conditions: 25°C for 10 min, 37°C for 120 min, 85°C for 5 s, and held at 4°C.
Quantitative PCR analysis was performed using POWER SYBR green PCR master mix (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was specifically amplified using GAPDH forward (5′-ACTCCACTCACGGCAAATTC-3′) and GAPDH reverse (5′-TCTCCATGGTGGTGAAGACA-3′) primers, which specifically amplified a 171-bp target sequence in mouse GAPDH cDNA. Utrophin cDNA was amplified using utrophin forward (5′-GGGGAAGATGTGAGAGATTT-3′) and utrophin reverse (5′-GTGTGGTGAGGAGATACGAT-3′) primers as previously described (Jasmin et al., 1995). These primers specifically amplified a 548-bp target of the mouse utrophin sequence. All primers were used at a 10-μM concentration. cDNA was diluted 1:10 in RNase/DNase-free water. 1 μl of diluted cDNA was added to 12 μl of 2× PCR master mix, 11 μl RNase/DNase-free water, and 0.5 μl of forward and reverse primers. Quantitative PCR analysis was performed using a single color real-time PCR detection system (MyiQ iCycler; Bio-Rad Laboratories). An initial denaturation cycle was performed at 94°C for 2 min. A total of 40 cycles consisting of denaturation at 94°C for 30 s, primer annealing at 60°C for 30 s, and extension at 72°C for 30 s were performed. A final extension at 72°C was performed for 10 min. Data analysis was performed using MyiQ Optical System software version 1.0 (Bio-Rad Laboratories). Levels of utrophin mRNA were normalized to GAPDH controls, and SSPN-Tg:mdx samples are represented relative to mdx controls (100%). Statistical analysis was performed using the standard t test.
Online supplemental material
Fig. S1 shows a reduction of atrophic fibers upon SSPN overexpression. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200808027/DC1.
Supplementary Material
Acknowledgments
We kindly thank Drs. M. Spencer and S. Lopez for their expertise with whole quadriceps imaging and quantitative PCR analysis.
A.K. Peter was supported by the Molecular, Cellular, and Integrative Physiology predoctoral training fellowship (National Institutes of Health grant T32 GM65823), the Edith Hyde Fellowship, the Ursula Mandel Fellowship, and the Harold and Lillian Kraus American Heart Predoctoral Fellowship. J.L. Marshall was supported by the Genetics Predoctoral Training Fellowship (U.S. Public Health Service National Research Service Award GM07104). R.H. Crosbie was supported by the National Institutes of Health (grant AR48179-01).
Abbreviations used in this paper: DG, dystroglycan; DGC, dystrophin–glycoprotein complex; DMD, Duchenne muscular dystrophy; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; H&E, hematoxylin and eosin; hSSPN, human SSPN; SG, sarcoglycan; SSPN, sarcospan; Tg, transgene; UGC, utrophin–glycoprotein complex.
References
- Barresi, R., and K.P. Campbell. 2006. Dystroglycan: from biosynthesis to pathogenesis of human disease. J. Cell Sci. 119:199–207. [DOI] [PubMed] [Google Scholar]
- Campbell, K.P., and S.D. Kahl. 1989. Association of dystrophin and an integral membrane glycoprotein. Nature. 338:259–262. [DOI] [PubMed] [Google Scholar]
- Crawford, G.E., J.A. Faulkner, R.H. Crosbie, K.P. Campbell, S.C. Froehner, and J.S. Chamberlain. 2000. Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J. Cell Biol. 150:1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosbie, R.H., J. Heighway, D.P. Venzke, J.C. Lee, and K.P. Campbell. 1997. Sarcospan: the 25-kDa transmembrane component of the dystrophin-glycoprotein complex. J. Biol. Chem. 272:31221–31224. [DOI] [PubMed] [Google Scholar]
- Crosbie, R.H., H. Yamada, D.P. Venzke, M.P. Lisanti, and K.P. Campbell. 1998. Caveolin-3 is not an integral component of the dystrophin-glycoprotein complex. FEBS Lett. 427:279–282. [DOI] [PubMed] [Google Scholar]
- Crosbie, R.H., C.S. Lebakken, K.H. Holt, D.P. Venzke, V. Straub, J.C. Lee, R.M. Grady, J.S. Chamberlain, J.R. Sanes, and K.P. Campbell. 1999. Membrane targeting and stabilization of sarcospan is mediated by the sarcoglycan subcomplex. J. Cell Biol. 145:153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelloRusso, C., J.M. Scott, D. Hartigan-O'Connor, G. Salvatori, C. Barjot, A.S. Robinson, R.W. Crawford, S.V. Brooks, and J.S. Chamberlain. 2002. Functional correction of adult mdx mouse muscle using gutted adenoviral vectors expressing full-length dystrophin. Proc. Natl. Acad. Sci. USA. 99:12979–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbeej, M., and K.P. Campbell. 2002. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr. Opin. Genet. Dev. 12:349–361. [DOI] [PubMed] [Google Scholar]
- Ervasti, J.M., and K.P. Campbell. 1991. Membrane organization of the dystrophin-glycoprotein complex. Cell. 66:1121–1131. [DOI] [PubMed] [Google Scholar]
