Abstract
In mammals, reproduction is dependent on specific neurons secreting the neuropeptide gonadotropin hormone–releasing hormone-1 (GnRH-1). These cells originate during embryonic development in the olfactory placode and migrate into the forebrain, where they become integral members of the hypothalamic–pituitary–gonadal axis. This migratory process is regulated by a wide range of guidance cues, which allow GnRH-1 cells to travel over long distances to reach their appropriate destinations. The Semaphorin4D (Sema4D) receptor, PlexinB1, is highly expressed in the developing olfactory placode, but its function in this context is still unknown. Here, we demonstrate that PlexinB1-deficient mice exhibit a migratory defect of GnRH-1 neurons, resulting in reduction of this cell population in the adult brain. Moreover, Sema4D promotes directional migration in GnRH-1 cells by coupling PlexinB1 with activation of the Met tyrosine kinase (hepatocyte growth factor receptor). This work identifies a function for PlexinB1 during brain development and provides evidence that Sema4D controls migration of GnRH-1 neurons.
Introduction
Gonadotropin hormone–releasing hormone-1 (GnRH-1) regulates anterior pituitary gonadotropes and it is essential for reproduction. GnRH-1–secreting neurons originate from the nasal placode (Wray, 2002) during embryonic development and migrate to the hypothalamus apposed to olfactory-vomeronasal nerves (Schwanzel-Fukuda and Pfaff, 1989; Wray et al., 1989). In humans, several monogenic disorders leading to idiopathic hypogonadotropic hypogonadisms (IHH) are caused by disruption of GnRH-1 neuronal ontogeny/migration (Gonzalez-Martinez et al., 2004). Unraveling new genetic pathways involved in the regulation of GnRH-1 system development is relevant for understanding the basis of pathogeneses leading to human IHH disorders. However, the full repertoire of molecular cues regulating the migratory process, and the correct targeting of GnRH-1 neurons to the hypothalamus have not been elucidated. The underlying mechanisms are believed to involve different classes of signaling molecules. In recent years, pleiotropic factors have been added to the list of molecules influencing the development of the GnRH-1 neuroendocrine compartment. Among these, hepatocyte growth factor (HGF; Giacobini et al., 2007) and secreted-class 3 semaphorins (Cariboni et al., 2007) have been shown to play a role in the control of GnRH-1 migratory process. Semaphorins constitute one of the largest protein families of phylogenetically conserved guidance cues (Tran et al., 2007). Although originally identified as embryonic axon guidance cues, secreted and membrane-bound semaphorins are now known to regulate multiple, distinct processes crucial for neuronal network formation, including axon growth, dendritic morphology, and neuronal migration (Casazza et al., 2007; Zhou et al., 2008). Accumulating evidence shows that semaphorins can repel or attract a wide range of neuronal and nonneuronal cells depending on the cellular targets and on the expression of different subunits of the receptor complexes (Giordano et al., 2002; Pasterkamp et al., 2003; Conrotto et al., 2005; Swiercz et al., 2007; Chen et al., 2008).
It has been shown that the main transducing semaphorin receptors belong to the plexin family (Tran et al., 2007). Plexins can associate with other membrane receptors, leading to activation of different biological programs (Giordano et al., 2002; Conrotto et al., 2004, 2005; Swiercz et al., 2004, 2007). In fact, we have previously shown that Semaphorin4D (Sema4D), other than being a collapsing signal for axonal growth cones (Swiercz et al., 2002), may also induce chemotaxis of epithelial and endothelial cells, and that it functions as a proangiogenic factor through coupling its receptor PlexinB1 with the Met tyrosine kinase (Giordano et al., 2002; Conrotto et al., 2004, 2005). Yet, the potential role of Sema4D in regulating neuronal cell migration has not been investigated so far. Moreover, although Sema4D high-affinity receptors, PlexinB1 and PlexinB2, are highly expressed in the developing olfactory structures (Perala et al., 2005; Deng et al., 2007), their functions in the development of the olfactory and GnRH-1 systems are still unknown.
Here, we find that Sema4D expression is present along the GnRH-1 migratory route, with a peak of expression in the hypothalamic target area, and that GnRH-1 cells express the Sema4D receptor PlexinB1, but not PlexinB2, in a temporal window associated with their migratory process. Analysis of PlexinB1-deficient mice revealed a migratory defect of GnRH-1 cells, leading to reduced size of this neuronal population in adult brains. Using different experimental approaches, we demonstrated that Sema4D promotes the migratory activity of immortalized GnRH-1 cells through the activation of PlexinB1 and the associated Met receptor.
Collectively, our data reveal a novel function of Sema4D in the development of GnRH-1 neurons and identify the PlexinB1–Met receptor complex as a fundamental asset for neuronal cell migration and guidance.
Results
Sema4D/PlexinB1 expression in the developing nasal regions
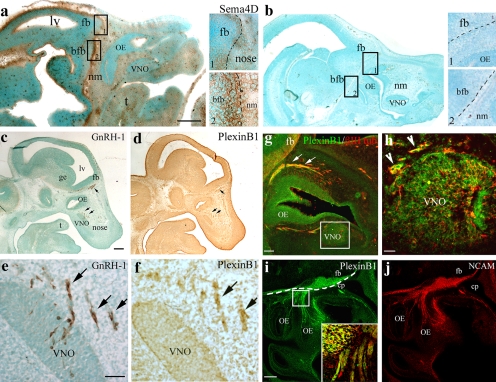
Sema4D is a membrane-bound semaphorin that is also proteolytically released in diffusible form in the extracellular space. To gain further insights into the function of this molecule in the developing GnRH-1–olfactory systems, we determined the spatiotemporal expression pattern of Sema4D protein during mouse embryonic development. At embryonic day 12.5 (E12.5), a robust Sema4D immunoreactivity was detectable in the rostral aspect of the developing forebrain (Fig. 1 a, see high-magnification view in box 1) and in the basal forebrain (Fig. 1 a, see high-magnification view in box 2), which, respectively, represent the intermediate and final targets of GnRH-1 migratory process (Fig. 1 a). Sema4D was also found to be expressed in the same regions at E14.5 and E17.5 (unpublished data). Inclusion of a staining control (cntr) confirmed the specificity of the Sema4D staining (Fig. 1 b; no primary antibody).
Figure 1.
Sema4D and PlexinB1 are expressed along the GnRH-1 migratory route. (a) Sema4D immunoreactivity is detected in the nasal mesenchyme (nm), surrounding the OE and VNO epithelia. A robust staining is evident at the nasal forebrain junction and rostral forebrain (see higher magnification view of box 1) as well as in the basal forebrain (bfb; see higher magnification view of box 2). (b) The specificity of the staining was confirmed by omission of the primary antibody incubation (see high-power view images in boxes 1 and 2). Sections in panels a and b were counterstained with the nuclear dye methyl green to allow visualization of the background tissue. (c–f) Consecutive sagittal sections of an E12.5 mouse immunostained, respectively, for GnRH-1 and PlexinB1. (c and e) GnRH-1–immunoreactive cells emerged from the developing VNO and migrate through the olfactory mesenchyme (arrows) toward the forebrain (fb). GnRH-1–labeled sections were counterstained (methyl green) to allow the identification of the different anatomical structures. (d and f) PlexinB1 immunoreactivity is distributed in the developing OE, in the VNO, and in a group of cells migrating out of the presumptive VNO (see arrows in f). These cells are distributed in a spatial and temporal pattern that parallels GnRH-1 immunostaining (arrows in e and f). (g and h) A sagittal section of an E12.5 mouse nose double-stained for PlexinB1 (green) and βIII-tubulin (red) indicates coexpression of these antigens in the OE, VNO, and in cells (h, arrowheads) and fibers (g, arrows) emerging from these structures. (i and j) A sagittal section of an E12.5 mouse nose double-stained for PlexinB1 (i) and NCAM (j) indicates coexpression of these antigens along the olfactory/vomeronasal axons, as shown by high-power confocal analysis (i, inset, which is an enlarged view of the boxed region). Broken lines indicate the boundary between the nose and the forebrain. cp, cribriform plate; fb, forebrain; lv, lateral ventricle; ge, ganglionic eminence; t, tongue. Bars: (a and b) 500 μm; (a and b, insets) 125 μm; (c and d) 250 μm; (e and f) 50 μm; (g) 50 μm; (h) 30 μm; (i and j) 100 μm; (i, inset) 30 μm.
It was previously found that the PlexinB1 transcript is highly expressed in the developing vomeronasal organ (VNO) and olfactory epithelium (OE; Perala et al., 2005). Immunohistochemistry for PlexinB1 and GnRH-1 was performed on consecutive sagittal sections of mouse embryos. At E12.5, the PlexinB1 expression pattern paralleled GnRH-1 neuronal distribution in the nasal region (Fig. 1, c–f). Indeed, GnRH-1–expressing cells migrating through the nasal mesenchyme (Fig. 1 e) likely coincide with those positive for PlexinB1 (Fig. 1 f). At this developmental stage, the receptor was also expressed in the developing OE and in the presumptive VNO, as demonstrated by simultaneous PlexinB1/βIII tubulin immunofluorescence experiments (Fig. 1, g and h). βIII tubulin is a neuron-specific marker that strongly labels the early olfactory and vomeronasal structures during embryogenesis (De Carlos et al., 1996; Roskams et al., 1998). Double-labeled cells and fibers emerging from the VNO were also detected (Fig. 1 h, arrowheads).
