Abstract
Actin assembly at the cell front drives membrane protrusion and initiates the cell migration cycle. Microtubules (MTs) extend within forward protrusions to sustain cell polarity and promote adhesion site turnover. Memo is an effector of the ErbB2 receptor tyrosine kinase involved in breast carcinoma cell migration. However, its mechanism of action remained unknown. We report in this study that Memo controls ErbB2-regulated MT dynamics by altering the transition frequency between MT growth and shortening phases. Moreover, although Memo-depleted cells can assemble the Rac1-dependent actin meshwork and form lamellipodia, they show defective localization of lamellipodial markers such as α-actinin-1 and a reduced number of short-lived adhesion sites underlying the advancing edge of migrating cells. Finally, we demonstrate that Memo is required for the localization of the RhoA guanosine triphosphatase and its effector mDia1 to the plasma membrane and that Memo–RhoA–mDia1 signaling coordinates the organization of the lamellipodial actin network, adhesion site formation, and MT outgrowth within the cell leading edge to sustain cell motility.
Introduction
Cell motility is the result of a complex series of events that must be integrated both spatially and temporally (Ridley et al., 2003). Efficient motility requires the coordinated regulation of actin, microtubules (MTs), and adhesion sites throughout the migration process (Lauffenburger and Horwitz, 1996; Rodriguez et al., 2003). While investigating signaling pathways involved in ErbB2-driven breast carcinoma cell migration, we have identified Memo, a protein that does not belong to any known family of signaling molecules, as a novel ErbB2 effector (Marone et al., 2004). Memo knockdown cells showed reduced migration despite their preserved ability to extend lamellae/lamellipodia. Interestingly, Memo appears to be involved in MT outgrowth within cell protrusions (Marone et al., 2004). We have investigated the mechanisms underlying Memo's function in ErbB2-driven motility in living breast carcinoma cell lines. We found that Memo regulates MT dynamics, formation of adhesion sites, and the organization of the lamellipodial actin network by contributing to localize the small G protein RhoA and its effector mDia1 at the plasma membrane.
Results and discussion
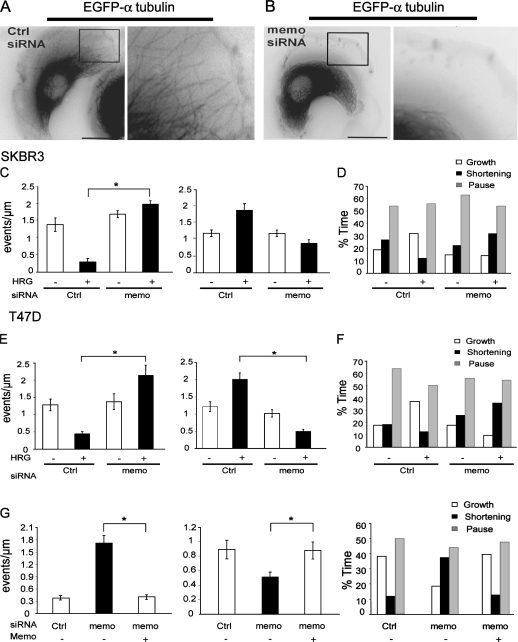
In migrating cells, MTs are usually organized radially with the minus ends anchored at the centrosome and the plus ends probing the cytoplasm. Because MT plus ends are dynamically unstable and switch abruptly between phases of growth, pause, and shortening (Mitchison and Kirschner, 1984), MT net growth depends on the tight regulation of MT dynamic instability. Upon heregulin-β1 (HRG)–induced ErbB2 activation, breast carcinoma cells extend wide membrane protrusions in the direction of cell displacement invaded by outgrowing MTs (Fig. 1 A). Incorporation of EGFP–α-tubulin into MTs allowed the tracking of individual MT plus ends in the lamellae/lamellipodia and the analysis of MT dynamics (Fig. 1 A and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200805107/DC1). We found that HRG treatment did not affect MT growth or shortening rates (not depicted) but strongly decreased the frequency of transitions from growth or pause to shortening (catastrophes) and increased the frequency of transitions from shortening to growth or pause (rescues; Fig. 1, C and E). As a consequence, MTs spent more time growing than shortening, in contrast to what was observed in the absence of stimulation (Fig. 1, D and F). Expression of Memo was knocked down via siRNA (Fig. S1). Memo knockdown cells were still capable of forming protrusions of the same size and at the same rate as control cells (unpublished data). However, despite the fact that MTs were still dynamic (Video 2), they failed to enter lamellipodia (Fig. 1 B). Analysis of plus end dynamics showed that in Memo knockdown cells, HRG treatment failed to decrease catastrophes or increase rescues (Fig. 1, C and E) and no longer affected the time MTs spent in the growth/shortening phases (Fig. 1, D and F) in SKBR3 and T47D breast carcinoma cells. Importantly, reexpression of Memo restored all ErbB2 effects on MTs (Fig. 1 G). Interestingly, Memo depletion did not significantly affect basal MT dynamics. Thus, Memo is an essential effector of ErbB2-regulated MT dynamics.
