Abstract
Proteins in the tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein family (YWHA; also known as 14-3-3) are involved in the regulation of many intracellular processes. We have examined the interaction of YWHA with peptidylarginine deiminase type VI (PADI6), an abundant protein in mammalian oocytes, eggs, and early embryos. Peptidylarginine deiminases catalyze the posttranslational modification of peptidylarginine to citrulline. PADI6 is associated with oocyte cytoplasmic sheets, and PADI6-deficient mice are infertile because of disruption of development beyond the two-cell stage. We found that PADI6 undergoes a dramatic developmental change in phosphorylation during oocyte maturation. This change in phosphorylation is linked to an interaction of PADI6 with YWHA in the mature egg. Recombinant glutathione S-transferase YWHA pull-down experiments and transgenic tandem affinity purification with liquid chromatography-mass spectrometry demonstrate a binding interaction between YWHA and PADI6 in mature eggs. YWHA proteins modulate or complement intracellular events involving phosphorylation-dependent switching or protein modification. These results indicate that phosphorylation and/or YWHA binding may serve as a means of intracellular PADI6 regulation.
Keywords: egg, gamete biology, gametogenesis, mouse, oocyte, oocyte development, oocyte maturation, ovum, PADI6, PAD6, peptidylarginine deiminase, phosphorylation, YWHA, 14-3-3
INTRODUCTION
Members of the tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein family (YWHA; also known as 14-3-3) are involved in the regulation of a number of intracellular processes. YWHA proteins are a highly conserved, ubiquitously expressed family of proteins that exist in all eukaryotic organisms, and seven isoforms of YWHA have been identified in mammals [1–4]. YWHA proteins complement or supplement intracellular events involving phosphorylation-dependent switching or protein-protein interaction [5, 6]. YWHA may alter the activity of bound proteins, change the associations or interactions of the bound proteins with other proteins, protect protein phosphorylation, promote protein stability, or alter the intracellular localization or destination of the bound protein. YWHA proteins form homodimers or heterodimers that generally bind to amino acid motifs in target proteins containing a phosphorylated serine residue and arginine at positions −3 or −4 (i.e., ArgXXpSer or ArgXXXpSer, where pSer is phosphorylated serine and X is any amino acid).
Proteomic and biochemical approaches have identified more than 200 proteins that may interact with YWHA [1, 6–8]. The functional role of YWHA has been studied in a variety of cellular processes, including signal transduction, trafficking, apoptosis, stress response, and malignant transformation [5, 6, 9]. Of particular interest, YWHA has a role in vertebrate development and cell-cycle regulation. For example, experimental reduction of specific YWHA isoforms causes gastrulation and axial patterning defects in Xenopus embryos [10] and defects in neural cell migration in mammals [11]. YWHA proteins interact with cell-cycle control proteins. For example, YWHA controls the localization of CDC25B in somatic cells [12], and YWHA is associated with the meiotic arrest of Xenopus oocytes [13, 14]. (For a review of the role of YWHA proteins in cell-cycle regulation, see Hermeking and Benzinger [15].) To our knowledge, the role of YWHA in mammalian oocyte maturation, fertilization, and early development has not been examined.
Peptidylarginine deiminase type VI (PADI6) is an abundant protein in mammalian oocytes, eggs, and early embryos [16]. It also is the most recently characterized member of the peptidylarginine deiminase enzyme family (PAD; EC 3.5.3.15). As a class, peptidylarginine deiminases catalyze the conversion of peptidyl or protein L-arginine to L-citrulline with the generation of NH3. Citrulline is a nonstandard amino acid that is not introduced during translation and is only present in proteins as a result of this posttranslational modification, which is known as citrullination. The PAD enzymes do not convert free arginine to citrulline. Citrullination causes a decrease in the net positive charge of a protein [17], which may affect protein-protein intermolecular interactions or alter the three-dimensional folding of proteins, thereby potentially changing the function or activity of substrate proteins. Five PAD isoforms have been described: PADIs 1–4 and 6. (For a review of the gene structure, protein distribution, and functions of the PAD isoforms, see Vossenaar et al. [17].) The primary substrates for PAD enzymes are structural proteins, such as the intermediate filaments keratin (PADI1, in keratinocytes), vimentin (PADI2, in skeletal muscle and macrophages), and neurofilament associated proteins, such as myelin basic protein (PADI2, in the brain). The PADs also citrullinate intermediate filament-associated proteins, such as filaggrin (PADI1, in keratinocytes) and trichohyalin (PADI3, in hair follicles). PADI4 is unique, because it is found in the nucleus, mainly in white blood cells, where it may function in citrullination of histone proteins, thereby altering chromatin structure and serving as a transcriptional coregulator [18]. Moreover, PAD-dependent conversion of methyl-arginine to citrulline residues could alter the patterns of histone methylation, which may influence gene regulation. Although some evidence suggests that conversion of methylated arginine to citrulline may occur in vivo [19], PAD enzymes are unable to deiminate methylated arginine in vitro [20], and this process needs further examination [18].
Human PADI6 mRNA transcripts are found predominately in ovarian tissue and also in the testis and peripheral blood leucocytes [21, 22]. Similarly, in the mouse, PADI6 is expressed in the mouse ovary (specifically in oocytes), ovulated egg, and early embryo [16]; this isoform is not known to be present in other mouse tissues. The human PADI6 protein is approximately 65% identical to the mouse PADI6 [22]. Analysis of mouse PADI6 reveals approximately 40% homology to the other known PAD isoforms (PADIs 1–4) [16, 22]. In mouse oocytes, PADI6 was found to be colocalized with keratin-containing intermediate filaments contained in cytoplasmic sheets [16], which suggested a potential interaction or substrate for the PADI6 protein. Evidence for the role of PADI6 affecting cytoplasmic sheets as well as subsequent development comes from analysis of PADI6-deficient mice [23]. Absence of PADI6 and, presumably, its citrullination activity appears to prevent the formation of the keratin-containing cytoplasmic sheets. Oocyte growth, ovulation, maturation, and fertilization are normal in PADI6-deficient mice, but development stops at the two-cell stage, suggesting that PADI6 is required for normal development beyond the two-cell stage.
In the present study, we show a cell cycle-dependent change in phosphorylation of PADI6. This stage-specific change in phosphorylation during oocyte maturation enables the interaction of YWHA with PADI6 in the mature egg. Evidence for this interaction comes from a number of biochemical tests using oocyte and egg extracts as well as the recently developed proteomic method of tandem affinity purification (TAP) followed by liquid chromatography-mass spectrometry (LCMS) to identify proteins interacting with a protein of interest. To our knowledge, this is the first YWHA binding interaction to be described for the mouse egg, and this information may provide insight regarding PADI6 activity, its regulation, and its interactions with other cellular proteins.
MATERIALS AND METHODS
Collection of Oocytes, Eggs, and Two-Cell Embryos
Mouse oocytes and eggs were collected in a minimal essential medium (MEM) as described previously [24, 25]. In brief, females were injected with 7.5 IU of eCG, and 44–48 h later, the ovaries were removed and repeatedly punctured with a 26-gauge needle to rupture follicles. Cumulus cell-enclosed oocytes were isolated, and the cumulus cells were removed by repeated pipetting though a small-bore pipette. Fully grown, germinal vesicle-intact oocytes with a diameter of approximately 80 μm (measured with an ocular reticle) were collected. The oocytes were transferred through several MEM washes in drop cultures under mineral oil. All oocytes were collected and cultured in MEM containing 0.1 mg/ml of dibutyryl cAMP (dbcAMP) to prevent spontaneous oocyte maturation except those used for in vitro maturation experiments, which were incubated without dbcAMP and allowed to mature.
Mature, metaphase II-arrested eggs were obtained from mice following superovulation, and the cumulus cells were removed with 0.3 mg/ml of hyaluronidase (type IV-S). Following cumulus cell dispersal, eggs were collected and washed through several drops of MEM. To obtain two-cell embryos, superovulated females were caged with male mice for 24 h. The oviducts were removed from the females 2 days after caging, and the embryos were flushed out with a syringe filled with MEM and tipped with a 30-gauge needle. Recovered embryos were collected and cultured in MEM. Except when noted, all media and chemicals were obtained from Sigma-Aldrich. The transgenic mice were produced in a licensed animal facility in accordance with the German Animal Welfare Act (Tierschutzgesetz) following the guidelines of the European Convention for Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (1986). Transgenic and all other mice used in the present experiments were housed and used at Kent State University under an approved Institutional Animal Care and Use Committee protocol following the National Research Council’s publication Guide for the Care and Use of Laboratory Animals.