To further confirm that PlexinB1 was expressed along the developing olfactory axons, double-label immunofluorescence was performed for PlexinB1 (Fig. 1 i) and neural cell adhesion molecule (NCAM; Fig. 1 j), another marker of the olfactory–vomeronasal system (Calof and Chikaraishi, 1989). PlexinB1 and NCAM expressions overlapped along olfactory axon bundles crossing the nasal mesenchyme at E12.5, as shown by single confocal planes (Fig. 1 i, inset). Hence, the PlexinB1 receptor demarcated the olfactory system and the migratory GnRH-1 cell population. Because of low signal-to-noise levels in the developing brain, we were unable to detect specific immunoreactivity for PlexinB1 along the vomeronasal caudal nerve that GnRH-1 cells follow into the ventral forebrain (Yoshida et al., 1995) nor in GnRH-1 neurons located in the brain at later developmental stages (E14.5 and E17.5; unpublished data).
The developmental expression patterns of Sema4D and PlexinB1 are consistent with a role of Sema4D as guidance cue for migratory GnRH-1 cells and/or developing olfactory axons.
Primary GnRH-1 neurons express PlexinB1 but not PlexinB2 during their migratory process
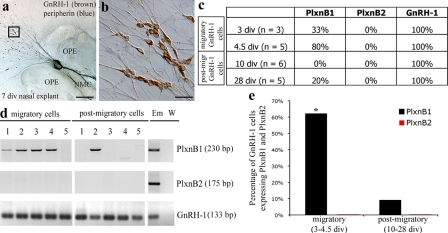
It has been previously shown that the migration of GnRH-1 neurons observed in vivo is consistently recapitulated in nasal explants ex vivo (Fig. 2, a and b; Fueshko and Wray, 1994). We obtained nasal explants and isolated single GnRH-1 cells at 3, 4.5, 10, and 28 d in vitro (div). From 3 to 7 div, GnRH-1 cells display high cell motility and migrate off the explant tissue mass associated to the olfactory axons (Fig. 2, a and b); whereas, at 10 and 28 div, GnRH-1 cells display characteristics of postmigratory and fully differentiated neurons (Fueshko and Wray, 1994). The expression pattern of PlexinB1 and PlexinB2 transcripts, the only known Sema4D receptors expressed in the central nervous system, was examined within individual GnRH-1 neurons (Fig. 2, c and d). A higher percentage of GnRH-1 neurons expressed PlexinB1 between 3 and 4.5 div compared with 10 and 28 div (P < 0.005; Fig. 2 e), whereas all examined cells were negative for PlexinB2 (Fig. 2 e). These results indicate that PlexinB1 is expressed by GnRH-1 cells in a temporal window limited to their migratory phase.
Figure 2.
PlexinB1 expression changes as a function of age within GnRH-1 neurons. (a and b) Photomicrograph of a nasal explant labeled for GnRH-1 (brown) and peripherin (blue) at 7 div. From 3 to 7 div, GnRH-1 neurons migrate from the olfactory pit epithelia (OPE), following olfactory axons to the midline and off the explant into the periphery. At 7 div, a large population of GnRH-1 neurons is located in the periphery of the explant (b). Panel b is an enlarged view of the box in panel a. (c) Summary of the percentage of GnRH-1 neurons expressing each transcript (PlexinB1, PlexinB2, and GnRH-1) by single-cell RT-PCR analysis. (d) Representative gel of PCR products from individual GnRH-1 cells at different in vitro time points. (e) Approximately 60% of the examined migratory GnRH-1 neurons (n = 8) expressed PlexinB1, whereas only 9% of the postmigratory cells (n = 11) were positive for PlexinB1. PlexinB2 mRNA was not detected in GnRH-1 neurons at any stage analyzed. *, significantly different from other time points (P < 0.005). Statistical significance was determined using Wilcoxon-Mann-Whitney nonparametric test. Em, E17.5 embryo; NMC, nasal midline cartilage; W, water. Bars: (a) 200 μm, (b) 20 μm.
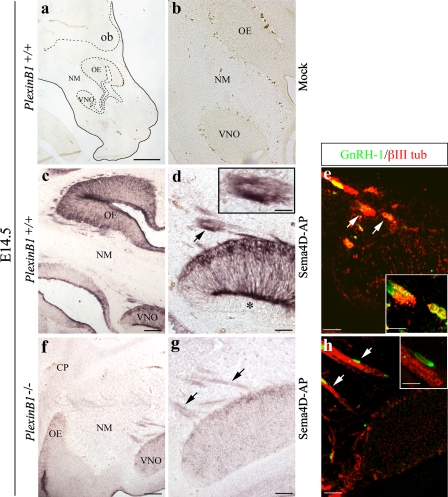
Sema4D binds the developing olfactory–vomeronasal system and migratory GnRH-1 cells
To assay whether cells in the olfactory–GnRH-1 system could interact with Sema4D, we performed binding studies by incubating mouse embryonic sections with Sema4D recombinant protein fused to AP (Sema4D-AP; Barberis et al., 2004). In brain sections derived from E14.5 PlexinB1+/+ mice, Sema4D-AP protein tightly bound to the VNO, OE, and olfactory/vomeronasal axons (Fig. 3, c and d), whereas no background binding was observed when Sema4D-AP incubation was omitted (Fig. 3, a and b). To determine the relative contribution of PlexinB1 and PlexinB2 in Sema4D binding, we performed the same experiments on sections of PlexinB1-null mice (Fazzari et al., 2007). Interestingly, the AP signal was much weaker in PlexinB1-deficient embryos (Fazzari et al., 2007) compared with wild-type (WT) animals (Fig. 3, f and g), which indicates that Sema4D binds the developing VNO–olfactory system, mainly through PlexinB1 receptor, even though a residual binding to PlexinB2 is also detectable. Moreover, an intense AP positivity was observed in clusters of cells emerging from the VNO in WT embryos (Fig. 3 d, arrow, see inset). We confirmed that these elements were indeed migratory GnRH-1 cells by double-staining an adjacent section for GnRH-1 and βIII tubulin (Fig. 3 e, arrows, see inset). Instead, in PlexinB1 knockout (KO) embryos, Sema4D was bound, though lightly, to the VNO cells and fibers, but not to migratory cell clusters (Fig. 3, g and h, arrows).
Figure 3.
Recombinant Sema4D-AP binds the embryonic vomeronasal–olfactory systems and GnRH-1 cells. (a and b) Sagittal sections of an E14.5 PlexinB1+/+ head incubated with mock-conditioned medium. No labeling is detectable in these samples. Dashed lines indicate, respectively, the presumptive VNO and OE, and the border between the nose and the forebrain. (c and d) Sema4D-AP binding on a sagittal section of an E14.5 PlexinB1+/+ forehead. Sema4D-AP strongly binds the OE as well as the olfactory nerves. (d) Sema4D-AP is also bound to the vomeronasal neurons, whereas the nonsensory VNO epithelium is depleted of any staining (asterisk). Intense labeling is evident along vomeronasal axons and migratory cells emerging out of the VNO (arrow, see inset). (e) An adjacent section, double-stained for GnRH-1 (green) and βIII-tubulin (red), indicates coexpression of these antigens in migratory cells (arrows), as shown by high-power confocal analysis (see inset), and suggests that these cells are distributed in a spatial pattern that parallels the Sema4D-AP staining (see d). (f and g) Sema4D-AP weakly binds the VNO–OE systems in E14.5 PlexinB1−/− foreheads. (g) High magnification of the VNO showing that the AP signal in the VNO and along the vomeronasal axons (arrows) is less intense in mutant than in WT embryos. (h) An adjacent section was double-stained for GnRH-1 (green) and βIII-tubulin (red) to show that in PlexinB1 KO embryos, Sema4D is not bound to GnRH-1 cells (arrows; see inset). CP, cribriform plate; NM, nasal mesenchyme. Bars: (a) 600 μm; (b) 160 μm; (c and f) 150 μm; (d, e, g, and h) 50 μm; (d, e, and h, insets) 20 μm.
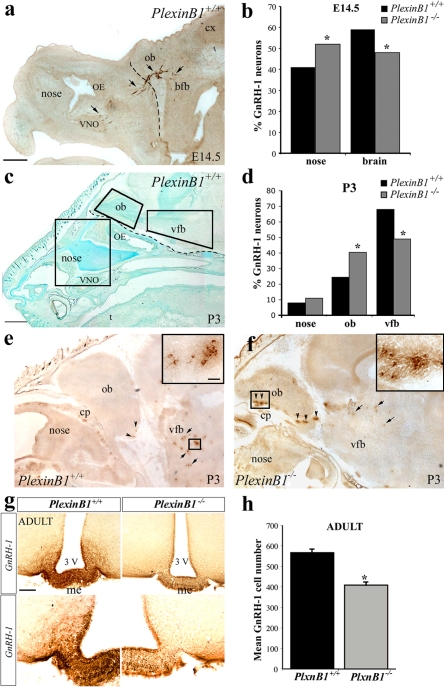
Altered migratory process of GnRH-1 neurons in PlexinB1−/− mice
To investigate the functional relevance of PlexinB1 in the development of the GnRH-1 neuroendocrine compartment, we examined the phenotype of PlexinB1-deficient mice as compared with WT. We analyzed the GnRH-1 cell distribution in the nasal compartment and in the brain of E14.5 embryos (Fig. 4, a and b) and of postnatal day 3 (P3) mice (Fig. 4, c and d). At E14.5, PlexinB1-null mice revealed a mild but statistically significant accumulation of GnRH-1 neurons in the nasal compartment (Fig. 4 b). Concomitantly, in PlexinB1−/− embryos, fewer GnRH-1 cells reached the brain compared with their WT counterparts (Fig. 4 b), in spite of a similar total number of GnRH-1–positive cells (WT: 1180 ± 56, n = 13; PlexinB1−/−: 1318 ± 69, n = 10; t test, P = 0.1). The migratory defect was more evident at P3. Brains of P3 PlexinB1−/− mice (n = 3) contained 20% fewer cells (P < 0.05) dispersed in the ventral forebrain compared with WT littermates (n = 4; Fig. 4, d–f, arrows). However, a 16% accumulation of GnRH-1 cells was observed at the level of the olfactory bulb in null animals (Fig. 4 f, arrowheads, see inset). The total number of GnRH-1–positive cells was not statistically different between WT mice and mutants (WT: 870 ± 92; PlexinB1−/−: 777 ± 84; t test, P = 0.5). The development of GnRH-1 and the olfactory systems are intimately entwined (Wray, 2002). If the course of the olfactory axons is disrupted, so is the migratory process of GnRH-1 cells. Evaluation with peripherin (Fueshko and Wray, 1994) immunohistochemistry of the olfactory/vomeronasal axons, along which the GnRH-1 neurons migrate, revealed no abnormalities in these transgenic mice (unpublished data).