Figure 1.
Memo is required for ErbB2-regulated MT dynamics. (A and B) SKBR3 cells expressing EGFP-tubulin and control (Ctrl; A) or Memo siRNA (B) were treated with 5 nM HRG for 10 min. Still images from Videos 1 and 2 (available at http://www.jcb.org/cgi/content/full/jcb.200805107/DC1) show limited MT outgrowth in Memo-depleted cells relative to control cells. Right panels show enlargement of boxed areas. (C–F) Individual MT plus ends were tracked over time after addition of HRG to SKBR3 (C and D) and T47D (E and F) cells, and dynamic instability was analyzed. Frequencies of rescues (right) and catastrophes (C and E, left) and the percentage of time MTs spent growing, shortening, or paused (D and F) were calculated. (G) Reexpression of Memo into HRG-treated Memo-depleted SKBR3 cells restores normal MT dynamics. Left, catastrophes; middle, rescues. Values are means ± SD from three independent experiments (30 MTs and nine cells). *, P < 0.05 by permutation analysis; n = 30. Bars, 10 μm.
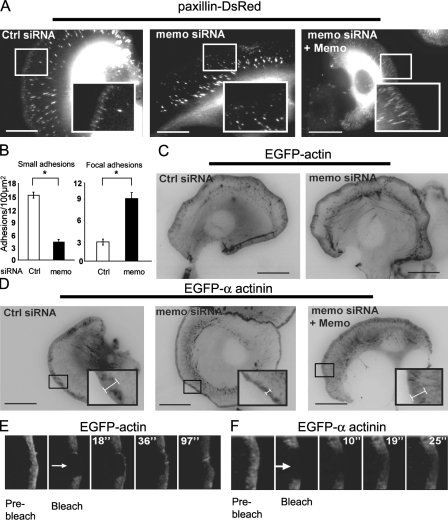
MTs have been shown to target adhesion complexes and promote their disassembly (Kaverina et al., 1999). Thus, we investigated the consequences of Memo knockdown on adhesion complexes formed by migrating cells, using paxillin as a marker. We observed that DsRed-paxillin was incorporated into two discrete populations of adhesions: small short-lived adhesion sites (mean lifetime of 12.3 ± 0.4 s; n = 70 individual adhesions in seven cells) underlying the advancing leading edge and large stable sites, the focal adhesions ([FAs] mean lifetime of 11.6 ± 1.2 min; n = 20 individual adhesions in four cells) deeper within the lamella, which derive from a subpopulation of the former (Fig. 2 A and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200805107/DC1). Interestingly, Memo knockdown resulted in a striking decrease in the number of small adhesion sites, whereas the number of FAs increased (Fig. 2, A and B; and Video 4). In accordance with the known role of MTs on FA turnover, we found that the lifetime of adhesion sites was increased in the absence of Memo (unpublished data). Reintroduction of Memo in knockdown cells restored small adhesions (Fig. 2 A). We propose that the reduction in short-lived adhesion sites that underlie membrane protrusions combined with the increased number of FAs that anchor cells to the substratum contributes to the motility defect in Memo knockdown cells.
Figure 2.
Memo is involved in small adhesion site formation and α-actinin lamellipodial localization. (A) Still images from Videos 3 and 4 showing paxillin-DsRed–labeled adhesion sites in SKBR3 cells. Paxillin incorporates into two populations of adhesion sites with distinct localizations, sizes, and lifespan (left). Memo is required for the formation of small peripheral adhesion (middle vs. left). Reexpression of Memo restores small adhesion sites (right). (B) Number of small and large (focal) adhesions in control and Memo-depleted cells. Values are means per 100 μm2 ± SEM from three independent experiments (four cells and three regions per cell). *, P < 0.05 by Mann-Whitney. (C and D) Still images from Videos 5–8 showing EGFP-actin (C) or EGFP–α-actinin-1 (D) expression in control and Memo-depleted migrating cells. Width of α-actinin–labeled lamellipodia (brackets) is reduced in Memo-depleted cells (middle) and restored upon Memo reexpression (right). Actin labeling is unchanged. Insets show enlargements of leading edges. (E and F) EGFP-actin (E) or EGFP–α-actinin-1 (F) fluorescence was bleached (arrows) within the lamellipodia of migrating cells, and fluorescence recovery was monitored over time (images from Videos 9 and 10, available at http://www.jcb.org/cgi/content/full/jcb.200805107/DC1). Actin fluorescence recovers from the cell membrane reflecting topical actin assembly, whereas α-actinin fluorescence recovers at a faster rate throughout the lamellipodia. Bars, 10 μm.