Antibodies
An affinity-purified rabbit phospho-specific antibody was obtained from Cell Signaling Technology (9601). This antibody was generated against synthetic peptides containing the phosphorylated serine YWHA binding motif. The antibody has been shown to recognize the YWHA binding motif in several putative YWHA binding proteins [26–31]. We refer to this antibody as the anti-p(S)YWHA binding motif antibody. An affinity-purified, polyclonal antibody recognizing PADI6 was generously provided by Dr. Scott Coonrod (Cornell University, Ithaca, New York). This antibody was generated in guinea pig against a peptide comprised of the N-terminal 200 amino acids of recombinant PADI6. Anti-PADI6 was used for Western blot experiments. A protein G-purified rat monoclonal that recognizes HSPA8 (Hsc-70; ab19136; Abcam) was used in immunoprecipitation experiments. The immunogen for this antibody was the hamster full-length native heat shock 70-kDa protein 8 (HSPA8). Affinity-purified polyclonal rabbit anti-YWHA (anti-14-3-3; 51–0700; Invitrogen) was used to detect YWHA protein. This antibody was raised against a 20-amino-acid synthetic peptide based on the N-terminus of the human YWHA and is cross-reactive with a broad range of YWHA isoforms.
One-Dimensional Protein Gel Electrophoresis and Western Blot Analysis
Oocytes and eggs were collected as described above, and the zonae pellucidae were removed by a brief treatment in acid Tyrode solution (0.14 M NaCl, 3 mM KCl, 1.6 mM CaCl2·2H2O, 0.5 mM MgCl2·6H2O, 5.5 mM glucose, and 0.1% polyvinyl alcohol [PVA], pH 2.5). Cells were rinsed in MEM, counted, and transferred to Tris-buffered saline (TBS; 25 mM Tris-HCl [pH 7.5] and 150 mM NaCl) containing 0.1% PVA in a microcentrifuge tube. The TBS was removed, lysing buffer added, and the cell lysates quick-frozen in ethanol/dry ice and stored at −70°C until use. The lysis buffer contained 10 mM Tris-HCl [pH 7.2], 1 mM EDTA, 1 mM EGTA, 0.1% (v/v) β-mercaptoethanol, 1% (v/v) Triton X-100, protease inhibitors (1 mM PMSF, 0.1 mM TPCK [Tosyl Phenylalamyl Chloromethul Ketone], 10 μM leupeptin, 1 μM pepstatin A, and 75 nM aprotinin), and phosphatase inhibitors (1 mM Na3 VO4, 100 nM calyculin A, 10 mM β-glycerophosphate, and 5 mM sodium pyrophosphate). Immediately before polyacrylamide gel electrophoresis, 1 μl of bromophenol blue (0.1% v/v) was added to each tube to track progression through the gel.
Oocyte, egg, and two-cell embryo proteins were electrophoretically separated by SDS-PAGE using a 4% stacking, 12% resolving polyacrylamide gel. The proteins were examined by immunoblot analysis after electrophoretic transfer for 1 h at 100 V to an Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore Corp.). The membranes were washed with TBS blocking buffer (TBS with 5% nonfat milk and 0.1% Tween-20) for at least 30 min before overnight (4°C) incubation with one of the primary antibodies diluted in blocking buffer (anti-PADI6, diluted 1:10 000; anti-p(S)YWHA binding motif, diluted 1:1000; or anti-HSPA8 diluted to 0.1 μg/ml). Following primary antibody incubation, the membranes were thoroughly washed with TBS containing 0.1% Tween-20 (TTBS) and incubated for 1 h with a horseradish peroxidase-conjugated secondary antibody (1:2000 donkey anti-rabbit immunoglobulin [Ig] G [Amersham, now GE Healthcare Life Sciences] or 1:10 000 goat anti-guinea pig IgG [Jackson ImmunoResearch Laboratories, Inc.). Blots were then washed with TTBS twice for 15 min each and four times for 5 min each. Proteins were visualized using an enhanced chemiluminescence kit and film according to the manufacturer’s instructions (GE Healthcare Life Sciences).
Two-Dimensional Electrophoresis
Mouse eggs and oocytes, collected as described above, were lysed, quick-frozen in ethanol/dry ice, and stored at −70°C until use. The lysis buffer for two-dimensional electrophoresis contained 8 M Urea, 4% (w/v) CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate), 4 mM Tris-HCl (pH 7.5), protease inhibitors (protease inhibitor tablet; Qiagen), and phosphatase inhibitors (5 mM sodium pyrophosphate, 50 mM NaF, 1 mM Na3 VO4, 100 nM calyculin A, and 10 mM β-glycerophosphate). Rehydration buffer (Bio-Rad ReadyPrep sequential extraction kit reagent 3; 5 M urea, 2 M thiourea, 2% CHAPS, 2% SB-3–10, 40 mM Tris, and 0.2% Bio-Lyte 3–10 ampholyte) containing 50 mM dithiothreitol (DTT) was added to lysates for 30 min at room temperature. The lysate, in rehydration buffer, was then used to actively rehydrate an immobilized pH gradient strip (pH 4–7, 7 cm; ReadyStrip 163-2001; Bio-Rad) following the manufacturer’s instructions at 50 V for 12 h using Bio-Rad Protean Isoelectric Focusing (IEF) Cell. Immediately following active rehydration, rapid automated IEF was performed for a total of approximately 14 000 volt-hours. The procedure for second-dimension protein separation and blotting was identical to that described above using the Bio-Rad Protean IEF system according to the manufacturer’s instructions.
Protein Microsequencing
Eggs were collected and lysed, and proteins were resolved by one-dimensional SDS-PAGE as described above. In the initial experiment to determine the protein identified by the anti-p(S)YWHA binding motif antibody, proteins from mature eggs were run on a gel and stained with Coomassie blue, and the major band at approximately 75 kDa was excised for analysis. This band corresponded exactly to the band labeled by the p(S)YWHA binding motif antibody in corresponding Western blots. In the TAP-YWHA purification (described below), the band was excised from a silver-stained gel. Processing of the gel band and microsequencing was performed in the laboratory of Dr. Michael Kinter (Lerner Research Institute Mass Spectrometry Laboratory for Protein Sequencing, Cleveland Clinic Foundation). In brief, gel pieces were destained, dehydrated (acetonitrile), reduced (DTT), and alkylated (iodoacetamide) before overnight trypsin digestion. Peptides extracted from the polyacrylamide were analyzed by LC-MS. The LC-MS system was a ThermoFisher LTQ linear ion-trap mass spectrometer system. The HPLC column was a self-packed, 8-cm × 75-μm (inner diameter) Phenomenex Jupiter C18 reversed-phase capillary chromatography column. The digest was analyzed to provide full-scan mass spectra for peptide molecular weights and product ion spectra to determine amino acid sequence. The collisionally induced dissociation spectra collected were used to search the National Center for Biotechnology Information nonredundant database with the search program Mascot (Matrix Science Ltd.). All matching spectra were verified by manual interpretation.
Dephosphorylation of Blotted Proteins with Lambda Protein Phosphatase
Eggs were collected, and proteins were extracted, resolved by SDS-PAGE, and electrophoretically transferred to PVDF membrane as described above. The membrane was then washed twice with deionized water and blocked with TBS containing 1% BSA and 0.1% Triton X-100 for 1 h. Dephosphorylation of PVDF-bound proteins was achieved by incubation in TBS containing 1% BSA, 0.1% Triton X-100, 2 mM MnCl2, and 400 U/ml of recombinant Lambda Protein Phosphatase (14–405; Upstate/Millipore) overnight at 4°C. Lambda Protein Phosphatase has activity against phosphorylated serine, threonine, tyrosine, and histidine residues. Following dephosphorylation treatment, the membrane was washed in PBS containing 0.1% Tween-20, then washed four times in deionized water. Western blot analysis, as outlined above, was performed after phosphatase treatment.