Figure 4.
Migration of GnRH-1 neurons in PlexinB1−/− mice is delayed. (a and b) Photomicrograph showing GnRH-1 (brown) immunoreactivity in a sagittal section of an E14.5 WT embryo. The areas of analysis for GnRH-1 neuron location along the migratory pathway were the nasal compartment and the brain, respectively. The broken line in panel a indicates the boundary between these regions, and the arrows point to GnRH-1–labeled cells dispersed from the VNO to the basal forebrain (bfb). (b) Analysis of GnRH-1 neurons location revealed a significant accumulation of cells in the nasal region of PlexinB1 KO mice compared with WT. Consistently, fewer GnRH-1 cells were located in the brain of mutant embryos as compared with WT. Quantitative analysis of GnRH-1 neurons was performed as a function of location (nasal region and brain). GnRH-1 cell distribution is presented as the mean percentage of labeled cells located in the nose or in the brain of WT and PlexinB1 KO embryos (E14.5 WT: n = 13; E14.5 PlexinB1−/−: n = 10). Statistical significance was determined using Kruskal-Wallis nonparametric test (P < 0.05). (c) At P3, the GnRH-1 migratory process is over and the great majority of GnRH-1 neurons have reached the ventral forebrain (vfb). The distribution of these cells was analyzed in the nasal region, in the olfactory bulb (ob), and in the vfb at this developmental stage. (d) Approximately 16% of the entire population is accumulated in the ob, whereas fewer cells (∼20% less) are dispersed in the vfb compared with WT littermates. Quantitative analysis of GnRH-1 neurons was performed as a function of location (nose, ob, and vfb). GnRH-1 cell distribution is presented as the mean percentage of labeled cells in each compartment (P3 WT: n = 3; P3 PlexinB1−/−: n = 4). Statistical significance was determined using Kruskall-Wallis nonparametric test (P < 0.05). (e and f) High-power views of sagittal sections of P3 WT and mutants stained for GnRH-1. (e) In WT animals, GnRH-1 neurons are mainly located in the vfb (see arrows and insets, which show enlarged views of the boxed regions), whereas few cells are distributed in the ob area (arrowheads). (f) In PlexinB1−/− mice, abnormal clusters of GnRH-1 cells are located at the level of the cribriform plate (cp; see inset) and of the ob (arrowheads), and fewer cells reached the vfb region (arrows). (g) Photomicrograph showing GnRH-1 immunoreactivity in coronal sections of adult WT and PlexinB1-null mice. The principal fibers' projections of GnRH-1 neurons are to the median eminence (me), and this region showed a dramatic loss of GnRH-1 fibers in mutant mice (g, right) compared with WT littermates (left). The bottom photomicrographs show high-power views of the median eminence in WT and KO brains. (h) Quantitative analysis of GnRH-1 neuronal population in adult brains revealed a 30% reduction of these cells in brains of PlexinB1 mutants as compared with WT (WT: n = 3; PlexinB1−/−: n = 5). The total number of cells was calculated and combined to give group means ± SEM. *, statistical significance was determined using a paired t test (P < 0.0005). cx, cortex. Bars: (a) 500 μm, (c) 600 μm; (e and f) 300 μm, (e and f, insets) 30 μm; (g, top) 125 μm; (g, bottom) 62.5 μm.
To investigate whether the delay in the migratory process would be reflected in the size of the GnRH-1 neuronal population in adult animals, we collected alternating coronal sections from adult mutants and WT mice and examined them for GnRH-1 expression by immunohistochemistry. Our analysis showed that although in WT animals many GnRH-1–immunoreactive fibers innervated the median eminence, where these cells release the hormone into the pituitary portal capillary system, the density of these fibers was significantly reduced in PlexinB1−/− animals (Fig. 4 g). Moreover, a marked reduction in the total number of GnRH-1 neurons in the brains of PlexinB1−/− mice was detected (Fig. 4 h).
Sema4D induces Met activation in immortalized GnRH-1 cells
The manipulation of the GnRH-1 migratory system and functional studies on these neurons have been challenging because of their limited number (800 in mice and 1,000–2,000 in primates) and widely dispersed distribution in the olfactory system. The generation of immortalized GnRH-1 neurons has permitted the study of more immature, migratory neurons (NLT and GN11 cell lines; Radovick et al., 1991). In particular, GN11 cells display a remarkable motility in vitro, and they have been largely used to investigate the molecular mechanisms controlling the directional migration of GnRH-1 neurons (Giacobini et al., 2002; Cariboni et al., 2005, 2007).
We previously demonstrated that migratory GnRH-1 cells (both primary GnRH-1 neurons [Giacobini et al., 2007] and GN11 cells [Giacobini et al., 2002]) express Met and are functionally regulated by its ligand HGF. To exploit these cells for functional studies in response to Sema4D, we first verified that GN11 cells retained coexpression of Met and PlexinB1 receptors, as seen for their counterparts in vivo.
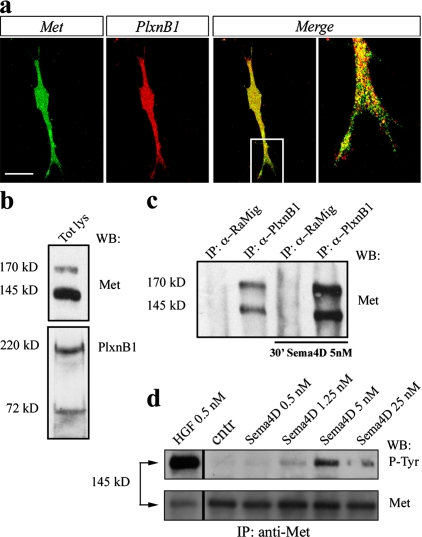
Double-labeling experiments followed by confocal microscopy analysis showed that Met and PlexinB1 receptors are codistributed throughout the cell surface of GN11 cells (Fig. 5 a) and often colocalize in dot-like clusters, as shown by single confocal plane images (Fig. 5 a, merge). Western blot analysis confirmed Met and PlexinB1 coexpression in GN11 cells (Fig. 5 b), and coimmunoprecipitation studies demonstrated PlexinB1–Met association in a molecular complex that increased upon Sema4D stimulation (Fig. 5 c). Moreover, when GN11 cells were treated for 30 min with either HGF (0.5 nM) or soluble Sema4D (0.5, 1.25, 5, and 25 nM), the Met receptor became tyrosine phosphorylated, which indicates activation of its signal transduction (Fig. 5 d). Interestingly, as known for other receptors, we observed a bell-shaped curve of tyrosine kinase activation in response to increasing concentrations of Sema4D (Fig. 5 d).
Figure 5.
Met and PlexinB1 associate in a complex, and Sema4D activates Met in GN11 cells. (a) Double immunofluorescence was performed on GN11 cells using antibodies to Met (green) and PlexinB1 (red) receptors. Confocal microscopy analysis revealed that a fraction of Met and PlexinB1 colocalize in dot-like clusters on the cell surface (see yellow dots in merged images). The far right panel is an enlarged view of the box region. Bar: (left three panels) 10 μm; (far right) 3 μm. (b) GN11 cell whole lysates were analyzed by WB with antibodies against Met (top) and PlexinB1 (bottom). GN11 cells endogenously express both receptors. We detected two specific bands corresponding to single-chain and cleaved heterodimeric forms of PlexinB1, as described previously in human and murine cells (Artigiani et al., 2003; Fazzari et al., 2007). (c) GN11 cells were treated for 30 min with 5 nM Sema4D (derived from Sema4D-expressing cells). Cell lysates were immunoprecipitated using anti-PlexinB1 or rabbit anti–mouse Ig (RaMig) as a control. Western blots were then probed with anti-Met antibodies. Data show that endogenous Met and PlexinB1 receptors basally coprecipitate in GN11 cells, and this association is increased after a 30-min stimulation with Sema4D. (d) GN11 cells were treated for 30 min with 0.5 nM HGF (∼50 ng/ml) or with semipurified conditioned medium containing Sema4D (at the indicated concentrations), and subjected to immunoprecipitation (IP) with anti-Met antibody. Probing the blot with antiphosphotyrosine antibodies showed that Sema4D treatment induces Met tyrosine phosphorylation at 1.25 and 5 nM concentrations, whereas 25-nM Sema4D stimulation did not activate Met. The control of the amount of the immunoprecipitated protein is shown on the bottom. Experiments were performed at least three times with similar results. The black lines indicate that intervening lanes have been spliced out.