Because of the known link between adhesion sites and actin filaments, we examined Memo's impact on the assembly of the filamentous-actin network in migrating cells. Actin filaments assemble at the leading edge of migrating cells in a Rac1- and Arp2/3-dependent manner to form a branched network that pushes the cell membrane forward (Pollard and Borisy, 2003). EGFP-actin labeling was concentrated in several structures, including the cell cortical area and actin stress fibers, and appeared similar in control and Memo-depleted cells. In particular, the extent of the EGFP-actin–labeled cortical actin meshwork (Fig. 2 C and Videos 5 and 6, available at http://www.jcb.org/cgi/content/full/jcb.200805107/DC1) was similar in control and Memo knockdown cells (width of cortical actin = 1.32 ± 0.11 μm and 1.42 ± 0.13 μm in control and Memo siRNA–expressing cells, respectively; 18 cells, 10 regions per cell). Similarly, the lamellipodial localization of Arp3 was unaffected by Memo depletion (Fig. S1). A recent study proposed that a dense actin network comprising the actin-bundling protein α-actinin might be associated with initiation of adhesion sites (Giannone et al., 2007). We observed that α-actinin–EGFP labeled a structure that overlapped with EGFP-actin–labeled lamellipodia. Interestingly, we observed that the α-actinin–labeled lamellipodia were almost 50% thinner in Memo knockdown relative to control cells (Fig. 2 D, Fig. 4 A, and Videos 7 and 8). Importantly, the width of α-actinin–labeled lamellipodia was restored to normal upon Memo reexpression (Fig. 2 D and Fig. 4 A). The effect was even more drastic for endogenous α-actinin-1, as immunolabeling appeared to be decreased throughout the lamellipodia of Memo-deficient cells (Fig. S1).
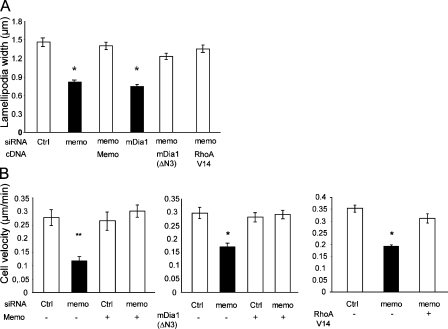
Figure 4.
RhoA and mDia1 restore lamellipodial α-actinin and cell motility in Memo-depleted cells. SKBR3 cells were transfected with Memo, mDia1, or control siRNA, and constructs coding for Memo, active mDia1 (mDia1[ΔN3]), or active RhoA (RhoAV14) as indicated. (A) α-Actinin-1 labeling was analyzed as in Fig. 2 D. Width of α-actinin–labeled lamellipodia was measured along the cell leading edge. Values are means ± SEM (10 regions per cell for 18 cells; three independent experiments). *, P < 0.025 by Kruskal-Wallis. (B) Cell velocity was determined by tracking cells every 5 min for 150 min. 90–150 cells were tracked per condition. Values are means ± SEM from three independent experiments. *, P < 0.05; **, P = 0.07 (note that P = 0.002 when n = 9) by Kruskal-Wallis.
We have further explored the organization of the lamellipodial network using FRAP experiments. After photobleaching of EGFP-actin, the whole bleached area flowed back until disappearance in the lamella, with little recovery through diffusion (Fig. 2 E and Video 9, available at http://www.jcb.org/cgi/content/full/jcb.200805107/DC1). This observation illustrates that the lamellipodial actin network is assembled at the cell membrane as a stable structure that is carried back by retrograde flow until it reaches the lamellipodium/lamella border, where it is disassembled (Ponti et al., 2004). Arp3 displayed the same type of recovery pattern (unpublished data). In contrast, recovery of fluorescence after bleaching of α-actinin–EGFP occurred throughout lamellipodia (Fig. 2 F and Video 10), reflecting fast α-actinin-1 association/dissociation to the actin network. Interestingly, Memo-depleted cells recovered fluorescence at the same rate as control cells (recovery half-time of t1/2 = 8.7 ± 1.1 s and 9.8 ± 1.8 s, respectively, for control and Memo-depleted cells; 45 regions in 15 cells; three independent experiments), indicating that Memo depletion does not directly affect the α-actinin association/dissociation rate. Instead, it could affect the actin network structure, preventing α-actinin binding/cross-linking. This is supported by the fact that lamellipodial localization of cortactin, another actin filament–binding protein, is also dependent on Memo. Although cortactin was present in the lamellipodia of control cells, it was restricted to the cell membrane in Memo knockdown cells (Fig. S1).