Immunoprecipitation of HSPA8
Egg extracts were incubated with anti-HSPA8 (10 μg/ml) or control rabbit serum (10–12 μg/ml) with rotation for 1 h at 4°C. Protein G-Sepharose beads (GE Healthcare Life Sciences) were washed once with distilled water and twice with TTBS. The Protein G-Sepharose beads were added to the antibody/egg extract preparation, and the mixture was incubated with rotation for an additional 1 h at 4°C. The immunoprecipitated complexes were pelleted by centrifugation (1500 rpm, 5 min), and the supernatant (flow-through) was retained. The pellet was washed by resuspension and centrifugation (1500 rpm, 5 min) twice with TTBS and twice with buffer containing 10 mM Tris-HCl, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 0.1 mM TPCK, 50 mM NaF, 5 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 0.1% (v/v) β-mercaptoethanol, and 1% NP-40, pH 7.2. The pellet and the original supernatant samples were divided equally by volume for analyses by SDS-PAGE and immunoblotting with anti-HSPA8 or anti-p(S)YWHA binding motif antibodies.
Subcloning, Expression, and Purification of Glutathione S-Transferase-YWHA and Glutathione S-Transferase
The YWHAZ (YWHAζ) cDNA, subcloned in pcDNA 3.1/V5-His-TOPO, was a generous gift from Dr. Helen Piwnica-Worms (Washington University). The cDNA was amplified by sense primer 5′-CTCGGATCCATGGATAAAAATGAGCTGGTTCA-3′ containing a BamHI restriction site and 3′antisense primer 5’-CTCGAATTCTTAATTTTCCCCTCCTTCTCCTG-3′ containing a stop codon followed by an EcoRI restriction site. The PCR was followed by digestion of the insert and a glutathione S-transferase (GST)-tag vector pGEX 4T-2 (GE Healthcare Life Sciences) with BamHI and EcoRI. One-hundred nanograms of digested vector were incubated at 4°C overnight with 30 ng of the digested PCR product (digested in the presence of DNA ligase). Escherichia coli (Nova blue strain) were transformed with ligation reaction product, and bacterial colonies containing YWHA cDNA were identified by PCR. The pGEX 4T-2 plasmid containing YWHA cDNA was amplified and purified using a Qiagen midi prep kit. The resulting subcloned product was sequenced to ensure that the sequence was correct.
Escherichia coli (BL21) cells were transformed with plasmid containing GST (empty vector pGEX 4T-2) and GST-YWHA. Cultures were incubated at 37°C in 2X YTA (16 g of tryptone, 10 g of yeast extract, and 5 g NaCl, with 100 mg/ml of ampicillin) until the optical density at 600 nm reached 0.5–2. Induction was done by adding isopropyl-1-thio-β-D-galactopyranoside at a final concentration of 75 mM, and the incubation was continued for another 4 h. The culture was centrifuged at 6000 × g at 4°C to sediment the cells. The pellet was resuspended in 50 ml of PBS containing protease inhibitors (Complete Protease Inhibitor Cocktail Tablet; Roche Applied Sciences). Cells were disrupted by sonication on ice with short bursts (six bursts for 6 sec each). Lysed bacteria were centrifuged at 16000 × g for 10 min. The supernatant was collected and incubated with Glutathione Sepharose 4B beads (GE Healthcare Life Sciences), washed three times with PBS, for 2 h. Beads, along with supernatant, were transferred to a disposable column (Bio-Rad). The beads were washed with PBS (threefold the bed volume) to remove any proteins binding nonspecifically to the beads. The GST and GST-YWHA that bound to beads was eluted with GST elution buffer (10 mM Tris-HCl [pH 8.0] and 20 mM glutathione). Both GST alone and GST-YWHA purified as described above were dialyzed overnight in PBS containing 1 mM phenylmethanesulfonyl fluoride. The SDS-PAGE analysis of purified GST and GST-YWHA showed a single band at approximately 27 kDa and approximately 55 kDa, respectively, after Coomassie blue staining. The concentrations of GST and GST-YWHA were estimated by comparison to BSA standard after SDS-PAGE analysis and Coomassie blue staining.
GST-YWHA Pull-Down Assay
Glutathione Sepharose 4B beads were prepared according to the manufacturer’s specifications. The beads were resuspended in buffer A (10 mM Tris-HCl, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 0.1 mM TPCK, 50 mM NaF, 5 mM sodium pyrophosphate, and 10 mM β-glycerophosphate, pH 7.2) to obtain a 50:50 slurry. The GST-YWHA or GST was added, in equal molar amounts, to 50 μl of bead slurry and incubated with rotation at 4°C for 3 h. The beads were washed with 400 μl of buffer A and centrifuged (1500 rpm, 5 min) to remove the supernatant. Egg and oocyte extracts were prepared as described in One-Dimensional Protein Gel Electrophoresis and Western Blot Analysis, added to the beads, and incubated with rotation at 4°C overnight. Beads were washed four times with 1 ml of buffer B (10 mM Tris-HCl, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 0.1 mM TPCK, 50 mM NaF, 5 mM sodium pyrophosphate, 10 mM β-glycerophosphate, and 0.1% [v/v] β-mercaptoethanol, pH 7.2). The GST fusion proteins and any bound proteins were eluted directly into Laemmli buffer and boiled for 5 min before resolving through 12% SDS-PAGE. Eluted proteins (pellet or bound) and supernatant (flow-through or unbound) samples were analyzed by Western blotting with anti-p(S)YWHA binding motif and anti-PADI6 antibodies.
Dephosphorylation of Egg Proteins and GST-YWHA Pull-Down Assay
Eggs were collected and extracted with 0.1% NP-40, then added to lambda phosphatase reaction buffer (50 mM Tris-HCl, 100 mM NaCl, 0.1 mM EGTA, 2 mM DTT, 0.01% Brij 35, and 2 mM MnCl2, pH 7.5) containing 14.8 U/μl of lambda phosphatase (Lambda Protein Phosphatase; New England BioLabs) and protease inhibitors (Complete Protease Inhibitor Cocktail Tablet) and incubated at 30°C for 20 min. The total reaction volume was transferred to a tube containing prepared Glutathione Sepharose 4B beads, and the GST-YWHA pull-down analysis was performed as described in GST-YWHA Pull-Down Assay.
YWHA Tandem Affinity Purification
The technique of tandem affinity purification (TAP) coupled with LC-MS permits the identification of proteins forming endogenous complexes within cells. The TAP method is highly sensitive, selective, and particularly suited for detecting multimeric protein complexes in their native environment within cells [32]. This method was used to demonstrate that YWHA interacts with PADI6 in the mature egg. The TAP tag consists of dual tag of protein A from Staphylococcus aureus (Prot A) and calmodulin binding peptide (CBP). The CBP tag is fused to the bait protein (for these experiments, YWHA), and its interacting partners are purified by two sequential steps. The CBP tag allows the purification on a calmodulin affinity column of the YWHA together with any proteins that are bound to it. Before purification, however, the complexes are first bound to IgG. The Prot A tag binds to IgG-sepharose and normally would be released only under denaturing conditions at low pH; however, a specific tobacco etch virus (TEV) protease recognition sequence (TEV cleavage site) is inserted between the Prot A and CBP tags. Treatment with TEV protease releases the CBP-protein complex from IgG-sepharose under mild native conditions. Thus, the fusion protein and its interacting partners are purified by two sequential steps. This purification is under native conditions, preserving protein-protein interactions, and the two-step purification dramatically reduces nonspecific binding of background proteins.
Transgenic mice expressing TAP-tagged YWHA in oocytes and eggs were used to examine endogenous YWHA binding interaction with PADI6. (The generation of these transgenic animals has been described by Angrand et al. [33]). In brief, the TAP cassette, composed of Prot A, a specific TEV protease recognition sequence, the calmodulin binding protein, and the YWHAZ (YWHAζ) protein sequence, was inserted between the human ubiquitin C promoter and the polyadenylation signal. The gel-purified ubiquitin C-TAPYWHAZ transgene was used to produce transgenic mice. Founders and progeny were genotyped by PCR using primers complementary to the human ubiquitin C promoter (5′-ACCCGTTCTGTTGGCTTAT-3′) and TAP cassette (5′-TGGCTGCTGAGACGGCTATGA-3′) sequences. The original transgenic founders expressing the TAP-YWHAZ construct were mated with outbred CD1 mice to produce a line expressing TAP-YWHA in all tissues examined. Only animals expressing the transgene were used in subsequent TAP experiments.
