Abstract
An estimated 1-3% of individuals within the United States are diagnosed with mental retardation (MR), yet the cause is unknown in nearly 50% of the patients. While several environmental, genetic and combined teratogenetic etiologies have been identified, many causative genes remain to be identified. Furthermore, the pathogenetic mechanisms underlying MR are known for very few of these genes. Males have a much higher incidence of MR implicating genes on the X-chromosome. We have recently identified a novel gene, SIZN1, on the X-chromosome and showed that it functions in modulating the BMP signaling pathway. Furthermore, we have shown this gene is necessary for basal forebrain cholinergic neuron (BFCN) specific gene expression. Given that cognitive function is impaired when BFCNs are lost or functionally disrupted, we undertook a screen of cognitively impaired males for SIZN1 mutations. We report on four different sequence variants in SIZN1 in 11 individuals with nonsyndromic X-linked mental retardation. Our data implicate SIZN1 as a candidate gene for X-linked mental retardation and/or as a neurocognitive functional modifier.
Keywords: BMP, SIZN1(ZCCHC12), forebrain cholinergic neuron, X-linked mental retardation
INTRODUCTION
One to 3% of children in the United States are diagnosed with mental retardation (MR) which is defined as having an IQ below 70 [Leonard and Wen, 2002; Roeleveld et al., 1997; Ropers, 2006; Ropers and Hamel, 2005]. Although the etiology of MR is heterogeneous, a genetic basis or predisposition is assumed in the majority of cases [Leonard and Wen, 2002]. Given that the incidence of MR is higher in males when compared to females [Leonard and Wen, 2002], it has been assumed many genes causing or contributing to cognitive function reside on the X-chromosome [Lehrke, 1972]. To date, over 80 X-linked mental retardation (XLMR) genes have been identified on the X-chromosome and this list continues to grow [Ropers, 2006; Ropers and Hamel, 2005]. Despite this rapid identification of XLMR genes, the biological function is incompletely understood for most and the neurobiological significance known for even fewer.
We have recently identified a new gene that positively modulates bone morphogenetic protein (BMP) signaling [Cho et al., 2008]. This gene, which we named SIZN1(Smad-interacting zinc finger protein 1;also annotated in GenBank as ZCCHC12), is expressed in the developing basal forebrain cholinergic neurons (BFCN) which are known to play an essential role in cognition [Baxter and Chiba, 1999]. The loss of BFCNs has been correlated with cognitive decline in some neurodegenerative diseases such as Alzheimer disease [Gallagher and Colombo, 1995; Granholm et al., 2000; Sarter and Bruno, 2004]. Furthermore, the neurotrophin tyrosine kinase receptor is necessary for BFCN axon projection development and mutations in this gene result in the congenital insensitivity to pain syndrome, a syndrome that includes mental retardation as a feature [Counts et al., 2004; Ginsberg et al., 2006; Verpoorten et al., 2006]. These findings, along with its localization on the X-chromosome, make SIZN1 an excellent XLMR candidate gene.
To test this hypothesis, we studied a large cohort of males with MR. Mutational analysis revealed two missense mutations and two deletions in the 3’ UTR of SIZN1. The known function of SIZN1 in modulating BMP signaling along with its functional role in BFCN gene expression provide further evidence for BFCN in cognition and a likely influence of SIZN1 in MR.
METHODS
Patient information
A group of 729 unrelated male patients with MR of unknown cause, all of whom receive services through the South Carolina Department of Disabilities and Special Needs (DDSN), were screened for mutations in the SIZN1 gene. This group was comprised of 328 blacks, 340 whites, and 61 from other backgrounds or with unknown racial or ethnic status. All patients had IQs ranging from 36 to 63. All patients had undergone both clinical genetics and dysmorphology examinations and 97% had been karyotyped. In addition, 86% of patients had previously undergone molecular testing for the trinucleotide repeat expansion in the FMR1 gene and were found to be negative. An additional 7% were negative by cytogenetic testing for Fragile X.