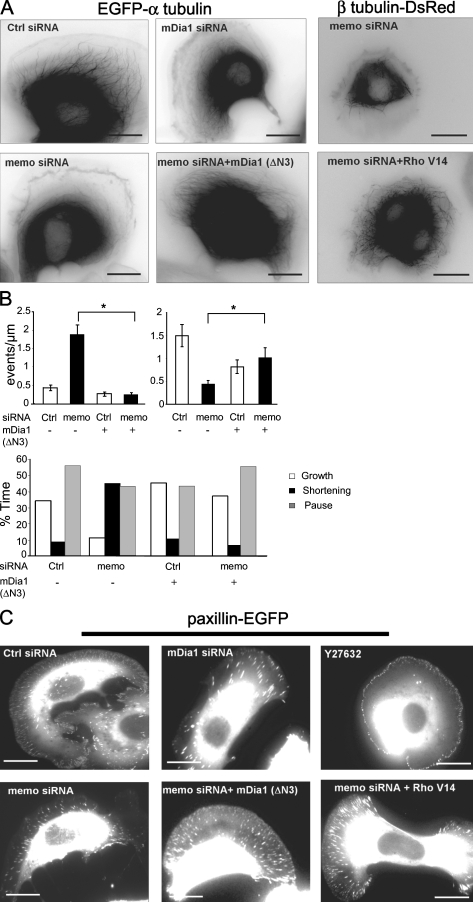
Our results show that Memo regulates MT dynamics, organization of the lamellipodial actin network, and initiation of adhesion sites. To position Memo in a signaling pathway, we investigated mDia1 because mDia formins are known to control actin polymerization (Ishizaki et al., 2001; Kovar et al., 2006) and the alignment and stability of a subpopulation of MTs (Palazzo et al., 2001; Wen et al., 2004). mDia1 knockdown (Fig. S1) prevented MT outgrowth (Fig. 3 A), inhibited formation of small adhesion sites (Fig. 3 C), and decreased the thickness of actinin-labeled lamellipodia (Fig. 4 A) to the same extent as Memo knockdown. Importantly, expression of a constitutively active form of mDia1, mDia1(ΔN3) (Ishizaki et al., 2001), in Memo knockdown cells compensated for the loss of Memo and restored Memo MT outgrowth toward the cell cortex (Fig. 3 A) in Memo knockdown cells. Analysis of MT dynamics showed that in the presence of active mDia1, Memo knockdown had no impact on the frequency of MT rescues or catastrophes, nor was the time MTs spent growing versus shortening affected (Fig. 3 B). Expression of active mDia1 also restored formation of small adhesion sites (in 92% of the cells vs. 12% of Memo knockdown cells; n = 69 and 74, respectively; Fig. 3 C) and normal actinin-labeled lamellipodia (Fig. 4 A) in Memo knockdown cells. Because mDia1 is an effector of RhoA (Watanabe et al., 1997), we tested the role of RhoA in Memo signaling. Expression of moderate levels of an active form of RhoA (RhoV14) in Memo knockdown cells induced thick stress fibers and large FAs but also restored Memo functions: MT outgrowth (in 89% of the cells vs. 5% of Memo-depleted cells and 90% of control cells; n = 73, 61, and 87, respectively; Fig. 3 A), formation of small adhesion sites (in 87% of the cells vs. 8% of Memo-depleted cells and 92% of control cells; n = 78, 63, and 79, respectively; Fig. 3 C), and normal lamellipodial α-actinin labeling (Fig. 4 A). These results demonstrate that RhoA and mDia1 are important Memo effectors that control MTs, the actin network, and adhesion sites. Finally, we evaluated the impact of the Memo–RhoA–mDia1 signaling pathway on cell motility using time-lapse microscopy. We found that Memo knockdown decreased velocity of SKBR3 cells by 40–50% (Fig. 4 B). Reintroduction of Memo, active mDia1, or active RhoA in Memo knockdown cells restored normal cell velocity, revealing the significant contribution of the Memo–RhoA-mDia1 pathway to ErbB2-dependent breast carcinoma cell migration.
Figure 3.
RhoA and mDia1 restore MT dynamics and peripheral adhesions in Memo-depleted cells. (A and B) SKBR3 cells expressing Memo, mDia1, or control siRNA and constructs coding for Memo, active mDia1 (mDia1[ΔN3]), or active RhoA (RhoAV14) were analyzed for MT dynamic instability as in Fig. 1. (B) Top left, catastrophes; top right, rescues. Values are means ± SD from three independent experiments (30 MTs and nine cells). *, P < 0.01 by permutation test. (C) Formation of adhesion sites was analyzed as in Fig. 2 A. Where indicated, cells were treated with 10 μM Y27632 for 120 min before addition of HRG. Bars, 10 μm.