Mature eggs were collected as described in Collection of Oocytes, Eggs, and Two-Cell Embryos. The TAP purification buffer consisted of 50 mM Tris-HCl, 150 mM NaCl, 0.1% (v/v) NP-40, 1.5 mM MgCl2, 5% (v/v) glycerol, phosphatase inhibitors (5 mM sodium pyrophosphate, 10 mM β-glycerophosphate, and 50 mM sodium fluoride), and protease inhibitors (Complete Protease Inhibitor Cocktail Tablet), pH 7.5. After removal of zonae pellucidae, eggs were immediately lysed in the TAP purification buffer and frozen at −70°C. Before the first purification step, IgG beads (IgG Sepharose G Fast Flow; GE Healthcare Life Sciences) were washed three times in buffer. Extract from 2400 eggs (protein content, ∼68 μg/ml) was added to 100 μl of IgG bead slurry, and the TAP-YWHA complex was allowed to bind through Prot A to the IgG beads for 4 h at 4°C. The TAP-YWHA/IgG bead complex was centrifuged and then was washed three times with TEV buffer (50 mM Tris-HCl, 0.5 mM EDTA, and 1 mM DTT, pH 8). The TEV protease (40 U; AcTEV Protease; Invitrogen) was added, and the TAP-YWHA/IgG bead complex was incubated with TEV protease overnight at room temperature with rotation to cleave the complex at the TEV cleavage site, releasing the YWHA/CBP complex from the IgG beads. The eluate (calmodulin binding protein coupled with YWHA and bound proteins) was recovered and added to calmodulin binding buffer (CBB, Interplay TAP Purification Buffer Kit; Stratagene) supplemented with CaCl2 (200 μM). Calmodulin beads (calmodulin affinity resin; Stratagene) were washed with CBB. The YWHA/calmodulin binding protein complex was mixed with the calmodulin beads with rotation for 3 h at 4°C. The beads were pelleted by centrifugation (1500 rpm, 5 min) and washed twice with CBB. Calmodulin elution buffer (50 mM Tris-HCl and 20 mM EGTA, pH 8) was added to release bound protein complexes (1 h with rotation at room temperature). The supernatant (final eluate) containing YWHA (with calmodulin binding protein) and proteins bound to YWHA was collected for gel electrophoresis. Following electrophoresis on a 12% polyacrylamide gel, the gel was silver-stained.
The silver-staining protocol was optimized for subsequent protein analysis using LC-MS. Gels were fixed for 30 min in fixer (50% [v/v] ethanol and 10% [v/v] acetic acid), washed for 1 h, then incubated for 5 min with reducersensitizer (1.5 g/L of potassium hexacyanoferrate III, 3.0 g/L of sodium thiosulfate, and 0.5 g/L of sodium carbonate). Gels were washed three times for 10 min each, then silver-stained (0.5 g/L of silver nitrate) for 2 min. Developer (3.0 g/L of sodium carbonate and 200 ml/L of formalin) was added and incubated until protein bands were distinctly visible. To stop further development, gels were incubated in stop solution (1% [v/v] acetic acid) for a minimum of 10 min. Protein microsequencing was performed as indicated in the section above.
RESULTS
To investigate the role of YWHA in oocyte maturation and early development, we collected extracts of immature oocytes, mature eggs, and two-cell embryos, then separated the proteins by polyacrylamide gel electrophoresis and probed with an anti-p(S)YWHA binding motif antibody in Western blot analysis. The anti-p(S)YWHA binding motif antibody labels proteins that might interact with YWHA. Immunoreactivity at a single, discrete band (∼75 kDa) was present in mature eggs and two-cell embryos but not in immature, germinal vesicle-intact oocytes (Fig. 1A, left). These results were confirmed by several more experiments in which fresh extracts were prepared and SDS-PAGE was followed by immunoblot analysis. One gel containing the extracts of mature eggs was stained with Coomassie blue, and the major band at approximately 75 kDa was excised and microsequenced to determine what proteins might be present. The excised band corresponded exactly to the band labeled by the p(S)YWHA binding motif antibody in corresponding Western blots. Sequencing data indicated that two proteins may be present in this band, one being the HSPA8 (also known as Hsc70, UniProt primary accession no. P63107) and the other PADI6 (UniProt primary accession no. Q8K3V4). We first focused on PADI6. The recovered sequenced peptides matched 18% of the full PADI6 sequence with 100% identity (Fig. 2).
FIG. 1.
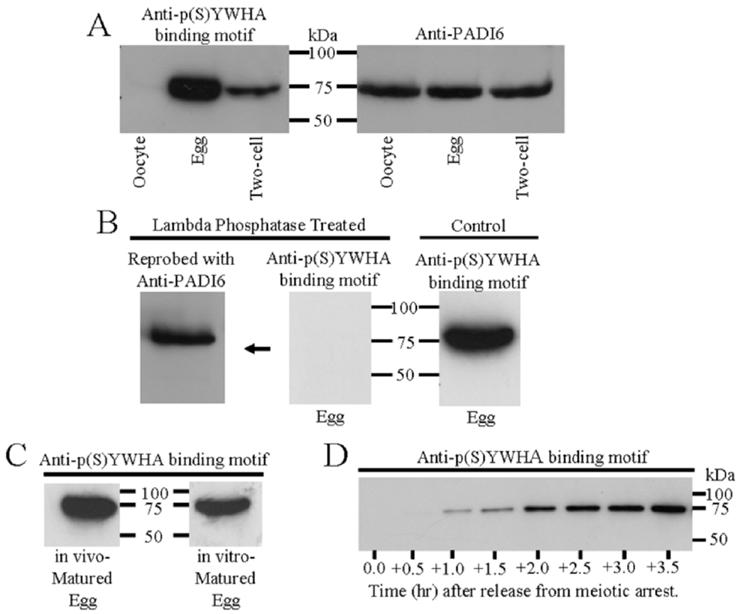
Western blots of proteins from zona-free immature oocytes, mature eggs, and two-cell embryos. Proteins were extracted, resolved by SDS-PAGE, and immunoblotted with the p(S)YWHA binding motif antibody or the PADI6 antibody following procedures outlined in Materials and Methods. All lanes were loaded with total protein extract from an equal number of cells or embryos (n 100 in A—C and 50 in D). These figures are representative of two or three repeated experiments. A) Comparative Western blots of oocytes, eggs, and two-cell embryos with the anti-p(S)YWHA binding motif or anti-PADI6 antibodies. The p(S)YWHA binding motif antibody labels a single band in eggs and two-cell embryos (but not in oocytes) that has the same molecular weight as the band labeled by the PADI6 antibody in oocytes, eggs, and embryos. No other bands were labeled by either of these antibodies. B) The p(S)YWHA binding motif antibody labels only phosphorylated proteins. No proteins are detected when a blot containing egg proteins was probed with the anti-p(S)YWHA binding motif antibody after lambda protein phosphatase treatment (center). The center blot shown was reprobed with anti-PADI6 (left) showing the presence of immunoreactive PADI6 protein at approximately 75 kDa. The control blot (right) was not treated with lambda phosphatase and was probed with the p(S)YWHA binding motif antibody showing an immunoreactive band at approximately 75 kDa. C) The p(S)YWHA binding motif antibody labels the same 75-kDa band in both in vivo- and in vitro-matured eggs. D) Phosphorylation of the 75-kDa protein begins within 1 h of release from meiotic arrest. Each lane of the Western blot was loaded with lysate from 50 prophase I oocytes (time 0) or lysate from 50 cells collected at the times indicated after the cells were washed free of dbcAMP. In this batch of cells, germinal vesicle breakdown was completed in most cells by two hours.
FIG. 2.