Incorporation PCR SSCP (IPS)
Genomic DNA was isolated from lymphocytes [Schwartz et al., 1990]. IPS was performed using 10 ng of genomic DNA in a total volume of 10 μl containing 1 × PCR reaction buffer with 1 μmol/l of each primer, 50 μmol/l of dNTPs, 0.05 μCi of α-32P dCTP (3000 Ci/mmol, PE), and 0.5 units of Go Taq DNA polymerase (Promega) [Sossey-Alaoui et al., 1999]. The single coding exon of the gene was covered with 4 overlapping primer pairs providing complete coverage of the coding sequence (SIZN1-1F/SIZN1-1R: 5′-TACCCTATCGTCGTCAGT-3′ and 5′-ATGGCTCGCAAGAAATC-3′; SIZN1-2F/SIZN1-2R: 5′-GGCGACCAACCCTAACCTAAGT-3′ and 5′-GCCGCTCCTGCTCATTTG-3′; SIZN1-3F/SIZN1-3R: 5′-TAAGGATTTTCTCAGGATGTATG-3′ and 5′-GGAATCAATGACCAGCACTT-3′; SIZN1-4F/SIZN1-4R: 5′-TCAGAGACAGGGCCAGACC-3′ and 5′-GGCCACTCTTGACCTCTCCAT-3′). PCR conditions were as follows:(94° C, 3 min), followed by 30 cycles of denaturation (94° C, 20 s), annealing (60° C or 55° C, 30 s), extension (72° C, 30 s), and final extension (72° C, 5 min). Following PCR, 10 μl of IPS loading dye (95% formamide, 10 mmol/l NaOH, 0.25% bromophenol blue, 0.25% xylene cyanol) was added, the samples were denatured at 95° C for 5 minutes and loaded on a 0.5 × MDE gel (FMC) prepared in 0.6 × TBE. The gel was run at 8 W for 20-24 hours at room temperature, dried and visualized using the BIOMAXTM MS films (Eastman Kodak).
Metabolic labelling of proteins and immunoprecipitation
Sizn1 half-life was determined by immunoprecipitation of pulse-labeled proteins. After transfection of expression constructs into C2C12 cells in a 100 mm dish, cells were split onto six 60 mm dishes one day later. Cells were incubated with 50 μCi of 35S-labelled methionine/cystine (Amersham) in 1 ml of methionine/cystine-free DMEM (Invitrogen) for 2 hr at 37°C to pulse label Sizn1 proteins. After this time, the medium was replaced with fully supplemented, unlabelled DMEM and an initial sample taken to determine the incorporation of the 35S-labelled methionine/cystine at the start of the experiment. Five further samples were taken during the next 18 h chase period as showed in Fig. 2. Immunoprecipitations were carried out as described[Aicardi]. Gels were dried and exposured on X-ray film. The band intensities corresponding to Sizn1 or its mutant were calculated with NIH image software. The Sizn1 half-life was calculated by linear regression analysis of log (intensity per band) against time.
Figure 2.