Sema4D induces chemotaxis of GnRH-1 cells
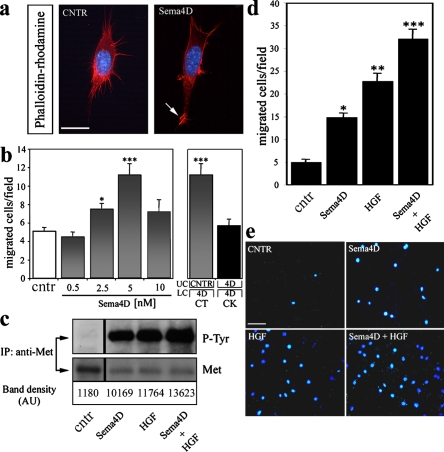
Semaphorins are known to regulate cytoskeletal dynamics (Tran et al., 2007). Interestingly, we observed that GN11 cells developed membrane ruffles and lamellipodial extensions within 10 min of Sema4D treatment, as visualized by F-actin staining (Fig. 6 a). Moreover, in treated cultures, cells extended a leading edge with actin ruffles (Fig. 6 a, arrow), a typical sign of local Rac activation, and depolymerized actin fibers at the rear edge (Fig. 6 a). These data indicated that Sema4D stimulation triggers cytoskeletal remodeling that is typical of migratory cells. To assess whether Sema4D can actually regulate the directional migration of GN11 cells, we performed microchemotaxis assays using Boyden's chamber. As shown in Fig. 6 b, soluble Sema4D induced a clear chemoattraction on GN11 cells as compared with cntr conditions. Sema4D exerted its effect in a concentration-dependent manner, with maximal response at 5 nM (Fig. 6 b). To elucidate whether the effect of Sema4D on GN11 motility was directional chemotaxis or simply the induction of random motility (i.e., chemokinesis), the cells were exposed to the same concentration of Sema4D (5 nM), added either in the lower chamber only, or in both upper and lower compartments of the Boyden's chamber. We observed a clear directional chemotaxis when the cells were exposed to a gradient of Sema4D diffusing from the lower chamber but only a modest induction of random cell motility when the factor was evenly distributed in both compartments (Fig. 6 b).
Figure 6.
Soluble Sema4D triggers GN11 cell motility. (a) Soluble Sema4D (5 nM) exposure (10 min) triggered dramatic cytoskeletal rearrangements in GN11 cells. Under such stimulation, we observed the formation of membrane ruffles and lamellipodial extensions (arrow) as visualized by F-actin staining. No morphological changes were observed when cells were stimulated with the concentrated mock-transfected supernatants (cntr, left). After the treatment, cells were stained with phalloidin-rhodamine (red) and with the nuclear dye DAPI (blue). (b) Chemotactic response of GN11 cells cultured for 24 h in serum-deprived medium before the chemomigration assay. A Boyden's chamber assay was performed in the absence (cntr, n = 11) or presence of increasing concentrations of soluble Sema4D (0.5 nM, n = 13; 2.5 nM, n = 11; 5 nM, n = 10; 10 nM, n = 7). Chemotactic (CT; n = 10) and chemokinetic (CK; n = 18) responses of GN11 cells were calculated in the presence of 5 nM Sema4D placed in the lower chamber (LC) or in the upper chamber (UC) and LC, respectively. (c) Sema4D and HGF cooperate to induce tyrosine phosphorylation of Met. GN11 cells were stimulated for 30 min with 5 nM Sema4D and 0.5 nM HGF at the doses known to induce the maximal biological responses in these cells, either alone or added together. Cells were subsequently extracted and immunoprecipitated with anti-Met antibodies. Blots were probed with anti-phosphotyrosine (top) and anti-mMet antibodies (middle). Quantification of tyrosine phosphorylation is shown on the bottom. Black lines indicate that intervening lanes have been spliced out. (d) A Boyden's chamber assay was performed in the presence of Sema4D and HGF, either alone or in combination. When 5 nM Sema4D and 0.5 nM HGF were added together, the migrated cells were increased as compared with all other treatments (cntr, n = 17; Sema4D, n = 26; HGF, n = 12; HGF + Sema4D, n = 19). (e) Representative images of a Boyden's chamber assay showing that GN11 cells migrated through 8-μm membrane pores, attracted by 5 nM Sema4D, 0.5 nM HGF, or both ligands added to the lower chamber. Cells that migrated to the other side of the filter were stained with DAPI and counted. Quantification of DAPI-stained nuclei is shown in panel d for comparison. Quantitative data of Boyden's chamber experiments (b and d) are mean ± SEM. ANOVA followed by Fisher's LSD post hoc analysis was used to compare groups. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. Bars: (a) 10 μm; (e) 50 μm.
During embryonic development, HGF expression has been documented in the nasal mesenchyme and in the developing olfactory bulb (Thewke and Seeds, 1996; Giacobini et al., 2007), with a spatiotemporal pattern that mimics Sema4D localization. Therefore, we examined if these two ligands could functionally cooperate or act in parallel in biological assays. We treated GN11 cells with optimal doses of Sema4D (5 nM) and HGF (0.5 nM), either alone or in combination. An additive effect for Met activation was observed when the two molecules were added together (Fig. 6 c). Moreover, the migratory response of GN11 cells in chemotaxis experiments was significantly increased, even though they were not additive, compared with the single ligand stimulations, which supports the notion that these molecules can act in parallel to induce neuronal cell migration through direct (HGF) and indirect (Sema4D) activation of Met (Fig. 6, d and e).
Sema4D-induced migration is mediated by Met
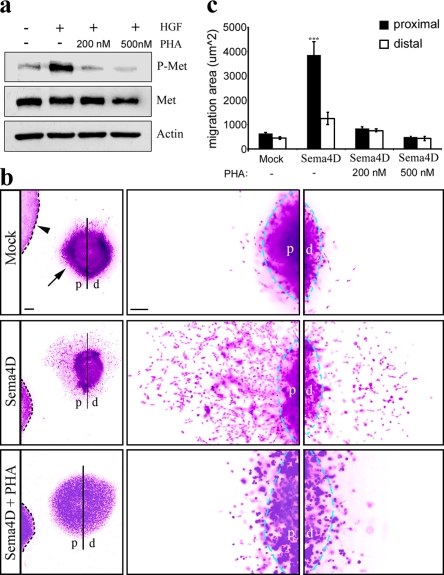
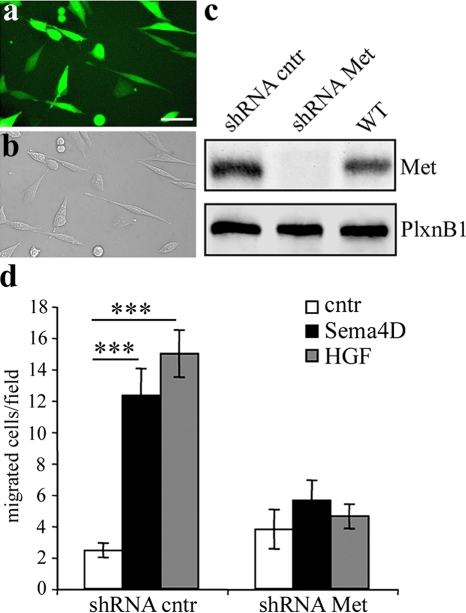
To evaluate whether Met is functionally required for Sema4D-induced motogenic effect, we performed three-dimensional matrix assays by coculturing for 72-h aggregates of GN11 cells together with aggregates of Sema4D-secreting or mock-transfected COS cells in the presence or absence of the specific Met kinase inhibitor PHA-665752 (PHA; Fig. 7 b). This highly specific small-molecule inhibitor has an IC50 for Met in the nanomolar range, >1,000 times higher than that for other receptor tyrosine kinases (Christensen et al., 2003). Treatment of GN11 cells with 200 nM and 500 nM PHA, respectively, curbed the phosphorylation of Met down to levels comparable to cntr conditions (Fig. 7 a). In coculture experiments, many more GN11 cells migrated out of the aggregate in the presence of a source of Sema4D, as compared with mock treatment (Fig. 7, b and c). Moreover, asymmetrical directional migration of GN11 cells pointed to a chemoattractive activity of Sema4D (Fig. 7, b and c). Such an attractive effect was prevented when the Met inhibitor PHA was applied to the culture medium (Fig. 7, b and c). These results were further confirmed by blocking Met activation with RNA interference technology. GN11 cells were infected with a lentiviral construct expressing both a Met-specific short hairpin RNA (shRNA) sequence (shRNA Met; Pennacchietti et al., 2003) and the marker GFP, under control of a separate promoter. As a specificity control, we used a construct expressing a mismatched Met shRNA sequence (shRNA cntr). Infection efficiency was confirmed by visualization of GFP expression under a fluorescent microscope (Fig. 8, a and b), and Met silencing was verified by Western blot analysis (Fig. 8 c). As shown in Fig. 8 c, infection with Met shRNA impaired the expression of Met in these cells, whereas cntr shRNA did not decrease Met expression compared with GN11 WT cells (WT). PlexinB1 expression was not affected after infection (Fig. 8 c). To determine the biological significance of Met in Sema4D-mediated GN11 cell migration, we performed migration assays using the Boyden's chamber. As expected, cntr cells migrated toward a source of HGF as well as of Sema4D (Fig. 8 d). In contrast, Met-silenced cells displayed impaired ability in responding to either of the two chemoattractive factors (Fig. 8 d). These results show that Sema4D-induced motogenic activity of GnRH-1 cells is mediated by PlexinB1–Met complex.