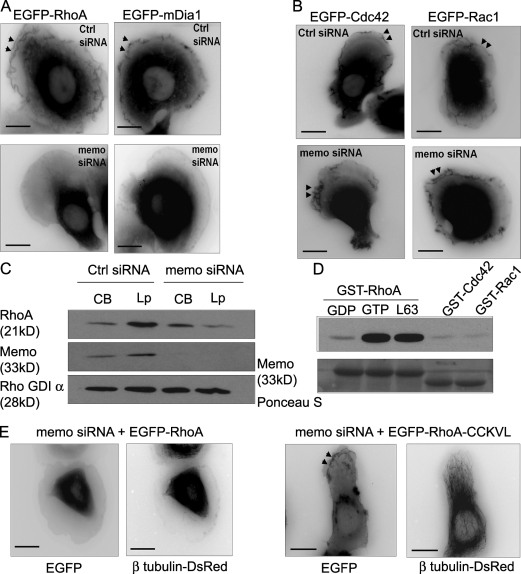
Activation of RhoGTPases is associated with their relocalization from the cytosol to the cell membrane (Kurokawa and Matsuda, 2005; Pertz et al., 2006). We investigated the possibility that Memo controls the recruitment of RhoA and mDia1 to the cell membrane. HRG treatment induced the recruitment of Memo from the cytosol to the plasma membrane (Marone et al., 2004). EGFP-mDia1, EGFP-RhoA, and the respective endogenous proteins were similarly recruited to lamellipodial membranes and ruffles (Fig. 5 A). RhoA and mDia1 labeling is not simply caused by increased cell volume at ruffles because cytoplasmic markers (e.g., the mCherry fluorescent protein) show limited labeling of ruffles (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200805107/DC1). Importantly, Memo knockdown prevented recruitment of EGFP-tagged and endogenous RhoA and mDia1 to the plasma membrane (Fig. 5 A and Fig. S2). This was not caused by decreased ruffling activity, as other RhoGTPases such as Rac1 and Cdc42 are still associated with plasma membrane and ruffles in the absence of Memo (Fig. 5 B and Fig. S3). Biochemical analysis of lamellipodia- and cell body–enriched fractions (Cho and Klemke, 2002) confirmed that RhoA and Memo were enriched in the cell leading edge and that decreased expression of Memo led to a specific reduction of RhoA in lamellipodia (Fig. 5 C). In fact, the functional interaction between Memo and RhoA could be the consequence of a physical interaction, as bacterially expressed Memo interacted with purified RhoA in pull-down experiments. Interestingly, Memo interacted preferentially with the active forms of RhoA (GTP-bound and constitutively active L63QRhoA). In contrast, Memo did not bind GTP-loaded Cdc42 or Rac1, which is indicative of the specificity of the interaction (Fig. 5 D). Thus, we propose that Memo retains active RhoA at the plasma membrane to allow spatially and temporally restricted activation of RhoA effectors such as mDia1 at the cell leading edge.
Figure 5.
Memo is required for localization of RhoA and mDia1 to membrane ruffles and lamellipodia. (A and B) SKBR3 cells expressing EGFP-RhoA or EGFP-mDia1 (A) or EGFP-Cdc42 or EGFP-Rac1 (B) constructs and control or Memo siRNA, as indicated, were treated with 5 nM HRG for 10 min. EGFP-RhoA and EGFP-mDia1 were recruited to cell membrane and ruffles (arrowheads) in control but not in Memo-depleted cells, whereas localization of EGFP-Cdc42 and EGFP-Rac1 was independent of Memo. (C) Cell body (CB) and lamellipodia (Lp) of control and Memo siRNA–expressing cells were fractionated. Equal amounts of proteins were analyzed by Western blotting to reveal the relative amounts of RhoA in the respective fractions. RhoA is decreased in the lamellipodia-enriched fraction but is unchanged in the cell body of Memo knockdown cells. (D) Bacterially produced Memo was incubated with GDP-loaded GST-RhoA or GTPγS-loaded GST-RhoA, GST-RhoA L63, GST-Cdc42, or GST-Rac1 bound to glutathione beads. RhoGTPase-associated Memo was visualized by Western blotting. Ponceau S protein staining (bottom) shows equivalent amounts of GST fusion proteins. (E) Cells were transfected with Memo siRNA and EGFP-RhoA or EGFP-RhoA-CCKVL. MTs were visualized by incorporation of β-tubulin–DsRed. RhoA-CCKVL localizes to membrane ruffles (arrowheads) and allows MT outgrowth in Memo-depleted cells. Bars, 10 μm.
If Memo's main role is to localize RhoA to the plasma membrane, a membrane-targeted form of RhoA should compensate for the loss of Memo in Memo knockdown cells. Expression of a fusion protein between wild-type RhoA and the palmitoylation motif of RhoB (RhoA-CCKVL; Michaelson et al., 2001) promoted constitutive membrane localization independently of Memo expression levels (Fig. 5 E). Expression of RhoA-CCKVL in Memo-deficient cells restored MT outgrowth (in 81% of the cells vs. 10% of RhoA-expressing cells; n = 65 and 61 cells, respectively) but also normal lamellipodial localization of α-actinin-1 and cell motility (unpublished data), showing that forcing RhoA to the plasma membrane recapitulates Memo's functions. The data also suggest that Memo is not directly required for activation of RhoA because wild-type membrane-associated RhoA was fully functional in the absence of Memo.