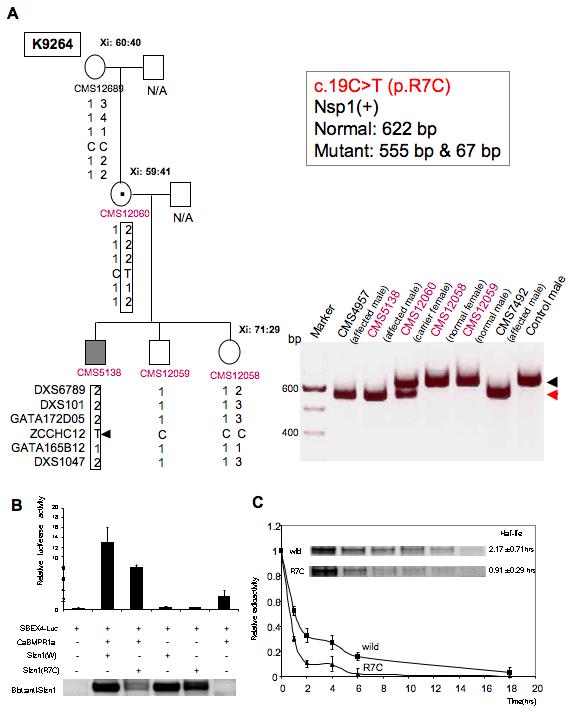
X chromosome marker analysis and X inactivation (Xi) data are shown for the c.19C>T(p.R7C) sequence variation in SIZN1. (A) c.19C>T (p.R7C) was found in family, K9264. Mutation analysis in family K9264 and two unrelated patients with MR (CMS4957 and CMS7492). A 622-bp SIZN1 exon 4 PCR product amplified from genomic DNA was digested with Nsp1 restriction enzyme to distinguish between the normal (622-bp black arrowhead) and the mutant (555-bp red arrowhead) alleles. A 67-bp fragment generated by the restriction digestion in affected individuals is not shown. Restriction digestion analysis shows affected males carry the c.19C→T mutation. (B) The p.R7C mutant Sizn1 shows reduced activity in SBEx4 reporter assay. Western blot analysis of the SIZN1 protein shows the reduced activity correlates with reduced protein levels (approximately 40% reduction; n=8). (C) Pulse-chase analysis of Sizn1 Proteins (n=3). Error bar are standard deviation (SD) in luciferase assay.
Allele Specific PCR
In order to test if the change was a rare polymorphism, DNA from normal controls were screened using Allele Specific PCR [Papp et al., 2003]. Primer SIZN1-MUT (5′-TGGCTAGCATCATTGGAT-3′) was utilized in conjunction with primer SIZN1-1R (see primer pairs above) to amplify only the allele containing the alteration (c.19C>T). In primer SIZN1-MUT, in addition to having a T as the last nucleotide, the third to last nucleotide was changed from a C to a G. This increased the specificity of the amplification of the mutant allele. Additionally, control primers were designed at the ACE2 locus on the X chromosome (5′-CACAAAGGTGACAATGG-3′ and 5′- GTTCTGTGAGACTGTTCATC-3′). PCR conditions were as given above with an annealing temperature of 55° C.
Cell culture and luciferase assay
C2C12 cell lines were cultured in DMEM containing 15% fetal bovine serum (FBS;Hyclone) at 37°C and 5% CO2. Luciferase assay was performed as described in previous paper [Cho et al., 2008]. pMIW/Sizn1-myc (each 0.5 μg) was used (wild type or p.R7C). Transfection efficiency was standardized using β-galactosidase activity. All assays were performed in duplicate.
RESULTS
Based on the mouse Sizn1 sequence, a homologous gene search with EnsEmbl human genome browser identified SIZN1 at Xq24 between IL13RA and RNF127 (also annotated in GenBank as LONRF3) (Fig.1). SIZN2, a SIZN1 homolog, is localized at Xq22.2 (Fig.1). SIZN1 was found to share 74 % sequence identity with mouse Sizn1 (Fig. 1) and 84 % sequence identity with SIZN2 [Cho et al., 2008]. Based on its chromosomal locations and our biochemical and functional data, we postulated SIZN1 to be a candidate XLMR gene.
Figure 1.