Identification of PADI6 by microsequencing. The 75-kDa band was excised from a gel containing egg proteins, and peptides were analyzed by LC-MS. Microsequenced peptides covered 18% of the mouse PADI6 sequence shown (UniProt primary accession no. Q8K3V4) with 100% identity (underlined portions). Bold and dashed underline portions indicate putative YWHA binding domains in PADI6 as indicated by a YWHA motif search in Scansite [34]. Both of the regions contain the ArgXXSer sequence associated with YWHA binding. In these sequences, Ser-219 and Ser-400 (asterisks) also are highly probable serine phosphorylation sites (NetPhos 2.0 Server [35]). Ser-219 and Ser-400 are putative targets of PKA and RPS6KA1, and Ser-400 also may be a PKC site as well as a AKT1 site (Scansite and NetPhosK 1.0 Server [34, 35]).
Analysis of the complete PADI6 protein sequence revealed two potential YWHA binding motifs (ArgXXSer) contained within the PADI6 protein (Fig. 2, bold lettering), which could account for the observation that this 75-kDa protein is labeled by the anti-p(S)YWHA binding motif antibody. Subsequent Western blot analysis with anti-PADI6 in oocytes, eggs, and two-cell embryo extracts confirmed that this band at approximately 75 kDa contained PADI6. In agreement with the initial description of PADI6 in these cells [16], the protein seemed to be approximately equal in amount within oocytes, eggs, and two-cell embryos (Fig. 1A, right). Our data suggested that PADI6, though present in oocytes, is not phosphorylated at sites recognized by the anti-p(S)YWHA binding motif antibody; however, it does become phosphorylated during oocyte maturation. As indicated by the anti-p(S)YWHA binding motif antibody experiments, PADI6 remains phosphorylated in two-cell embryos, though apparently to a lesser degree than in mature eggs (Fig. 1A).
To confirm that the anti-p(S)YWHA binding motif antibody was recognizing a phosphorylated protein, egg proteins were extracted, separated by electrophoresis, and blotted onto a membrane. The membrane was then treated with lambda protein phosphatase before probing with the anti-p(S)YWHA binding motif antibody (Fig. 1B). No reactivity with the anti-p(S)YWHA binding motif antibody was seen in blots treated with lambda protein phosphatase, which served to remove phosphate groups from proteins in the blot. When the same blot was reprobed with anti-PADI6 antibody, however, immunoreactivity was present at precisely the same molecular weight as in a control blot (not treated with phosphatase) probed with anti-p(S)YWHA binding motif antibody. These results indicate that the anti-p(S)YWHA antibody selectively recognizes a phosphorylated motif in PADI6 in extracts of mature eggs.
Oocyte maturation is induced in vivo by LH, which results in lowered oocyte cAMP levels. Mammalian oocytes, however, mature spontaneously in vitro, in the absence of LH, when isolated from ovarian follicles. Proteins were extracted from both in vivo- and in vitro-matured oocytes, resolved, blotted, and probed with anti-p(S)YWHA binding motif antibody. Immunoreactivity was observed in both in vivo- and in vitro-matured eggs at 75 kDa (Fig. 1C). Therefore, PADI6 phosphorylation is associated with hormone-independent spontaneous oocyte maturation as well as with in vivo oocyte maturation. These results indicate that both in vivo- and in vitro-matured eggs can be used to study PADI6 phosphorylation and interactions with YWHA. Phosphorylation of PADI6 likely is caused by signaling events that are activated soon after maturation is initiated. Phosphorylated PADI6 is easily detected within 1 h of release from meiotic arrest by removal of dbcAMP from the in vitro maturation medium (Fig. 1D).
Sequencing data revealed the possible presence of two proteins in the 75-kDa excised and sequenced band, PADI6 and HSPA8. To rule out the possibility that HSPA8 and not PADI6 is phosphorylated, we examined the phosphorylation status of HSPA8 following immunoprecipitation in egg extracts using an anti-HSPA8 antibody. Anti-HSPA8 probed blots of the immunoprecipitation pellet sample showed a band at 75 kDa (Fig. 3, lane 3), confirming that the anti-HSPA8 antibody immunoprecipitated the HSPA8 protein; however, when proteins from the same pellet sample were separated by electrophoresis, blotted, and probed with anti-p(S)YWHA binding motif antibody, no immunoreactivity was seen (Fig. 3, lane 1). The presence of immunoreactive protein was observed in the supernatant or flow-though sample that was blotted and probed with anti-p(S)YWHA binding motif antibody (Fig. 3, lane 2). This demonstrated that the anti-p(S)YWHA binding motif antibody does not label the HSPA8 protein. Control immunoprecipitation experiments using equivalent amounts of rabbit serum instead of anti-HSPA8 antibody showed no nonspecific binding of HSPA8 in the pellet fraction. When control samples were blotted and probed with anti-HSPA8, no immunoreactivity was seen in the pellet (Fig. 3, lane 4); however, the presence of immunoreactive protein was detected in supernatant (Fig. 3 lane 5).
FIG. 3.
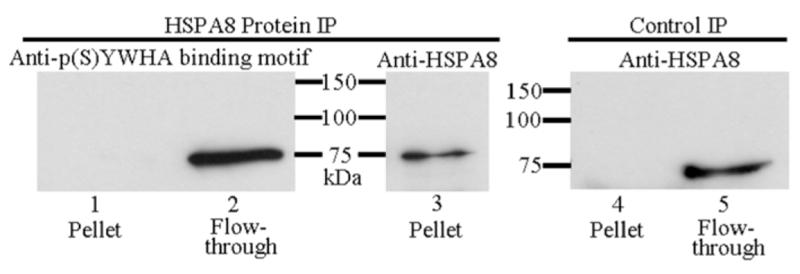
Immunoprecipitation (IP) of HSPA8 protein followed by SDS-PAGE and immunoblotting indicates that the p(S)YWHA binding motif antibody does not recognize HSPA8. Left) HSPA8 IP. Right) Control IP. The HSPA8 protein IP pellet sample was divided equally by volume, resolved by SDS-PAGE, blotted, and probed with the anti-p(S)YWHA binding motif antibody (lane 1) or anti-HSPA8 (lane 3). The HSPA8 IP pellet sample is not labeled by the anti-p(S)YWHA binding motif antibody, although the protein labeled by this antibody is in the flow-through fraction (lane 2). Control IP used rabbit serum in place of HSPA8 antibody. The HSPA8 protein was not immunoprecipitated (lane 4) though the protein was present in the flow-through protein faction (lane 5).
Two-dimensional PAGE analysis of egg and oocyte extracts with both the anti-p(S)YWHA binding motif and anti-PADI6 antibodies indicated an immunoreactive overlap in eggs. Specifically, both antibodies labeled an approximately 75-kDa, approximately 5.7-pI band in egg extract blots (Fig. 4, A and B). No reactivity was detected in oocyte extract blots probed with anti-p(S)YWHA (Fig. 4C), however, anti-PADI6 reactivity was observed (Fig. 4D). These results provide further evidence that the p(S)YWHA binding motif antibody selectively labeled PADI6 in extracts from mature eggs but not in extracts from immature oocytes. This information, combined with one-dimensional Western blot analysis, lambda protein phosphatase data, and the immunoprecipitation experiments ruling out HSPA8, indicate that the PADI6 protein may be phosphorylated at a YWHA binding site in eggs but in not oocytes. These experiments lead to the prediction that YWHA should bind to PADI6 in eggs (and not oocytes), because PADI6 may be phosphorylated at one or both of the two YWHA binding motifs present in the PADI6 protein.
FIG. 4.