It is noteworthy that Memo signals via RhoA but does not control known functions of RhoA such as formation of stress fibers or maturation of FAs (Burridge and Wennerberg, 2004). In fact, these are under the control of the Rho-associated kinase (ROCK), as Y27632 inhibition of ROCK activity drastically reduced stress fibers (not depicted) and FAs while preserving small peripheral adhesions (Fig. 3 C), which is in striking contrast to mDia1 inhibition. These results are in line with earlier work showing that ROCK is involved in the formation of central but not peripheral adhesions (Totsukawa et al., 2004). Moreover, an active form of ROCK induced thick stress fibers and large FAs but failed to restore small adhesion sites or lamellipodial α-actinin labeling in Memo-depleted cells (unpublished data). Thus, Memo knockdown provides a method to distinguish between discrete pools of RhoA with distinct functions: one controlling cortical actin and initiation of adhesion sites via the mDia1 effector at the cell leading edge, and the other triggering ROCK-dependent stress fiber formation, adhesion maturation, and cell contraction within the cell body.
Although the role of the Rac1–Scar–Arp2/3 pathway for the assembly of the branched actin meshwork was clearly demonstrated (Pollard and Borisy, 2003), models of lamellipodia protrusion need to take into account evidences for the coexistence within the lamellipodium of kinetically distinct yet overlapping actin networks (Ponti et al., 2004) and the role of other types of regulators of actin assembly such as formins (Yang et al., 2007) and Ena/VASP proteins (Bear et al., 2002). We propose that mDia1, as an effector of Memo–RhoA, contributes to the organization of the lamellipodial actin network. mDia1 controls the lamellipodial localization of both α-actinin-1 and cortactin, suggesting that an alteration of the network organization allows actin-binding protein association to the actin network. We are exploring the possibility that, as shown for mDia2 recently (Yang et al., 2007), mDia1 controls the length or branching of actin filaments. This could lead to a modification of the structure of the actin network and allow α-actinin-1 localization and bundling or scaffolding activity in lamellipodia (Otey and Carpen, 2004). As a recent study suggested that myosin-dependent tension exerted on the lamellipodial network controls formation of adhesion sites (Giannone et al., 2007), changes in actin filament length, branching, or bundling could in turn affect the response of the actin network to tension and formation of peripheral adhesions. Thus, our study showing that the Memo–RhoA–mDia1 pathway controls the cytoskeleton and adhesion sites at the cell leading edge adds a layer of complexity to the signaling network that controls cell motility.
Materials and methods
Cell transfection and plasmid constructs
SKBR3 and T47D breast carcinoma cells grown in DME and 10% fetal calf serum were transfected by nucleofection (Amaxa) with 2 μg siRNA for Memo (nt 1,438–1,458; within the 3′ untranslated region), mDia1 (nt 132–152), or LacZ (nt 2,289–2,309; Invitrogen) and the following plasmid constructs: GFP-Memo, Myc-Memo6, EGFP–α-tubulin (Clontech Laboratories, Inc.), DsRed–β-tubulin (provided by V. Homburger, Institut de Génomique Fonctionnelle, Montpellier, France), EGFP–γ-actin (provided by J. Wehland, Helmholtz Centre for Infection Research, Braunschweig, Germany), paxillin-DsRed, paxillin-EGFP (provided by A.R. Horwitz, University of Virginia, Charlottesville, VA), EGFP-mDia1, Flag-mDia1ΔN3 (provided by S. Narumiya, Kyoto University, Kyoto, Japan), EGFP-RhoA-CCKVL, EGFP-RhoA (provided by M.R. Philips, New York University, New York, NY), EGFP-RhoA-V14, EGFP-Rac1, EGFP-Cdc42 (provided by M.A. Schwartz, University of Virginia, Charlottesville, VA), or EGFP–α-actinin-1 (11908; Addgene; provided by C.A. Otey, University of North Carolina, Chapel Hill, NC).
Analysis of MT dynamic instability
Cells grown on collagen-coated glass coverslips were placed in a double coverslip chamber maintained at 37°C and observed upon addition of 5 nM HRG (R&D Systems) using the 100× plan Fluotar NA 1.3 or plan Apochromat NA 1.4 objective of a fluorescence microscope (DM-IRBE Microsystem [Leica]; Axiovert 200 [Carl Zeiss, Inc.]). 31 images per cell were acquired at 4-s intervals using a digital camera (charge-coupled device Coolsnap FX [Princeton Instruments]; Coolsnap HQ [Roper Scientific]). Plus ends of individual MTs were tracked with time using MetaMorph software (MDS Analytical Technologies). The number of catastrophes (transition from growth or pause to shortening) and rescues (transitions from shortening to pause or growth) was calculated as described previously (Pourroy et al., 2006). Means and SEM were calculated per MT.