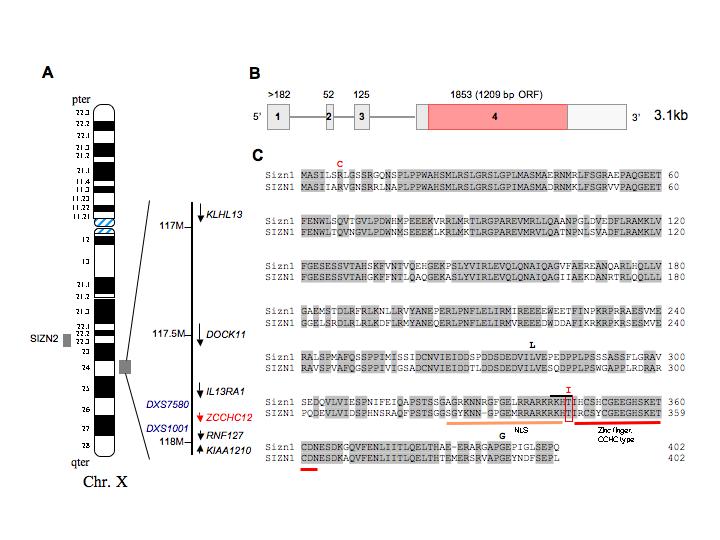
A. Schematic diagram of the human X chromosome showing the SIZN1(ZCCHC12) locus. B. Genome structure on human X-chromosome. SIZN1 has 4 exons with the entire coding region contained within exon 4. C. Alignment between mouse and human Sizn1 protein. Location of sequence variations, a putative NLS (nuclear localization sequence) and a CCHC-type zinc finger motif at the C-terminus are indicated.
To test this hypothesis, 729 males with MR (IQs ranging from 36 to 63) were screened by Incorporation PCR SSCP (Single Strand Conformational Polymorphism) [Sossey-Alaoui et al., 1999]. All suspicious alleles were directly sequenced. In addition, 494 males with normal intelligence were screened. Two missense sequence alterations were identified in 8 (8/729, 1.09%) males in this cohort of males with mental retardation(Table I). A missense variation, p.R7C (c.19C>T), was identified in a highly conserved arginine in four unrelated males; three were sporadic samples for which we do not have DNA on family members. In one small kindred (K9264) this variation segregated with MR (Fig. 2A). We speculate that the p.R7C alteration in the carrier mother is likely a de novo change as the grandmother was not the carrier of the alteration (Fig. 2A). Unfortunately, DNA is not available on the maternal grandfather, precluding definitive exclusion of inheritance through this individual. However, one of 494 (0.2%) control males also showed this sequence alteration raising the possibility of this being a rare polymorphism. Unfortunately, no additional information was available on this male control.
Table I.
SIZN1 variants observed in males with MR of unknown etiology
| Frequency in males with MR |
Frequency in control- males |
||
|---|---|---|---|
| Mutation or alteration |
Amino acid change |
Total(729) | Total(494) |
| c.19C>T | p.R7C | 4(0.55%) | 1(0.2%) |
| c.1031C>T | p.T344I | 4(0.55%) | 0(0%) |
| 3′UTR+73_77 del6 | n/a | 2(0.27%) | 0(0%) |
| 3′UTR+73_76 del5 | n/a | 1(0.14%) | 0(0%) |
In families K9264 and K8923, analysis of markers from the long arm of the X chromosome including the SIZN1 gene locus revealed that affected male(s) in family inherited the same X chromosomal region from marker DXS6789 to marker DXS1047 from the carrier mother (Figs. 2A and 3A). We also performed X inactivation studies in all available females in these families (Figs. 2 and 3). No specific skewing of X inactivation was noted. All females in family K9264 showed a random pattern of X inactivation. The carrier female in family K8923 showed moderately skewed X inactivation (84:16). To determine the functional consequence of the p.R7C variation, an expression vector with the p.R7C variant in SIZN1 was generated and the transcriptional co-activation activity were measured by using the SBE (Smad Binding Element) x4 luciferase reporter gene assay as previously described [Cho et al., 2008]. An approximately 40% reduction of Sizn1 function was found in comparison to wild type activity (Fig. 2B). In addition, we also found a significantly reduced protein level of the mutant protein in these assays, potentially accounting for the decreased activity (Fig. 2B). Based on these data, we hypothesized the p.R7C alteration might affect Sizn1 stability. To examine this possibility, pulse-chase experiments were performed to assess protein half-life. We found the half-life of the wild type protein to be approximately 2.17±0.71hrs. As expected, the p.R7C mutant protein had a reduced half-life of 0.91±0.29 hrs. Thus, the functional consequences of this mutation reflect changes in the protein stability with secondarily reduced activity in BMP pathway activity (Figs. 2B, C).