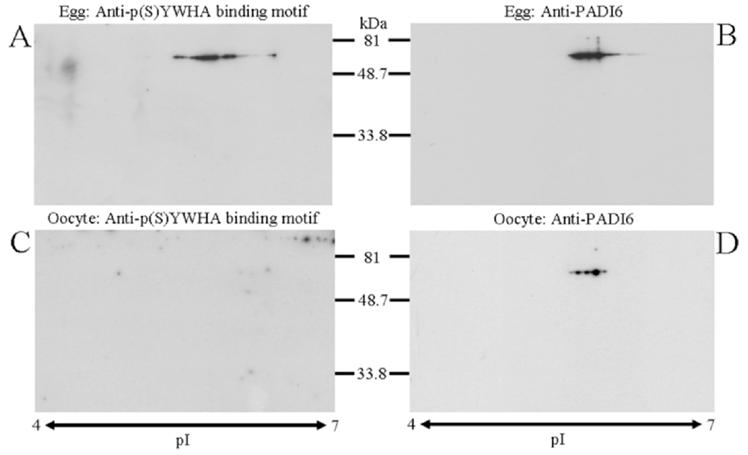
Western blots of egg and oocyte extracts after two-dimensional polyacrylamide gel electrophoresis provide additional evidence that anti-p(S)YWHA binding motif antibody labels PADI6 in eggs. For each blot shown, an equal number of cells were lysed, and the proteins were electrophoresed in two dimensions, blotted, and probed with the indicated antibody. Immunoreactivity for anti-PADI6 antibody was observed at approximately 75 kDa and an isoelectric point (pI) of approximately 5.7 in egg and oocyte blots (B and D), whereas anti-p(S)YWHA binding motif antibody reactivity was seen in a similar location only in eggs (A and C). The two-dimensional blots are representative of three experiments.
To explore the role of YWHA in these cells, we first confirmed the previous observation that YWHA can be detected by Western blot in eggs [36] and showed that it also is present in oocytes. Extracts of egg and oocytes were resolved by SDS-PAGE, blotted, and probed with anti-YWHA antibody. The YWHA protein was present in eggs and oocytes in approximately the same amount (Fig. 5A). We reprobed this blot with anti-p(S)YWHA binding motif antibody, which again clearly showed the PADI6 protein is phosphorylated in eggs but not oocytes.
FIG. 5.
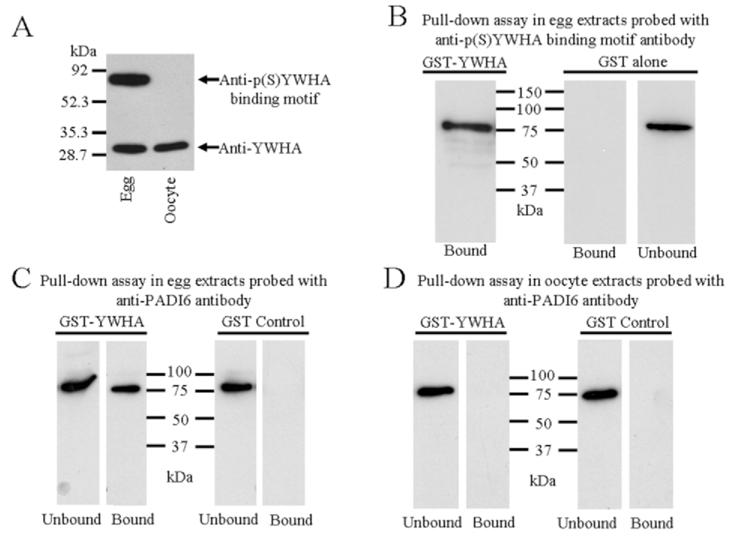
A) Immunoblotted egg and oocyte extracts probed with anti-YWHA and reprobed with anti-p(S)YWHA binding motif antibody, indicating the presence of the YWHA protein in both egg and oocyte. Confirming the previous experiments, the phosphorylated YWHA binding motif of PADI6 is recognized only in egg extracts. B—D) YWHA pull-down assays in eggs and oocytes. Triton X-100-extracted eggs or oocytes were incubated with GST-YWHA fusion protein attached to glutathione sepharose beads (lanes labeled GST-YWHA). In the control experiments, extracts were incubated with recombinant GST alone attached to the beads (lanes labeled GST alone or GST control). B) GST-YWHA bound a protein in egg extracts that was labeled with the anti-p(S)YWHA binding motif antibody (GST-YWHA, Bound lane). GST alone did not bind this protein (GST alone, Bound lane) but it was present in the flow-through or eluate (GST alone, Bound lane). C) A similar GST-YWHA pull-down experiment was performed egg extracts. Following polyacrylamide gel electrophoresis, the proteins were blotted and probed with anti-PADI6; GST-YWHA bound PADI6 (GST-YWHA, Bound lane). As expected, not all of the PADI6 was bound, and some could be detected in the eluate (GST-YWHA, Unbound lane). In control experiments with egg preparations, GST alone did not bind PADI6 (GST Control, Bound lane), but it was detected in the eluate (GST Control, Unbound). D) GST-YWHA did not bind PADI6 in oocyte extracts (GST-YWHA, Bound lane), nor did GST alone (GST Control, Bound), though PADI6 was in the eluate (Unbound lanes). B–D are representative of three separate experiments.
To test the hypothesis that YWHA interacts with PADI6 in eggs, a GST-YWHA fusion protein was used in pull-down experiments of both egg and oocyte extracts. Extracts of oocytes or eggs were incubated with GST-YWHA fusion protein bound to glutathione sepharose beads. In the control experiments, extracts were incubated with recombinant GST alone bound to the beads. The pellet sample containing the GST-YWHA (or GST alone) and any bound proteins was eluted directly into sample buffer, electrophoresed, blotted, and probed with the anti-p(S)YWHA binding motif antibody or anti-PADI6. The GST-YWHA bound an egg protein that was labeled with the anti-p(S)YWHA binding motif antibody (Fig. 5B, GST-YWHA, Bound lane). The GST alone did not bind this protein, but it was present in the supernatant or flow-through sample (Fig. 5B, GST Alone, Bound and Unbound lanes). Moreover, when the same pull-down experiment was repeated and the fractions were blotted and probed with anti-PADI6, the results were identical; GST-YWHA bound the protein recognized by anti-PADI6 (Fig. 5C , GST-YWHA, Bound lane). The GST alone did not bind PADI6, but PADI6 was present in the supernatant or flow-through sample (Fig. 5C, GST Control, Bound and Unbound lanes). This provides evidence that YWHA specifically binds to PADI6 in mature eggs, which also is phosphorylated as indicated by the anti-p(S)YWHA binding motif antibody.
When the same GST-YWHA pull-down experiment was performed using extracts of oocytes and the pellet protein fraction was electrophoresed, blotted, and probed with anti-PADI6, the results demonstrated that YWHA did not bind PADI6 in oocyte extracts (Fig. 5D, GST-YWHA, Bound lane), but it certainly was present in the flow-through (Fig. 5D, GST-YWHA, Unbound lane). Similar to the experiments with egg extracts, GST alone did not bind PADI6 in oocyte extracts, but it was present in the flow-through sample (Fig. 5D, GST Control, Bound and Unbound lanes), thus ruling out the chance of nonspecific interactions with GST.
To demonstrate that YWHA does not bind unphosphorylated PADI6, GST-YWHA pull-down analyses were performed after egg extracts were treated with lambda phosphatase to dephosphorylate egg proteins. Following the GST-YWHA pull-down procedure, bound and unbound (pellet and flow-through, respectively) fractions were electrophoresed, blotted, and probed with anti-p(S)YWHA binding motif antibody. As expected, neither fraction was labeled by this antibody (Fig. 6A), because the proteins were dephosphorylated. As shown previously, however, phosphorylated PADI6 was detected in extracts of eggs not treated with lambda phosphatase (Fig. 6A, control). When the same blot was reprobed with anti-PADI6, a band indicating the presence of PADI6 protein was observed in the unbound fraction but not in the bound fraction (Fig. 6B). These data provide further evidence for the phosphorylation-dependent interaction of PADI6 and YWHA.
FIG. 6.