- Ervasti, J.M., and K.P. Campbell. 1993. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 122:809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti, J.M., K. Ohlendieck, S.D. Kahl, M.G. Gaver, and K.P. Campbell. 1990. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 345:315–319. [DOI] [PubMed] [Google Scholar]
- Ervasti, J.M., S.D. Kahl, and K.P. Campbell. 1991. Purification of dystrophin from skeletal muscle. J. Biol. Chem. 266:9161–9165. [PubMed] [Google Scholar]
- Grady, R.M., H. Zhou, J.M. Cunningham, M.D. Henry, K.P. Campbell, and J.R. Sanes. 2000. Maturation and maintenance of the neuromuscular synapse: genetic evidence for roles of the dystrophin–glycoprotein complex. Neuron. 25:279–293. [DOI] [PubMed] [Google Scholar]
- Gregorevic, P., J.M. Allen, E. Minami, M.J. Blankinship, M. Haraguchi, L. Meuse, E. Finn, M.E. Adams, S.C. Froehner, C.E. Murry, and J.S. Chamberlain. 2006. rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat. Med. 12:787–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler, M.E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397–422. [DOI] [PubMed] [Google Scholar]
- Holt, K.H., and K.P. Campbell. 1998. Assembly of the sarcoglycan complex. Insights for muscular dystrophy. J. Biol. Chem. 273:34667–34670. [DOI] [PubMed] [Google Scholar]
- Jasmin, B.J., H. Alameddine, J.A. Lunde, F. Stetzkowski-Marden, H. Collin, J.M. Tinsley, K.E. Davies, F.M. Tome, D.J. Parry, and J. Cartaud. 1995. Expression of utrophin and its mRNA in denervated mdx mouse muscle. FEBS Lett. 374:393–398. [DOI] [PubMed] [Google Scholar]
- Judge, L.M., M. Haraguchiln, and J.S. Chamberlain. 2006. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J. Cell Sci. 119:1537–1546. [DOI] [PubMed] [Google Scholar]
- Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5:136–148. [DOI] [PubMed] [Google Scholar]
- Matsumura, K., J.M. Ervasti, K. Ohlendieck, S.D. Kahl, and K.P. Campbell. 1992. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 360:588–591. [DOI] [PubMed] [Google Scholar]
- Menke, A., and H. Jockusch. 1991. Decreased osmotic stability of dystrophin-less muscle cells from the mdx mouse. Nature. 349:69–71. [DOI] [PubMed] [Google Scholar]
- Michele, D.E., and K.P. Campbell. 2003. Dystrophin-glycoprotein complex: post-translational processing and dystroglycan function. J. Biol. Chem. 278:15457–15460. [DOI] [PubMed] [Google Scholar]
- Miller, G., E.L. Wang, K.L. Nassar, A.K. Peter, and R.H. Crosbie. 2007. Structural and functional analysis of the sarcoglycan-sarcospan subcomplex. Exp. Cell Res. 313:639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom, G.L., P. Gregorevic, and J.S. Chamberlain. 2007. Viral-mediated gene therapy for the muscular dystrophies: successes, limitations, and recent advances. Biochim. Biophys. Acta. 1772:243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendieck, K., and K.P. Campbell. 1991. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J. Cell Biol. 115:1685–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, A.K., and R.H. Crosbie. 2006. Hypertrophic response of Duchenne and limb-girdle muscular dystrophies is associated with activation of Akt pathway. Exp. Cell Res. 312:2580–2591. [DOI] [PubMed] [Google Scholar]
- Peter, A.K., G. Miller, and R.H. Crosbie. 2007. Disrupted mechanical stability of the dystrophin-glycoprotein complex causes severe muscular dystrophy in sarcospan transgenic mice. J. Cell Sci. 120:996–1008. [DOI] [PubMed] [Google Scholar]
- Petrof, B.J., J.B. Shrager, H.H. Stedman, A.M. Kelly, and H.L. Sweeney. 1993. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. USA. 90:3710–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, M.J., J.R. Guyon, H. Sorimachi, A. Potts, I. Richard, M. Herasse, J. Chamberlain, I. Dalkilic, L.M. Kunkel, and J.S. Beckmann. 2002. Stable expression of calpain 3 from a muscle transgene in vivo: immature muscle in transgenic mice suggests a role for calpain 3 in muscle maturation. Proc. Natl. Acad. Sci. USA. 99:8874–8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, V., J.A. Rafael, J.S. Chamberlain, and K.P. Campbell. 1997. Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J. Cell Biol. 139:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley, J.M., A.C. Potter, S.R. Phelps, R. Fisher, J.I. Trickett, and K.E. Davies. 1996. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 384:349–353. [DOI] [PubMed] [Google Scholar]
- Tinsley, J., N. Deconinck, R. Risher, D. Kahn, S. Phelps, J.M. Gillis, and K. Davies. 1998. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med. 4:1441–1444. [DOI] [PubMed] [Google Scholar]
- Weller, B., G. Karpati, and S. Carpenter. 1990. Dystrophin-deficient mdx muscle fibers are preferentially vulnerable to necrosis induced by experimental lengthening contractions. J. Neurol. Sci. 100:9–13. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., and E. Ozawa. 1990. Glycoprotein complex anchoring dystrophin to sarcolemma. J. Biochem. (Tokyo). 108:748–752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.