Figure 7.
GN11 cells migrate toward a source of Sema4D by activating Met. (a) Validation of the Met pharmacological inhibitor PHA. GN11 cells were stimulated for 20 min with HGF in the presence or absence of the indicated doses of PHA, then subjected to immunoblotting using antibodies against the phosphorylated form of Met (P-Met, top), Met (middle), and actin (bottom). As expected, HGF stimulation triggers a robust Met phosphorylation as compared with the untreated cell lysates. This effect was prevented when HGF was applied together with PHA. (b) Three-dimensional matrix assays were performed by coculturing GN11 cell aggregates (arrow) with COS cell aggregates (arrowhead) expressing or not expressing recombinant soluble Sema4D, and in the presence or absence of PHA. After 72 h, all cell aggregates were fixed and stained with the nuclear dye Sytox green, and photographs were taken for quantitative analysis. Photomicrographs have been digitally inverted, and a pseudocolor (purple) has been superimposed to highlight the cell distribution. GN11 cells migrate out of the aggregate symmetrically in the case of cntr COS cells (mock; top), whereas they are strongly attracted by Sema4D-expressing COS cell aggregates (Sema4D; middle). In these experimental conditions, GN11 cells migrate more than in cntr conditions, and they are more numerous in the quadrant facing the Sema4D source (proximal side). This effect was prevented when PHA was added in the medium (Sema4D + PHA; bottom). Bars: (left) 250 μm; (right) 50 μm. (c) Quantitative analysis was performed by measuring the area covered by GN11 cells outside the aggregates (mock, n = 5; Sema4D, n = 3; Sema4D + 200 nM PHA, n = 3; Sema4D + 500 nM PHA, n = 3). The migration area was measured in the proximal and distal quadrants as the surface covered by Sytox green nuclear staining, excluding the aggregate inner mass area, and expressed as mean ± SEM. ***, P < 0.0001 from two-way ANOVA analysis. d, distal side; p, proximal side.
Figure 8.
Met is required to elicit Sema4D-induced migration in GN11 cells. (a–c) Validation of the RNA interference lentiviral system. GN11 cells were infected with a lentiviral construct encoding for a Met shRNA sequence (shRNA Met) or for a mismatched Met shRNA (shRNA cntr), and for the GFP sequence. Infection efficiency was confirmed by visualization of GFP expression under the fluorescent microscope (a). Virtually all GN11 cells express GFP (a and b). Bar, 10 μm. (c) Western blot analysis of Met protein levels in total cell lysates demonstrated that, in Met shRNA–infected cells, Met was silenced (bottom). On the contrary, in shRNA cntr cells and in WT cells, Met levels were normal. PlexinB1 expression was comparable in all cell lysates (top). (d) shRNA cntr cells or shRNA Met cells were treated with 5 nM Sema4D or 0.5 nM HGF in Boyden's chamber assays (shRNA cntr: cntr, n = 28; Sema4D, n = 21; HGF, n = 23; shRNA Met: cntr, n = 13; Sema4D, n = 29; HGF, n = 26). Cells that migrated to the lower side of the filters were counted. GN11 cells in which Met was silenced were unable to migrate toward a Sema4D gradient. Results are expressed as mean ± SEM. ***, P < 0.0001 from two-way ANOVA.
Discussion
Although initially characterized as repulsive neuronal guidance cues, semaphorins are now considered versatile signals regulating cell migration (Casazza et al., 2007; Zhou et al., 2008). Semaphorin receptors falling into the PlexinB subfamily are expressed in striking patterns in the developing nervous system (Worzfeld et al., 2004; Perala et al., 2005; Pasterkamp et al., 2007); however, their functional relevance in vivo is poorly understood (Deng et al., 2007; Fazzari et al., 2007; Friedel et al., 2007).
We demonstrate here that Sema4D is robustly expressed along the GnRH-1 migratory route during mouse embryonic development, with higher levels in the final target areas. We found that the Sema4D receptor, PlexinB1, is expressed in migratory GnRH-1 cells exiting the olfactory neuroepithelium. These data were confirmed by single-cell RT-PCR analysis on primary GnRH-1 neurons isolated from nasal explants. PlexinB1 expression correlated with the migratory stage of these cells, being higher at 3 and 4.5 div and down-regulated in postmigratory GnRH-1 neurons (10 and 28 div). Recent work has shown a key requirement for PlexinB2 but not PlexinB1 in the patterning of the vertebrate nervous system in vivo (Deng et al., 2007). Our single-cell RT-PCR data provide evidence that GnRH-1 cells do not express PlexinB2 at any developmental stage analyzed, thus excluding a possible direct involvement of this receptor in the cellular response mediated by Sema4D. We further showed that Sema4D binds the developing VNO/OE epithelia, and that PlexinB1–Sema4D binding is much stronger than PlexinB2–Sema4D in these regions. Moreover, we demonstrated that in PlexinB1+/+ animals, Sema4D-AP–positive cell clusters migrating off the developing VNO are indeed GnRH-1 neurons.
Additional semaphorins and semaphorin receptors may play a role in this system. Recently, the Sema7A receptor, PlexinC1, was found to be expressed in migratory GnRH-1 cells in rats (Pasterkamp et al., 2007). Cariboni et al. (2007) reported a direct role for Sema3F as a repellent for the migration of GnRH-1 cells both in vivo and in vitro. Here, we find that PlexinB1-null mice show deficits consistent with reduced migration of GnRH-1 neurons. The development and organization of the olfactory axonal scaffold in PlexinB1−/− mice did not show abnormalities, which suggests that the migratory defect might be cell autonomous rather than dependent on alterations of the olfactory axonal pathway. In brains of adult mutant animals, the population of GnRH-1 cells was significantly reduced, by approximately one third compared with PlexinB1+/+ littermates. Consistently, the median eminence of these mice also appeared less innervated by GnRH-1 fibers as compared with WT. Our single-cell RT-PCR data revealed that GnRH-1 neurons differ in their PlexinB1 expression as a function of age. This might explain why only a fraction of neurons are affected by the lack of PlexinB1 gene. It is important to emphasize that a high degree of heterogeneity within the GnRH-1 neuron population has been documented extensively during the last years (Herbison et al., 2007; Schwarting et al., 2007). This seems to warrant the fact that no single genetic mutation may totally prevent these neurons from reaching their final destinations (Schwarting et al., 2007). Therefore, it is likely that failure of puberty in mammals may only occur when the majority of GnRH-1 neurons are affected by multigenic mutations (Herbison et al., 2007; Kim et al., 2007).
It is possible that in PlexinB1 mutant animals, the GnRH-1 cells that do not reach the final target areas at the right time may be eliminated by cell death. However, because of the small size of the GnRH-1 neuronal population and the limited temporal window during which apoptosis takes place, it was not possible to determine if this was the case. In other mutant mice, such as neuropilin 2−/−, netrin1−/−, and ephrin3–5−/− mice, there also were reported losses in immunoreactive GnRH-1 neuron numbers, but it was not feasible to determine the cause (Schwarting et al., 2004; Gamble et al., 2005; Cariboni et al., 2007).
In our study, we have also analyzed whether mice deficient for Sema4D would display a GnRH-1 phenotype similar to that seen in PlexinB1 KO mice. Sema4D-null mice do not show gross abnormalities in other tissues other than the immune system (Shi et al., 2000). Our quantitative analysis of the total number of GnRH-1 cells in adult brains revealed no differences in the size of this neuronal population between WT and mutant mice (WT: 654 ± 16.5, n = 4; Sema4D−/−: 660 ± 24, n = 3; t test, P = 0.8), which suggests that the absence of this semaphorin could be compensated by alternative, as yet unidentified, ligands for PlexinB1, or by the potentiation of other synergistic morphogenetic signals such as netrins, SDF-1, and HGF (Schwarting et al., 2001, 2004, 2006; Giacobini et al., 2007).
Sema4D-induced signaling pathways can use different mechanisms to elicit either repulsion/inhibition of cell motility or promigratory/chemoattractive responses (Zhou et al., 2008). In PC12 cells, Sema4D activation of PlexinB1 suppresses cell migration through inhibition of R-Ras and, subsequently, of β1-integrin activity (Oinuma et al., 2006). In other cellular systems, Sema4D effects seem to be gated through the association of the receptor PlexinB1 with tyrosine kinases such as Met, Ron, and ErbB2 (Giordano et al., 2002; Conrotto et al., 2004; Swiercz et al., 2004; Conrotto et al., 2005; Swiercz et al., 2007). Our data demonstrate for the first time in neurons a structural and functional association of PlexinB1 and Met receptors (holoreceptor complex) in GN11 cells. Moreover, Sema4D stimulation increases the holoreceptor complex formation. We observed a bell-shaped curve of Met tyrosine kinase activation in response to increasing concentrations of Sema4D, which parallels the bell-shaped curve of Sema4D-stimulated cell motility. One possible explanation for this observation is that Sema4D may be released by cells in monomeric and homodimeric forms (Elhabazi et al., 2001), this last being generally preferred when the semaphorin is present at high concentrations. It is not known which is the relative ability of the two forms in stimulating the formation of PlexinB1–Met complexes, but it is conceivable that their relative amount can modulate PlexinB1 homo- or heterodimerization.