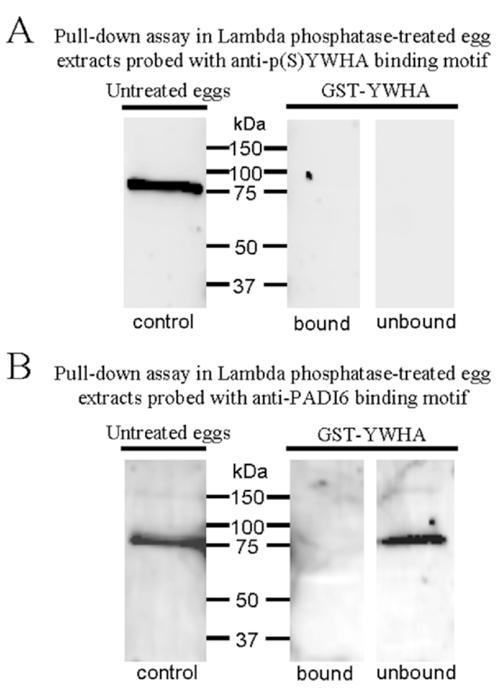
Western blots of GST-YWHA pull-down assay of egg extracts treated with lambda phosphatase. A) GST-YWHA bound and unbound fractions probed with anti-p(S)YWHA demonstrate that the phosphatase treatment removes phosphates in these extracts, because the antibody no longer recognizes any protein. The control lane consists of an extract of eggs not treated with lambda phosphatase and not used in the pull-down assay, and it confirms that the same antibody preparation is effective. B) The same blot as in A reprobed with anti-PADI6. The PADI6 protein was present in the GST-YWHA unbound fraction and in the untreated control but was undetectable in the GST-YWHA bound lane, providing additional evidence that the YWHA/PADI6 interaction is phospho-dependent. This experiment was repeated three times with identical results.
While results from the experiments described above indicate the PADI6 is a YWHA binding protein, when phosphorylated, there is some concern with these kinds of studies that the proteins of interest may not be interacting in vivo. We used the powerful technique of TAP to demonstrate an in vivo PADI6 and YWHA interaction in the mature mouse egg. Transgenic mice expressing TAP-tagged YWHA under the control of the ubiquitin C promoter were used to demonstrate an endogenous YWHA/PADI6 interaction. Before the purification was performed, we confirmed that mice carried the TAP-YWHA construct by PCR of genomic DNA and then confirmed [33] that such animals expressed the TAP-YWHA protein in reproductive tissues and cells. Extracts of ovaries and eggs were prepared from transgenic mice known to contain the TAP-tagged YWHA by PCR. Proteins from these extracts were resolved on a gel and probed in a Western blot with anti-YWHA, which recognizes YWHA. Ovaries contained TAP-YWHA (data not shown), and more importantly, as shown in Figure 7B, TAP-YWHA was detected in mature eggs.
FIG. 7.
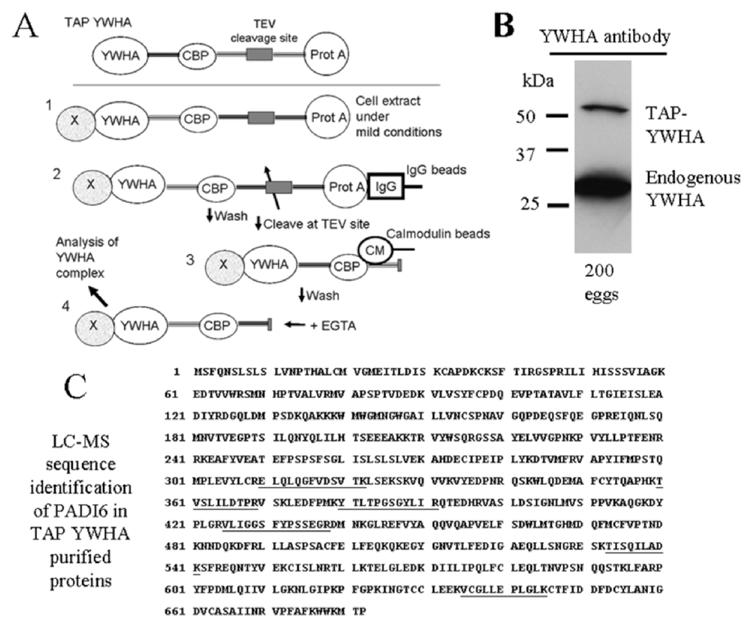
Procedures for the TAP of YWHA binding partners, TAP-YWHA expression, and LC-MS analysis. A) The TAP tag consists of dual tag of protein A from Staphylococcus aureus (Prot A) and calmodulin binding peptide (CBP): 1) In the first step of purification, cell extract containing TAP-YWHA and bound proteins is obtained under mild conditions. 2) The affinity for protein A to IgG is exploited to immobilize the TAP-YWHA to IgG-sepharose. Complexes are washed to remove proteins binding nonspecifically, and the CBP-YWHA is released by incubating the beads with TEV protease. 3) CBP is bound to calmodulin beads in presence of calcium. 4) Finally, the fusion protein, along with its binding partners, is eluted in a buffer containing EGTA, and the complex is characterized. B) Extracts of 200 eggs were prepared from transgenic mice known to contain the TAP-tagged YWHA by PCR. These extracts were resolved on a gel and probed in a Western blot with an anti-YWHA antibody to confirm that the eggs contained expressed TAP-YWHA. In these experiments, the anti-YWHA antibody labels any endogenous YWHA as well as the TAP-YWHA, which is detected at a higher molecular weight because it has the TAP construct associated with it. C) Results of the LC-MS indicate that the final eluate from the TAP-YWHA purification of 2400 mature eggs contained PADI6. The TAP-YWHA associated proteins were separated by SDS-PAGE, silver-stained, and sequenced by mass spectroscopic analysis. Sequencing data indicated that PADI6 was present in the band at approximately 75 kDa. Microsequenced peptides covered 10% of the mouse PADI6 sequence shown (UniProt primary accession no. Q8K3V4) with 100% identity (underlined portions).
The TAP procedure as described in Materials and Methods is outlined in Figure 7A. Mature eggs (n = 2400) were retrieved from TAP-YWHA-positive females. Proteins from the final eluate of the two-step purification were separated by polyacrylamide gel electrophoresis. The gel was stained with silver, and bands were excised for LC-MS as described in Materials and Methods. Analysis revealed that PADI6 was present, having bound to the TAP-YWHA produced in eggs of the transgenic animals. The recovered sequenced peptides matched 10% of the full PADI6 sequence with 100% identity (Fig. 7C). This experiment, using one of the most specific purification methods available today along with LC-MS, indicates in vivo binding of PADI6 to YWHA in mature eggs.
DISCUSSION
Mammalian oocytes become arrested at prophase I of meiosis, and viability of female gametes depends on the development of cellular and molecular competencies during oocyte maturation. As the oocyte resumes the cell cycle and forms a mature egg, the developing germ cell undergoes chromatin and cytoskeletal reorganization, develops mechanisms to block polyspermy, and acquires the ability to shift from maternal to zygotic gene expression if fertilized. The YWHA protein is known to function in amphibian oocyte maturation [11, 13] and plays a role in later amphibian development [10, 11]; however, the role of YWHA in mammalian oocyte maturation and development is not known. To determine the part that YWHA may play in mouse oocyte maturation and embryonic development, we searched for YWHA binding partners using an antibody that recognizes the YWHA binding motif in target proteins. We identified PADI6 as a YWHA-interacting protein. The results of the experiments presented here indicate that the interaction of YWHA and PADI6 in mature eggs is dependent on the phosphorylation status of PADI6. Thus far, PADI6 is the only protein that has been demonstrated to bind to YWHA in the mouse egg, but there may be others.
Evidence for the YWHA/PADI6 interaction in mature eggs is provided by the GST-YWHA pull-down experiments and by the transgenic TAP-YWHA purification procedure. The TAP method originally was developed in yeast [37], and its use has led to the discovery of new protein-protein interactions in studies of bacteria, yeast, and other cells in culture. The TAP method is highly sensitive, selective, and particularly suited for detecting multimeric protein complexes in their native cellular environment [32]. The key advantages of TAP purification over other affinity purification methods are, first, that the purification is under native conditions, preserving protein-protein interactions, and, second, that the two-step purification dramatically reduces the complexity of analysis by very efficient reduction of the nonspecific binding of background proteins. At present, TAP coupled with LC-MS is one of the best procedures for examining protein complexes in their native state within cells. Using this method, we confirmed the in vivo interaction of PADI6 with YWHA in the mature egg.