Figure 3.
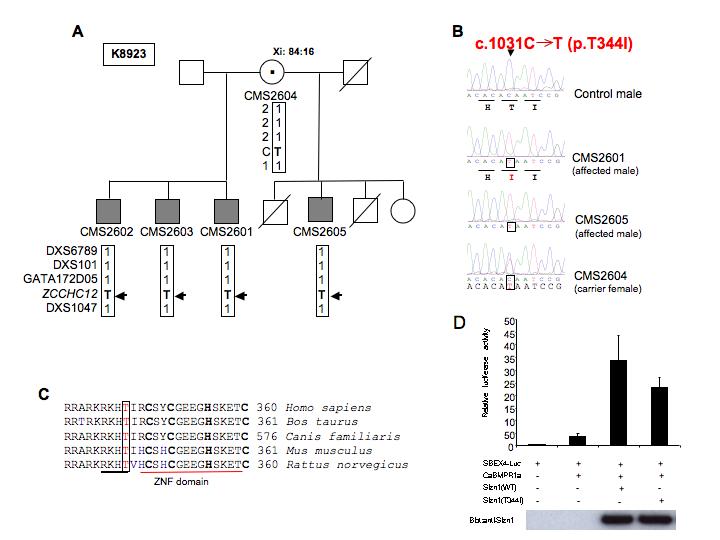
SIZN1 mutation c.1031C>T(p.T344I) in family K8923 (A). Partial pedigree of family K8923 with XLMR chromosome marker analysis and X inactivation (Xi) data are indicated. (B) Automated sequence chromatograms of SIZN1 exon 4 from a control male and from two affected males and a carrier female in family K8923 showing a “C” to “T” alteration at nucleotide 1031 (c.1031C→T). Carrier mother is heterozygous for the alteration. Mutant alleles are boxed. This alteration is predicted to cause a p.T344I missense change in SIZN1. (C) Sequence comparison across species shows the Thr 344 residue is highly conserved amino acid. (D) The Sizn1 related enhancement in BMP signaling is suppressed by the T344I mutant of Sizn1 (30%) in SBEx4 reporter assay. Error bar are standard deviation (SD) in luciferase assay.
The second sequence variant, c.1031C>T (p.T344I), was identified in four affected males in a family consisting of three brothers and a half brother (Fig. 3). This sequence alteration was not found in any individual from the control population (0/494). We also tested the p.T344I variant in our functional assay and found its activity was 70% of wild type (Fig. 3). While it is uncertain what a 30% reduction in activity translates to in terms of BMP signaling, Sizn1 may also be a coactivator for other signaling pathway such as Wnt [Cho et al., 2008], which are not measured in our functional assay. The last set of two additional sequence variations were all found in the 3′UTR (within exon 4). Two variations are deletions in repeat DNA sequences of the 3′UTR (GGGGTG x 3). No variation in the 3′ UTR was identified in any of the 494 control males tested. It is possible that these variations may lead to mRNA instability. However, confirmation of this possibly waits further testing.
Two additional sequence alterations, c.837G>A and c.1179C>A, were found in the coding sequence. However, these resulted in no change in the protein sequence (p.L279L and p.G393G respectively) and therefore were considered polymorphisms.
Finally to determine if SIZN1 might be associated with a specific syndrome we compared the phenotypic feature of patients with two different sequence alterations p.R7C and p.T344I (Table II). No consistent phenotype was observed in the affected males, suggesting SIZN1 mutations are associated with nonsyndromic XLMR..
Table II.