Quantification of adhesion sites and actin dynamics
Cells were observed as described in the previous paragraph except that images of paxillin-expressing cells were acquired at 4-s intervals for 2 min or 1-min intervals for 30 min. Background-corrected fluorescence intensity images were used to measure small adhesion sites and FA number and lifetime in lamellipodia and lamellae, respectively. MetaMorph software was used to measure the width of the actin network in the leading edge of migrating cells expressing EGFP-actin or EGFP–α-actinin-1. FRAP experiments were performed on a confocal microscope (LSM510; Carl Zeiss, Inc.) with a 63× plan Apo 1.4 NA objective. EGFP fluorescence was eliminated by 30 bleach cycles at 100% intensity of the 488-nm argon laser. Recovery curves generated from photobleached areas of the same sizes and locations were sampled every 0.5 s for 42 s and corrected for overall photobleaching and were used to calculate t1/2.
Motility assay
Cell motility was analyzed as described previously (Pourroy et al., 2006) except that pictures were collected for 150 min at 5-min intervals. Means of velocity were calculated using MetaMorph and Excel (Microsoft) software.
Pull-down assay, cell fractionation, and Western blotting
GST-RhoA, GST-Cdc42, GST-Rac1, and GST-RhoA L63 beads (Cytoskeleton, Inc.) were loaded with GDP or GTPγS according to the manufacturer's instructions before overnight incubation with 2 μg of bacterially produced Memo and Western blot analysis. Cell body or lamellipodia-enriched fractions were obtained as described previously (Cho and Klemke, 2002) after the addition of 5 nM HRG to the lower chamber of a 3-μm pore Transwell (Costar) for 1 h. Antibodies directed against Memo (monoclonal antibody to amino acids 25–43), mDia1, Shc, Gsk3β (BD Biosciences), RhoA, RhoGDIα (Santa Cruz Biotechnology, Inc.), and Flag (Sigma-Aldrich) were used for Western blotting.
Immunofluorescence microscopy
Cells grown on collagen I–coated coverslips were fixed in 4% formaldehyde and permeabilized in 0.2% Triton X-100 before the addition of antibodies directed toward Cdc42, RhoA, α-actinin-1 (Santa Cruz Biotechnology, Inc.), mDia1, Rac1, Arp3 (BD Biosciences), cortactin (provided by E. Van Obberghen-Schilling, National Center for Scientific Research, Nice, France), and α-tubulin (provided by W. Krek, Swiss Federal Institute of Technology, Zurich, Switzerland). Secondary antibodies and Alexa Fluor 546 phalloidin were obtained from Invitrogen. DNA was counterstained with Hoechst dye (Sigma-Aldrich). Images were recorded with a microscope (ApoTome ImagerZ1; Carl Zeiss, Inc.) with a 63× plan Apo 1.4 NA objective coupled to a camera (AxioCamMRm; Carl Zeiss, Inc.) and driven by AxioVision LE software (Carl Zeiss, Inc.).
Statistical analysis
Data are presented as mean ± SEM and were analyzed by Mann-Whitney, Kruskal-Wallis, or permutation tests using StatXact software (Cytel) as indicated. P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows the efficiency of the Memo, mDia1, and RhoA siRNAs and the effect of Memo depletion on lamellipodial markers. Fig. S2 shows that RhoA labeling of the cell membrane is not simply caused by ruffling and that Memo is required for plasma membrane localization of endogenous RhoA and mDia1. Fig. S3 shows that Memo is not required for plasma membrane localization of Rac1 and Cdc42. Video1 shows MT outgrowth upon HRG treatment. Video 2 shows the lack of MT outgrowth in Memo-depleted cells. Videos 3 and 4 show adhesion site dynamics in control and Memo-depleted cells. Videos 5–8 show actin and α-actinin-1 localization in control and Memo-depleted cells. Videos 9 and 10 show EGFP-actin and EGFP–α-actinin-1 FRAP. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200805107/DC1.
Acknowledgments
We thank V. Homburger, A.R. Horwitz, S. Narumiya, C.A. Otey, M.R. Philips, M.A. Schwartz, E. Van Obberghen-Schilling, and J. Wehland for sharing reagents; N. Hynes, J.-P. Borg, D. Birnbaum, F. Birg, and D. Job for critical reading of the manuscript; M. Sebbagh, J. Gillibert-Duplantier, and S. Aresta for discussions; and D. Salaun for technical support.
A. Badache was supported by the Avenir Program of the French National Institute for Health and Medical Research, Fondation pour la Recherche Médicale, Conseil Général des Bouches du Rhône, Institut National du Cancer, and Ligue contre le Cancer. K. Zaoui was supported by a studentship from Région Provence Alpes Côte d'Azur and the French National Institute for Health and Medical Research.