We recently showed that HGF is expressed in the nasal compartment at early stages of development and that it promotes the migration of Met-expressing immortalized and primary GnRH-1 neurons (Giacobini et al., 2002, 2007). Notably, HGF expression pattern in the nasal region of mouse embryos parallels that of Sema4D. In this paper, we identified an additive effect of HGF and Sema4D for Met activation and a consequent increase in cell motility as compared with the single ligands stimulations. These results suggest that HGF and Sema4D, in vivo, might act in a combinatorial manner to allow a fine spatial tuning of GnRH-1 migration.
Finally, we demonstrated the requirement for Met in Sema4D-induced directional migration on GN11 cells by three-dimensional coculture experiments in the presence of a Met-specific kinase inhibitor and by chemotaxis assays in which we interfered with Met expression through RNA interference technology.
It has now become clear that GnRH-1 migratory process is not only dependent on the expression of specific receptors in these neurons but also on their ability to process guidance information provided by their surroundings. Our results shed light on a novel function for Sema4D as a long-range guidance cue in the GnRH-1 neuronal migration and identify PlexinB1–Met interaction as an essential requirement for Sema4D promigratory effect in this system. These data advance our understanding on the molecular mechanisms controlling the development of GnRH-1 system and may represent an important asset to elucidate the etiology of numerous forms of IHH.
Materials and methods
Animals
Experiments were conducted in accordance with current European Union and Italian law, under authorization of the Italian Ministry of Health, No. 66/99-A. CD-1 embryos (Charles River Laboratories) were harvested at E11.5, E12.5, E14.5, and E17.5 (plug day, E0.5) and used for RNA isolation, or fixed overnight at 4% PFA in 0.1 M phosphate buffer, pH 7.4, cryoprotected, then frozen and stored (−80°C) until processing for immunocytochemistry. PlexinB1−/−, Sema4D−/−, and their WT littermates have been described previously (Shi et al., 2000; Fazzari et al., 2007). Sema4D KO mice (Shi et al., 2000) were provided by H. Kikutani (Research Institute for Microbial Diseases, Osaka University, Osaka, Japan). Adult mice were anesthetized with an intraperitoneal injection of 200 mg/kg ketamine and perfused with 4% PFA.
Immunocytochemistry
Primary antisera used were against GnRH-1: SW-1, and rabbit (Rb) polyclonal (Wray et al., 1988), provided by S. Wray (National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD); PlexinB1: IC-2 (Artigiani et al., 2003), Rb, and H300, Rb, (Santa Cruz Biotechnology Inc.); Semaphorin 4D: CD100, mouse monoclonal (BD Biosciences), H300 Rb (Santa Cruz Biotechnology, Inc.) and BMA-12, rat IgG2a (eBioscience, Inc.); peripherin (No. AB1530, Rb; Millipore); NCAM (No. C9672, mouse monoclonal IgG; Sigma-Aldrich); Met (No. SP260 and No. H-190, rabbit polyclonal; and No. B-2, mouse monoclonal IgG; Santa Cruz Biotechnology, Inc.). Immunohistochemistry was performed as described previously (Giacobini et al., 2007). For double immunoperoxidase staining, the chromogen for the first antigen–antibody complex was DAB (brown precipitate; Kramer and Wray, 2000), whereas the chromogen for the second antigen–antibody complex was SG substrate (blue precipitate; Vector Laboratories). For double-immunofluorescence experiments, Alexa Fluor 488– and Alexa Fluor 568–conjugated secondary antibodies (Invitrogen) were used. F-actin was stained with rhodamine-labeled phalloidin (Invitrogen). Fluorescent specimens were mounted in 1,4-diazabicyclo[2.2.2]octane (DABCO; Sigma-Aldrich).
Image analysis
Images were captured using a microscope (Eclipse 80i; Nikon) and 2×/0.06 NA, 10×/0.30 NA, and 20×/0.50 NA objectives (Nikon) equipped with a digital camera (CX 9000; mbf Bioscience). For the observation coupled with confocal analysis, a laser-scanning Fluoview confocal system (IX70; Olympus) and 10×/0.30 NA, 20×/0.70 NA, and 60×/1.25 NA objectives (Olympus) were used. For the subsequent analysis of the digitized pictures, ImageJ (National Institutes of Health) and Photoshop (Adobe) software were used.
Nasal explants
Embryos were obtained from timed pregnant animals in accordance with current European Union and Italian law, under authorization of the Italian Ministry of Health, No. 66/99-A. Nasal pits of E11.5 staged CD-1 embryos (Charles River Laboratories) were isolated under aseptic conditions in Gey's Balanced Salt Solution (Invitrogen) enriched with glucose (Sigma-Aldrich) and maintained at 4°C until plating. Nasal explants were placed onto glass coverslips coated with 10 μl of chicken plasma (Cocalico Biologicals, Inc.). 10 μl thrombin (Sigma-Aldrich) was then added to adhere (thrombin/plasma clot) the explant to the coverslip. Explants were maintained in defined serum-free medium (Fueshko and Wray, 1994) containing 2.5 mg/ml fungizone (Sigma-Aldrich) at 37°C with 5% CO2 for up to 30 div. From culture day 3 to day 6, fresh media containing floxuridine (8 × 10−5 M; Sigma-Aldrich) was given to inhibit proliferation of dividing olfactory neurons and nonneuronal explant tissue. The media was changed to fresh serum-free medium twice a week.
Single GnRH-1 cell isolation and PCR analysis
Cell isolation and single-cell RT-PCR were performed as described previously (Giacobini et al., 2007). In brief, nasal explants were washed twice with 1× PBS (without Mg2+ or Ca2+), and single GnRH-1 cells were identified by their bipolar morphology, association with outgrowing axons, and location, and then isolated from nasal explants using a micropipette controlled by a micromanipulator (Narishige) connected to an inverted microscope (IX51; Olympus). cDNA was produced, and PCR amplification was performed as described previously (Kramer et al., 2000). The resulting product was phenol-chloroform–extracted and then ethanol precipitated, and an aliquot was run on a 1.5% agarose gel. 500 ng of E17.5 total brain RNA served as a positive control. All cDNA pools were initially screened for GnRH-1 (correct cell phenotype), β-tubulin, and L19 (two housekeeping genes, microtubule and ribosomal) using PCR. All cells used in this study were positive for all three transcripts. Images were captured using a gel documentation system (Gel Doc) and analyzed using Quantity One software (both from Bio-Rad Laboratories).
Analysis of GnRH-1 neurons in PlexinB1- and Sema4D-deficient mice
Serial sections (16-μm thickness) from PlexinB1−/− and WT E14.5 and P3 mice were cut (cryostat; Leica). Immunohistochemistry for GnRH-1 and peripherin was performed as described (see “Immunocytochemistry”). Quantitative analysis of GnRH-1 neuronal numbers was performed as a function of location (see Fig. 2). Sagittal free-floating sections (30-μm thickness) from adult PlexinB1−/−, Sema4D−/−, and respective WT mice were cut and labeled for GnRH-1 as described (see “Immunocytochemistry”) (GnRH-1 immunoreactivity was visualized using a DAB substrate). The total numbers of GnRH-1 cells were counted in each brain.
Cell cultures
All cell lines were grown in monolayer at 37°C in a 5% CO2, in DME (Invitrogen) containing 1 mM sodium pyruvate, 2 mM glutamine (Invitrogen), 100 μg/ml streptomycin, 100 U/ml penicillin, and 4,500 mg glucose (MP Biomedicals), supplemented with 10% FBS (Invitrogen). The medium was replaced at 2-d intervals. Subconfluent cells were routinely harvested by trypsinization and seeded in 58-cm2 dishes (100,000 cells). Only cells within six passages were used throughout the experiments.
Production of recombinant Sema4D expression in COS cells
Recombinant Sema4D fused to AP (Sema4D-AP) was harvested from the conditioned medium (CM) of stably transfected COS cells. Semipurified size-fractionated preparations of the semaphorin were obtained from these CM, as described previously (Barberis et al., 2004), and their content of Sema4D was determined by SDS-PAGE and Coomassie blue staining analysis. The purified recombinant soluble GST-fused Sema4D were produced and purified from COS cells as described previously (Giordano et al., 2002).
Binding assays
AP-binding experiments were performed on E14.5 PFA 4% fixed sections (16-μm thick). After a blocking step in PBS with 10% fetal bovine serum (Invitrogen), sections were incubated with supernatant from Sema4D-AP–transfected COS7 cells for 2 h and then fixed in 4% PFA for 20 min. Endogenous phosphatase were inactivated at 65°C for 30 min in PBS. Ligand bound to sections was revealed in Tris, pH 9.5, 5 mM MgCl2, 100 mM NaCl, 0.3 g/liter 4-nitro blue tetrazolium chloride, and 0.25 g/liter 5-bromo-4-chloro-3-indolyl-phosphate (Promega).
Cell stimulation
For in vivo phosphorylation experiments, subconfluent cells were grown overnight in serum-free medium and then stimulated with mock or HGF (0.5 nM), purified recombinant glutathione S-transferase–fused Sema4D (Sema4D-GST), or concentrated Sema4D fused with AP (Sema4D-AP) conditioned medium at the indicate concentrations for 30 min. For the cytoskeletal remodeling assay, GN11 cells are treated with mock medium or 5 nM Sema4D-AP for 10 min. For cell stimulation with mock treatment, we used appropriate dilution of elution buffer used to purify the fused Sema4D-GST from Sepharose glutathione chromatography or of concentrated 48-h conditioned media from mock-transfected COS cells.