Clinical Phenotype of males with SIZN1 variants
| p.R7C | p.T344I | |
|---|---|---|
| Head circumference ≤ 25th centile | 0/4 | 4/4 |
| Dysmorphic facial features | 3/4 | 0/4 |
| 5th finger clinodactyly | 0/4 | 4/4 |
| Large hands | 0/4 | 4/4 |
| Mild hearing loss | 0/4 | 2/4 |
| Seizures | 3/4 | 0/4 |
| Schizophrenia | 2/4 | 0/4 |
| Behavior problems | 1/4 | 2/4 |
| Moderate mental retardation | 1/4 | 0/4 |
| Mild mental retardation | 3/4 | 4/4 |
DISCUSSION
We found SIZN1 sequence alterations in 11 males with MR. Two of the mutations were coding sequence missense mutations in eight individuals, four from one family. Another three patients had a small deletion involving the 3’UTR. As described above, the facts that the patients do not have similar phenotypes suggests no recognizable XLMR syndrome could be associated with SIZN1 variants.
We detected one p.R7C variant in an apparently normal male control. Thus it is possible that the phenotype associated with this mutation is not 100% penetrant. Alternatively, SIZN1 may be a modifier (or risk factor) that requires other polymorphisms or mutations for the full expression of the phenotype. Nonetheless, our functional data with this mutation are extremely intriguing and would seem to indicate that there may be cognitive impairment associated with mutations in this gene. Our data for the p.R7C alteration in SIZN1 is also curiously similar to that found with several mutations in ATRX [Gibbons and Higgs, 2000; McDowell et al., 1999; Tang et al., 2004]. Mutations in this gene result in the ARTX-syndrome characterized by MR, α-thalassemia and developmental abnormalities. Analyses of ATRX protein level by western blot also indicated that the level of ATRX expression was significantly reduced in patients with mutations [Gibbons and Higgs, 2000; McDowell et al., 1999; Tang et al., 2004]. Together these data suggest minor changes in the cellular level of functional proteins are potentially sufficient to perturb cognitive function.
Mutation of residue Thr 344, which is perfectly conserved from rodents to humans, is predicted to be significant. Residue Thr 344 is located in between the NLS (nuclear localization sequence) and the zinc finger motif. According to “Scansite”, Thr344 is also a strong candidate phosphorylation site for Akt. Although we observed only a 30% reduction in functional activity with the p.T344I variant, because it is a highly conserved residue, we expect Thr 344 is likely important for the regulation of transcriptional coactivation of SIZN1 through phosphorylation.
Finding of sequence variants in SIZN1 in the XLMR familial cases, along with our previous studies [Cho et al., 2008], further implicate the basal forebrain cholinergic neurons in cognition. We have shown SIZN1 modulates BMP signaling and that it is necessary for the normal expression of cholinergic nervous specific genes. An even greater understanding of pathogenic mechanism of SIZN1 mutations will come through the analysis of mutant mouse models, which we have generated, and the evaluation of brains from patients with specific mutations.
ACKNOWLEGMENTS
We would like to thank the members of the Golden Lab for helpful discussion and advice. We wish to express our gratitude to the patients and their families for cooperation. We thank Cindy Skinner for assistance in obtaining patients’ samples and D. Schultz for assistance in sequencing. This work was supported, in part, by NIH grants NS45034 to JAG, and HD26202 to CES and a grant from the South Carolina Department of Disabilities and Special Needs. This manuscript is dedicated to the memory of Ethan Francis Schwartz, 1996-1998.