Abbreviations used in this paper: FA, focal adhesion; HRG, heregulin-β1; MT, microtubule; ROCK, Rho-associated kinase.
References
- Bear, J.E., T.M. Svitkina, M. Krause, D.A. Schafer, J.J. Loureiro, G.A. Strasser, I.V. Maly, O.Y. Chaga, J.A. Cooper, G.G. Borisy, and F.B. Gertler. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521. [DOI] [PubMed] [Google Scholar]
- Burridge, K., and K. Wennerberg. 2004. Rho and Rac take center stage. Cell. 116:167–179. [DOI] [PubMed] [Google Scholar]
- Cho, S.Y., and R.L. Klemke. 2002. Purification of pseudopodia from polarized cells reveals redistribution and activation of Rac through assembly of a CAS/Crk scaffold. J. Cell Biol. 156:725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone, G., B.J. Dubin-Thaler, O. Rossier, Y. Cai, O. Chaga, G. Jiang, W. Beaver, H.G. Dobereiner, Y. Freund, G. Borisy, and M.P. Sheetz. 2007. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 128:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki, T., Y. Morishima, M. Okamoto, T. Furuyashiki, T. Kato, and S. Narumiya. 2001. Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat. Cell Biol. 3:8–14. [DOI] [PubMed] [Google Scholar]
- Kaverina, I., O. Krylyshkina, and J.V. Small. 1999. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J. Cell Biol. 146:1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar, D.R., E.S. Harris, R. Mahaffy, H.N. Higgs, and T.D. Pollard. 2006. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 124:423–435. [DOI] [PubMed] [Google Scholar]
- Kurokawa, K., and M. Matsuda. 2005. Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol. Biol. Cell. 16:4294–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger, D.A., and A.F. Horwitz. 1996. Cell migration: a physically integrated molecular process. Cell. 84:359–369. [DOI] [PubMed] [Google Scholar]
- Marone, R., D. Hess, D. Dankort, W.J. Muller, N.E. Hynes, and A. Badache. 2004. Memo mediates ErbB2-driven cell motility. Nat. Cell Biol. 6:515–522. [DOI] [PubMed] [Google Scholar]
- Michaelson, D., J. Silletti, G. Murphy, P. D'Eustachio, M. Rush, and M.R. Philips. 2001. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152:111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T., and M. Kirschner. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242. [DOI] [PubMed] [Google Scholar]
- Otey, C.A., and O. Carpen. 2004. Alpha-actinin revisited: a fresh look at an old player. Cell Motil. Cytoskeleton. 58:104–111. [DOI] [PubMed] [Google Scholar]
- Palazzo, A.F., T.A. Cook, A.S. Alberts, and G.G. Gundersen. 2001. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 3:723–729. [DOI] [PubMed] [Google Scholar]
- Pertz, O., L. Hodgson, R.L. Klemke, and K.M. Hahn. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 440:1069–1072. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and G.G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Ponti, A., M. Machacek, S.L. Gupton, C.M. Waterman-Storer, and G. Danuser. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786. [DOI] [PubMed] [Google Scholar]
- Pourroy, B., S. Honore, E. Pasquier, V. Bourgarel-Rey, A. Kruczynski, C. Briand, and D. Braguer. 2006. Antiangiogenic concentrations of vinflunine increase the interphase microtubule dynamics and decrease the motility of endothelial cells. Cancer Res. 66:3256–3263. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J., M.A. Schwartz, K. Burridge, R.A. Firtel, M.H. Ginsberg, G. Borisy, J.T. Parsons, and A.R. Horwitz. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709. [DOI] [PubMed] [Google Scholar]
- Rodriguez, O.C., A.W. Schaefer, C.A. Mandato, P. Forscher, W.M. Bement, and C.M. Waterman-Storer. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5:599–609. [DOI] [PubMed] [Google Scholar]
- Totsukawa, G., Y. Wu, Y. Sasaki, D.J. Hartshorne, Y. Yamakita, S. Yamashiro, and F. Matsumura. 2004. Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J. Cell Biol. 164:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, N., P. Madaule, T. Reid, T. Ishizaki, G. Watanabe, A. Kakizuka, Y. Saito, K. Nakao, B.M. Jockusch, and S. Narumiya. 1997. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16:3044–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Y., C.H. Eng, J. Schmoranzer, N. Cabrera-Poch, E.J. Morris, M. Chen, B.J. Wallar, A.S. Alberts, and G.G. Gundersen. 2004. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6:820–830. [DOI] [PubMed] [Google Scholar]
- Yang, C., L. Czech, S. Gerboth, S. Kojima, G. Scita, and T. Svitkina. 2007. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5:e317. [DOI] [PMC free article] [PubMed] [Google Scholar]