Protein analysis
Except for the immunoprecipitation experiment, cells were lysed and proteins were extracted in boiling extraction buffer (125 mM Tris-HCl, pH 6.8, and 2.5% SDS). For immunoprecipitations, cells were lysed with modified RIPA buffer (20 mM Tris-HCl, pH 7.4, 5 mM EDTA, 150 mM sodium chloride, 10% glycerol, and 1% Triton X-100) in the presence of 1 μg/ml leupeptin, 3 μg/ml aprotinin, 1 μg/ml pepstatin, 2 mM phenylmethylsulphonyl fluoride, and 1 mM sodium orthovanadate. 1–2-mg proteins were immunoprecipitated for 2 h at 4°C with the indicated antibodies. After immunoprecipitation, high-stringency washes were performed (modified RIPA buffer containing 1 M lithium chloride). Western blots were performed according standard methods, and nitrocellulose membranes were probed with phosphotyrosine PY20 monoclonal antibody (BD Biosciences), Met (B-2, mouse monoclonal IgG; Santa Cruz Biotechnology Inc.; mouse anti–human Met monoclonal antibody; Invitrogen), anti–phospho-Met (P-Met rabbit polyclonal antibody; Cell Signaling Technology), PlexinB1 (A-8, mouse monoclonal IgG; Santa Cruz Biotechnology, Inc.), and mouse monoclonal β-actin (Sigma-Aldrich).
Boyden's chamber assay
The assay was performed using a 48-well Boyden's microchemotaxis chamber according to manufacturer's instructions (Neuro Probe). In brief, the cells grown in complete medium until subconfluence were harvested, and the suspension (105 cells/50 μl serum-free DME) was placed in the open-bottom wells of the upper compartment. Each pair of wells was separated by a polyvinylpyrrolidone-free polycarbonate porous membrane (8-μm pores) precoated with gelatin (0.2 mg/ml in PBS). The lower chamber of the Boyden's apparatus was filled with DME FBS 0% in the absence or in the presence of 0.5 nM HGF/scatter factor and 0.5, 2.5, 5, and 10 nM Sema4D.
Chemokinesis (stimulation of increased random cell motility) was distinguished from chemotaxis by placing the same concentration of the factor in both the upper and lower wells of the Boyden's chamber, thereby eliminating the chemical gradient. After 4 h of incubation, cells attached to the upper side of the filter were mechanically removed. Cells that migrated to the lower side were fixed and stained with either DAPI nuclear dye or Diff Quick staining kit (Dade Behring AG) according to the manufacturer's instructions. The stained cells were photographed and counted.
Cell aggregates
Three-dimensional matrix assays were performed by coculturing GN11 cell aggregates with COS cell aggregates expressing or not expressing recombinant soluble Sema4D in the presence or absence of the PHA inhibitor. For the aggregates, cells were collected by trypsinization, resuspended in 5 μl (GN11) or 20 μl (COS) of growth factor–free Matrigel (BD Biosciences; 106 cells/ml for both GN11 and COS cells), and placed on the lid of a culture dish. As the droplets of cell aggregates were set, they were plated onto a growth factor–reduced Matrigel-coated Millicell inserts (Millipore) and kept in culture for 72 h. For analysis, the aggregates were fixed with PFA 4% for 40 min and stained with 167 nM of the nuclear marker Sytox green (Invitrogen) for 30 min at room temperature.
Generation of plasmids, lentivirus vector preparation, and in vitro cell transduction
The cntr/Met shRNA plasmids have been described previously (Corso et al., 2008). Vector stocks were produced as described previously (Vigna and Naldini, 2000). The concentration of viral p24 antigen was assessed using the HIV-1 p24 core profile ELISA kit (PerkinElmer) according to the manufacturer's instructions. Cells were transduced overnight with 0.2 μg/ml of p24 equivalents in the presence of 8 μg/ml polybrene (Sigma-Aldrich).
Statistical analysis
For comparison of multiple groups, statistical significance was determined using a two-way analysis of variance (ANOVA; for Gaussian distributed data) followed by Fisher's least significant difference post-hoc analysis or Kruskal-Wallis (nonparametric) tests. For comparison between paired normally distributed data, a paired t test was used. The significance level was set at P < 0.05. Data groups are indicated as mean ± SEM (for normally distributed data). A nonparametric unpaired t test (Wilcoxon-Mann-Whitney) was used to compare percentages of primary GnRH-1 cells at different in vitro stages expressing specific transcripts. The significance level was set at P < 0.01.
Acknowledgments
We are grateful to P. Fazzari and C. Giampietro for experimental advice and helpful discussions and to H. Kikutani for providing us with the Sema4D−/− mice.
The work was supported by Compagnia di San Paolo (Neurotransplant 2004.2019), Regione Piemonte Ricerca Scientifica Applicata (Comitato Interministeriale Programmazione Economica project No. A23), Programmi di ricerca di Rilevante Interesse Nazionale (PRIN 2007 to A. Fasolo), and Regione Piemonte Ricerca Sanitaria Finalizzata (to L. Tamagnone), Ricerca Finanziata Università di Torino 2007. The authors declare that there are no conflicts of interest regarding this paper.
P. Giacobini and A. Messina contributed equally to this paper.
Abbreviations used in this paper: ANOVA, analysis of variance; cntr, control; div, days in vitro; E, embryonic day; GnRH-1, gonadotropin hormone–releasing hormone-1; HGF, hepatocyte growth factor; IHH, idiopathic hypogonadotropic hypogonadisms; KO, knockout; NCAM, neural cell adhesion molecule; OE, olfactory epithelium; P, postnatal day; PHA, PHA-665752; Sema4D, Semaphorin4D; shRNA, short hairpin RNA; VNO, vomeronasal organ; WT, wild type.
References
- Artigiani, S., D. Barberis, P. Fazzari, P. Longati, P. Angelini, J.W. van de Loo, P.M. Comoglio, and L. Tamagnone. 2003. Functional regulation of semaphorin receptors by proprotein convertases. J. Biol. Chem. 278:10094–10101. [DOI] [PubMed] [Google Scholar]
- Barberis, D., S. Artigiani, A. Casazza, S. Corso, S. Giordano, C.A. Love, E.Y. Jones, P.M. Comoglio, and L. Tamagnone. 2004. Plexin signaling hampers integrin-based adhesion, leading to Rho-kinase independent cell rounding, and inhibiting lamellipodia extension and cell motility. FASEB J. 18:592–594. [DOI] [PubMed] [Google Scholar]
- Calof, A.L., and D.M. Chikaraishi. 1989. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 3:115–127. [DOI] [PubMed] [Google Scholar]
- Cariboni, A., S. Rakic, A. Liapi, R. Maggi, A. Goffinet, and J.G. Parnavelas. 2005. Reelin provides an inhibitory signal in the migration of gonadotropin-releasing hormone neurons. Development. 132:4709–4718. [DOI] [PubMed] [Google Scholar]
- Cariboni, A., J. Hickok, S. Rakic, W. Andrews, R. Maggi, S. Tischkau, and J.G. Parnavelas. 2007. Neuropilins and their ligands are important in the migration of gonadotropin-releasing hormone neurons. J. Neurosci. 27:2387–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casazza, A., P. Fazzari, and L. Tamagnone. 2007. Semaphorin signals in cell adhesion and cell migration: functional role and molecular mechanisms. Adv. Exp. Med. Biol. 600:90–108. [DOI] [PubMed] [Google Scholar]
- Chen, G., J. Sima, M. Jin, K.Y. Wang, X.J. Xue, W. Zheng, Y.Q. Ding, and X.B. Yuan. 2008. Semaphorin-3A guides radial migration of cortical neurons during development. Nat. Neurosci. 11:36–44. [DOI] [PubMed] [Google Scholar]
- Christensen, J.G., R. Schreck, J. Burrows, P. Kuruganti, E. Chan, P. Le, J. Chen, X. Wang, L. Ruslim, R. Blake, et al. 2003. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 63:7345–7355. [PubMed] [Google Scholar]
- Conrotto, P., S. Corso, S. Gamberini, P.M. Comoglio, and S. Giordano. 2004. Interplay between scatter factor receptors and B plexins controls invasive growth. Oncogene. 23:5131–5137. [DOI] [PubMed] [Google Scholar]
- Conrotto, P., D. Valdembri, S. Corso, G. Serini, L. Tamagnone, P.M. Comoglio, F. Bussolino, and S. Giordano. 2005. Sema4D induces angiogenesis through Met recruitment by Plexin B1. Blood. 105:4321–4329. [DOI] [PubMed] [Google Scholar]
- Corso, S., C. Migliore, E. Ghiso, G. De Rosa, P.M. Comoglio, and S. Giordano. 2008. Silencing the MET oncogene leads to regression of experimental tumors and metastases. Oncogene. 27:684–693. [DOI] [PubMed] [Google Scholar]
- De Carlos, J.A., L. Lopez-Mascaraque, and F. Valverde. 1996. Early olfactory fiber projections and cell migration into the rat telencephalon. Int. J. Dev. Neurosci. 14:853–866. [DOI] [PubMed] [Google Scholar]
- Deng, S., A. Hirschberg, T. Worzfeld, J.Y. Penachioni, A. Korostylev, J.M. Swiercz, P. Vodrazka, O. Mauti, E.T. Stoeckli, L. Tamagnone, et al. 2007. Plexin-B2, but not Plexin-B1, critically modulates neuronal migration and patterning of the developing nervous system in vivo. J. Neurosci. 27:6333–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhabazi, A., S. Delaire, A. Bensussan, L. Boumsell, and G. Bismuth. 2001. Biological activity of soluble CD100. I. The extracellular region of CD100 is released from the surface of T lymphocytes by regulated proteolysis. J. Immunol. 166:4341–4347. [DOI] [PubMed] [Google Scholar]