REFERENCES
- Aicardi J. The etiology of developmental delay. Semin Pediatr Neurol. 1998;5:15–20. doi: 10.1016/s1071-9091(98)80013-2. [DOI] [PubMed] [Google Scholar]
- Baxter MG, Chiba AA. Cognitive functions of the basal forebrain. Curr Opin Neurobiol. 1999;9:178–183. doi: 10.1016/s0959-4388(99)80024-5. [DOI] [PubMed] [Google Scholar]
- Cho G, Lim Y, Zand D, Golden JA. Sizn1 is a novel protein that functions as a transcriptional coactivator of bone morphogenic protein signaling. Mol Cell Biol. 2008;28:1565–1572. doi: 10.1128/MCB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counts SE, Nadeem M, Wuu J, Ginsberg SD, Saragovi HU, Mufson EJ. Reduction of cortical TrkA but not p75(NTR) protein in early-stage Alzheimer’s disease. Ann Neurol. 2004;56:520–531. doi: 10.1002/ana.20233. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Colombo PJ. Ageing: the cholinergic hypothesis of cognitive decline. Curr Opin Neurobiol. 1995;5:161–168. doi: 10.1016/0959-4388(95)80022-0. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Higgs DR. Molecular-clinical spectrum of the ATR-X syndrome. Am J Med Genet. 2000;97:204–212. doi: 10.1002/1096-8628(200023)97:3<204::AID-AJMG1038>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Che S, Wuu J, Counts SE, Mufson EJ. Down regulation of trk but not p75NTR gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J Neurochem. 2006;97:475–487. doi: 10.1111/j.1471-4159.2006.03764.x. [DOI] [PubMed] [Google Scholar]
- Granholm AC, Sanders LA, Crnic LS. Loss of cholinergic phenotype in basal forebrain coincides with cognitive decline in a mouse model of Down’s syndrome. Experimental neurology. 2000;161:647–663. doi: 10.1006/exnr.1999.7289. [DOI] [PubMed] [Google Scholar]
- Lehrke R. A theory of X-linkage of major intellectual traits. Response to Dr.Anastasi and to the Drs. Nance and Engel. Am J Ment Defic. 1972;76:626–631. [PubMed] [Google Scholar]
- Leonard H, Wen X. The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev. 2002;8:117–134. doi: 10.1002/mrdd.10031. [DOI] [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O’Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, Chapman L, Rhodes D, Higgs DR. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci U S A. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp AC, Pinsonneault JK, Cooke G, Sadee W. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques. 2003;34(5):1068–1072. doi: 10.2144/03345dd03. [DOI] [PubMed] [Google Scholar]
- Roeleveld N, Zielhuis GA, Gabreels F. The prevalence of mental retardation: a critical review of recent literature. Dev Med Child Neurol. 1997;39(2):125–32. doi: 10.1111/j.1469-8749.1997.tb07395.x. [DOI] [PubMed] [Google Scholar]
- Ropers HH. X-linked mental retardation: many genes for a complex disorder. Curr Opin Genet Dev. 2006;16:260–269. doi: 10.1016/j.gde.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6(1):46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- Sarter M, Bruno JP. Developmental origins of the age-related decline in cortical cholinergic function and associated cognitive abilities. Neurobiol Aging. 2004;25:1127–1139. doi: 10.1016/j.neurobiolaging.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Schwartz CE, Ulmer J, Brown A, Pancoast I, Goodman HO, Stevenson RE. Allan-Herndon syndrome. II. Linkage to DNA markers in Xq21. Am J Hum Genet. 1990;47:454–458. [PMC free article] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Lyon JA, Jones L, Abidi FE, Hartung AJ, Hane B, Schwartz CE, Stevenson RE, Srivastava AK. Molecular cloning and characterization of TRPC5 (HTRP5), the human homologue of a mouse brain receptor-activated capacitative Ca2+ entry channel. Genomics. 1999;60:330–340. doi: 10.1006/geno.1999.5924. [DOI] [PubMed] [Google Scholar]
- Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- Verpoorten N, Claeys KG, Deprez L, Jacobs A, Van Gerwen V, Lagae L, Arts WF, De Meirleir L, Keymolen K, Ceuterick-de Groote C, De Jonghe P, Timmerman V, Nelis E. Novel frameshift and splice site mutations in the neurotrophic tyrosine kinase receptor type 1 gene (NTRK1) associated with hereditary sensory neuropathy type IV. Neuromuscular disorders : NMD. 2006;16:19–25. doi: 10.1016/j.nmd.2005.10.007. [DOI] [PubMed] [Google Scholar]