- Fazzari, P., J. Penachioni, S. Gianola, F. Rossi, B.J. Eickholt, F. Maina, L. Alexopoulou, A. Sottile, P.M. Comoglio, R.A. Flavell, and L. Tamagnone. 2007. Plexin-B1 plays a redundant role during mouse development and in tumour angiogenesis. BMC Dev. Biol. 7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel, R.H., G. Kerjan, H. Rayburn, U. Schuller, C. Sotelo, M. Tessier-Lavigne, and A. Chedotal. 2007. Plexin-B2 controls the development of cerebellar granule cells. J. Neurosci. 27:3921–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fueshko, S., and S. Wray. 1994. LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev. Biol. 166:331–348. [DOI] [PubMed] [Google Scholar]
- Gamble, J.A., D.K. Karunadasa, J.R. Pape, M.J. Skynner, M.G. Todman, R.J. Bicknell, J.P. Allen, and A.E. Herbison. 2005. Disruption of ephrin signaling associates with disordered axophilic migration of the gonadotropin-releasing hormone neurons. J. Neurosci. 25:3142–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacobini, P., C. Giampietro, M. Fioretto, R. Maggi, A. Cariboni, I. Perroteau, and A. Fasolo. 2002. Hepatocyte growth factor/scatter factor facilitates migration of GN-11 immortalized LHRH neurons. Endocrinology. 143:3306–3315. [DOI] [PubMed] [Google Scholar]
- Giacobini, P., A. Messina, S. Wray, C. Giampietro, T. Crepaldi, P. Carmeliet, and A. Fasolo. 2007. Hepatocyte growth factor acts as a motogen and guidance signal for gonadotropin hormone-releasing hormone-1 neuronal migration. J. Neurosci. 27:431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano, S., S. Corso, P. Conrotto, S. Artigiani, G. Gilestro, D. Barberis, L. Tamagnone, and P.M. Comoglio. 2002. The semaphorin 4D receptor controls invasive growth by coupling with Met. Nat. Cell Biol. 4:720–724. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Martinez, D., Y. Hu, and P.M. Bouloux. 2004. Ontogeny of GnRH and olfactory neuronal systems in man: novel insights from the investigation of inherited forms of Kallmann's syndrome. Front. Neuroendocrinol. 25:108–130. [DOI] [PubMed] [Google Scholar]
- Herbison, A.E., R. Porteous, J.R. Pape, J.M. Mora, and P.R. Hurst. 2007. Gonadotropin-releasing hormone (GnRH) neuron requirements for puberty, ovulation and fertility. Endocrinology. 149:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., Y. Hu, S. Cadman, and P. Bouloux. 2007. Diversity in fibroblast growth factor receptor 1 regulation: learning from the investigation of Kallmann syndrome. J. Neuroendocrinol. 20:141–163. [DOI] [PubMed] [Google Scholar]
- Kramer, P.R., and S. Wray. 2000. Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev. 14:1824–1834. [PMC free article] [PubMed] [Google Scholar]
- Kramer, P.R., R. Krishnamurthy, P.J. Mitchell, and S. Wray. 2000. Transcription factor activator protein-2 is required for continued luteinizing hormone-releasing hormone expression in the forebrain of developing mice. Endocrinology. 141:1823–1838. [DOI] [PubMed] [Google Scholar]
- Oinuma, I., H. Katoh, and M. Negishi. 2006. Semaphorin 4D/Plexin-B1–mediated R-Ras GAP activity inhibits cell migration by regulating β(1) integrin activity. J. Cell Biol. 173:601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasterkamp, R.J., J.J. Peschon, M.K. Spriggs, and A.L. Kolodkin. 2003. Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature. 424:398–405. [DOI] [PubMed] [Google Scholar]
- Pasterkamp, R.J., S.M. Kolk, A.J. Hellemons, and A.L. Kolodkin. 2007. Expression patterns of semaphorin7A and plexinC1 during rat neural development suggest roles in axon guidance and neuronal migration. BMC Dev. Biol. 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchietti, S., P. Michieli, M. Galluzzo, M. Mazzone, S. Giordano, and P.M. Comoglio. 2003. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 3:347–361. [DOI] [PubMed] [Google Scholar]
- Perala, N.M., T. Immonen, and H. Sariola. 2005. The expression of plexins during mouse embryogenesis. Gene Expr. Patterns. 5:355–362. [DOI] [PubMed] [Google Scholar]
- Radovick, S., S. Wray, E. Lee, D.K. Nicols, Y. Nakayama, B.D. Weintraub, H. Westphal, G.B. Cutler Jr., and F.E. Wondisford. 1991. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc. Natl. Acad. Sci. USA. 88:3402–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskams, A.J., X. Cai, and G.V. Ronnett. 1998. Expression of neuron-specific beta-III tubulin during olfactory neurogenesis in the embryonic and adult rat. Neuroscience. 83:191–200. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda, M., and D.W. Pfaff. 1989. Origin of luteinizing hormone-releasing hormone neurons. Nature. 338:161–164. [DOI] [PubMed] [Google Scholar]
- Schwarting, G.A., C. Kostek, E.P. Bless, N. Ahmad, and S.A. Tobet. 2001. Deleted in colorectal cancer (DCC) regulates the migration of luteinizing hormone-releasing hormone neurons to the basal forebrain. J. Neurosci. 21:911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarting, G.A., D. Raitcheva, E.P. Bless, S.L. Ackerman, and S. Tobet. 2004. Netrin 1-mediated chemoattraction regulates the migratory pathway of LHRH neurons. Eur. J. Neurosci. 19:11–20. [DOI] [PubMed] [Google Scholar]
- Schwarting, G.A., T.R. Henion, J.D. Nugent, B. Caplan, and S. Tobet. 2006. Stromal cell-derived factor-1 (chemokine C-X-C motif ligand 12) and chemokine C-X-C motif receptor 4 are required for migration of gonadotropin-releasing hormone neurons to the forebrain. J. Neurosci. 26:6834–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarting, G.A., M.E. Wierman, and S.A. Tobet. 2007. Gonadotropin-releasing hormone neuronal migration. Semin. Reprod. Med. 25:305–312. [DOI] [PubMed] [Google Scholar]
- Shi, W., A. Kumanogoh, C. Watanabe, J. Uchida, X. Wang, T. Yasui, K. Yukawa, M. Ikawa, M. Okabe, J.R. Parnes, et al. 2000. The class IV semaphorin CD100 plays nonredundant roles in the immune system: defective B and T cell activation in CD100-deficient mice. Immunity. 13:633–642. [DOI] [PubMed] [Google Scholar]
- Swiercz, J.M., R. Kuner, J. Behrens, and S. Offermanns. 2002. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 35:51–63. [DOI] [PubMed] [Google Scholar]
- Swiercz, J.M., R. Kuner, and S. Offermanns. 2004. Plexin-B1/RhoGEF-mediated RhoA activation involves the receptor tyrosine kinase ErbB-2. J. Cell Biol. 165:869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz, J.M., T. Worzfeld, and S. Offermanns. 2007. ERBB-2 and met reciprocally regulate cellular signaling via plexin-B1. J. Biol. Chem. 283:1893–1901. [DOI] [PubMed] [Google Scholar]
- Thewke, D.P., and N.W. Seeds. 1996. Expression of hepatocyte growth factor/scatter factor, its receptor, c-met, and tissue-type plasminogen activator during development of the murine olfactory system. J. Neurosci. 16:6933–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, T.S., A.L. Kolodkin, and R. Bharadwaj. 2007. Semaphorin regulation of cellular morphology. Annu. Rev. Cell Dev. Biol. 23:263–292. [DOI] [PubMed] [Google Scholar]
- Vigna, E., and L. Naldini. 2000. Lentiviral vectors: excellent tools for experimental gene transfer and promising candidates for gene therapy. J. Gene Med. 2:308–316. [DOI] [PubMed] [Google Scholar]
- Worzfeld, T., A.W. Puschel, S. Offermanns, and R. Kuner. 2004. Plexin-B family members demonstrate non-redundant expression patterns in the developing mouse nervous system: an anatomical basis for morphogenetic effects of Sema4D during development. Eur. J. Neurosci. 19:2622–2632. [DOI] [PubMed] [Google Scholar]
- Wray, S. 2002. Development of gonadotropin-releasing hormone-1 neurons. Front. Neuroendocrinol. 23:292–316. [DOI] [PubMed] [Google Scholar]
- Wray, S., B.H. Gahwiler, and H. Gainer. 1988. Slice cultures of LHRH neurons in the presence and absence of brainstem and pituitary. Peptides. 9:1151–1175. [DOI] [PubMed] [Google Scholar]
- Wray, S., P. Grant, and H. Gainer. 1989. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc. Natl. Acad. Sci. USA. 86:8132–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K., S.A. Tobet, J.E. Crandall, T.P. Jimenez, and G.A. Schwarting. 1995. The migration of luteinizing hormone-releasing hormone neurons in the developing rat is associated with a transient, caudal projection of the vomeronasal nerve. J. Neurosci. 15:7769–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., R.A. Gunput, and R.J. Pasterkamp. 2008. Semaphorin signaling: progress made and promises ahead. Trends Biochem. Sci. 33:161–170. [DOI] [PubMed] [Google Scholar]