The PADI6 sequence analysis suggests that the protein contains two potential YWHA binding domains. The putative YWHA binding domains in PADI6 contain an arginine at position −3 with respect to serine (ArgXXpSer), which is thought to be important for serine phosphorylation [38] and is important for YWHA binding [39]. The manufacturer’s notes indicate that the p(S)YWHA binding motif antibody used in the present study binds to proteins containing the sequence ArgXXpSerXPro, a common YWHA binding motif, but proline at position +2 is not required for all YWHA interactions [38, 40]. Moreover, this particular phosphoserine-dependent antibody is known to bind to phosphorylated serine motifs that do not contain proline [26]. The anti-p(S)YWHA binding motif antibody does not bind to PADI6 if the egg proteins, before electrophoresis or membrane blots, are treated with a phosphatase to remove phosphates, confirming that the antibody is phospho-specific. The antibody recognizes PADI6 only in the mature egg. It is apparent that in the transition from oocyte to egg, PADI6 undergoes a change in phosphorylation, likely at YWHA binding sites that can be detected by the p(S)YWHA binding motif antibody. This stage-specific change in phosphorylation alone may influence the function of the PADI6 protein. In addition, the binding of YWHA, which occurs only in mature eggs, may modulate or alter PADI6 function as well. The YWHA antibody that we used recognizes several isoforms. We have not yet determined which YWHA isoform may be important in the egg; however, in many cells, YWHA isoforms may be interchangeable and have shared activities. The GST pull-down and TAP-YWHA experiments used a YWHAZ (YWHAζ) construct, and a specific isoform analysis for YWHA in the egg will be needed.
Based on some preliminary experiments using a general phosphoprotein probe (data not shown), we speculate that PADI6 may exist as a phosphorylated protein in both oocyte and egg; however, in the transition of oocyte to egg, a limited number of additional phosphorylation events or changes in the pattern of phosphorylation occur. We might expect that at least one phosphate is added, as evidenced by the anti-p(S)YWHA binding motif antibody experiments. We do not see a significant shift in the isoelectric point (pI) when comparing oocyte and egg PADI6 using two-dimensional electrophoresis; however, the addition of one phosphate may not be enough to produce a visible shift in pI. Moreover, it is possible that PADI6 is both dephosphorylated and phosphorylated at multiple sites during oocyte maturation. One could imagine phosphorylation at one site (a YWHA binding motif) with dephosphorylation at another location; such a situation would not alter the pI.
The PADI6 proteins in eggs of other species also may be phosphorylated and bind YWHA in a similar manner. Strong conservation exists within the PADI6 isotype for those species that have been examined. For example, mouse PADI6 shares high-sequence conservation with human PADI6 [21]. The human PADI6 protein contains putative YWHA binding motifs very similar to those we identified in mouse, although the arginine at position −3 (with respect to serine) may be replaced by lysine. This, however, also is a known YWHA binding motif. There may be a similar, stage-specific PADI6 phosphorylation and subsequent PADI6/YWHA interaction in human eggs and, perhaps, in other mammals.
The PADI6 first was described after its discovery in mouse oocytes, eggs, and early embryos [16] and subsequently was shown to be expressed as transcripts in human ovary, peripheral blood leukocytes, and testes by RT-PCR [21]. The homologous gene sequence is known for rat, dog, cat, chimpanzee, and several other species. The PADI6 shares some features with other PAD enzymes, and potential target proteins exist within the oocyte and egg (e.g., cytokeratins and histones). The specific targets and pathways for regulation of PADI6, however, are not known at this time. Moreover, PADI6 may have some unique properties that distinguish it from other PAD enzymes. The PADs 1–4 are regulated by calcium [41]. All mammalian PAD enzymes are highly conserved; however, whereas the catalytic region of PADI6 is similar to those of other PAD enzymes, the protein contains a number of amino acid substitutions that may be important in calcium binding. These substitutions suggest that PADI6 activity may not be regulated by calcium [42]. Thus far, enzymatic activity of PADI6 has not been demonstrated in an in vitro system. For example, unlike PADI4 (and all the other isoforms), PADI6 is unable to deiminate arginine in histone-derived synthetic peptides [20]. This could be a result of the absence of calcium-induced conformational changes in the protein that are critical for activation of the active site, or the peptides tested in vitro may not be similar to protein targets within the oocyte. Evidence for PADI6 catalytic activity in oocytes is indirect (see below) but suggests that PADI6 is active in growing oocytes. Studies of PADI6 enzymatic activity (both in vivo and in vitro) should now take into account the phosphorylation status of the protein.
It has been reported that PADI6, as determined by immunoelectron microscopy, is colocalized to keratin-containing intermediate filaments, also known as cytoplasmic sheets, in the mouse oocyte [16]. If an active enzyme, PADI6 may citrullinate oocyte or egg proteins, and it has been suggested that one target could be the cytokeratins in these cells. Recently, a PADI6 conditional knockout mouse was generated [23]. Female mice deficient in PADI6 are infertile but otherwise normal. Infertility results from a block of further development beyond the two-cell-stage embryo. This block is linked to an apparent lack of an unknown citrullinated protein or proteins in the oocytes. Furthermore, developing oocytes without PADI6 lack cytoplasmic sheets. The evidence that PADI6 is an active enzyme in the mouse oocyte is based on indirect immunohistochemical analysis with an antibody raised against a histone H4 peptide containing citrulline. The exact identity of the citrullinated protein or proteins in the oocyte is not known. Absence of some citrullinated protein in the PADI6 knockout mouse suggests that PADI6 may be active in growing oocytes even at low cytoplasmic calcium which might reflect a different calcium sensitivity, or because the calcium binding sites in PADI6 are not conserved compared to other PADs, that PADI6 activity is regulated in some other manner.
If it is confirmed that PADI6 has enzymatic activity in oocytes, it may be that this activity must be altered during the later stages of meiosis and during fertilization—for example, to prevent citrullination of histone proteins in the egg or sperm chromatin, which are exposed to the cytoplasmic enzymes in the mature egg. The regulation of PADI6 may depend on its phosphorylation status and/or interaction with YWHA, particularly if PADI6 is not regulated by calcium. In addition, because the calcium sensitivity of PADI6 has not been examined directly, we do not know if the large calcium transients that accompany fertilization [24] might be sufficient to activate the enzyme in the fertilized egg, despite the alterations in the calcium binding domain of this PAD isotype. This calcium increase may be enough to stimulate PADI6 citrullination activity. Again, the phosphorylation of PADI6 or its interaction with YWHA may modulate PADI6 activity if it is sensitive to high endogenous calcium. A direct test for enzymatic activity in the oocyte and egg is needed, and this test should take into account the phosphorylation status of PADI6 and YWHA binding. The interaction between YWHA and PADI6 may alter enzyme activity either directly or by protecting the phosphorylated site on PADI6. The YWHA protein has been shown to protect phosphorylated proteins from inopportune dephosphorylation [13]. Alternatively, YWHA proteins modify associations or interactions of the proteins to which they are bound. Interactions of PADI6 with other proteins may be altered when it is phosphorylated and bound to YWHA.
The identity of the kinases that may phosphorylate the potential YWHA binding motifs in PADI6 remains unknown. The YWHA binding domain kinases include PKA, AKT1 (PKB), PKC, PAK1 (p21 [CDKN1A]-activated kinase 1), RPS6KA1 (ribosomal protein S6 kinase polypeptide 1, also known as RSK1, MAP kinase-activated protein kinase 1a, or MAPKAPK1A), and MAPKAPK2 (MAP kinase-activated protein kinase 2) [1]. Examination of the two putative YWHA binding motifs in PADI6 reveals that these sites are potential phosphorylation targets for several of these kinases. For example, Ser-400 in one of the YWHA binding motifs is a potential target for PKA, PKC, RPS6KA1, and AKT1. Of these, it will be interesting to examine PKB and RPS6KA1, because the activity of each increases before or about the time of germinal vesicle breakdown during oocyte maturation [43, 44]. Phosphorylation of proteins may be dependent on the dynamic interaction of kinases and phosphatases, and the role of phosphatases will need to be examined as well. PADI6 appears to be a unique member of the PAD family. Future examination of PADI6 activity and function in the growing oocyte, egg, and two-cell embryo will need to take into account phosphorylation status and YWHA interaction.
ACKNOWLEDGMENTS
We thank Kimberly Meyers for advice on some of the experiments, Dr. Scott Coonrod (Cornell University) for generously supplying the PADI6 antibody and some initial discussions, Dr. Michael Kinter (Cleveland Clinic Foundation) for advice on sequencing methods, and Dr. Helen Piwnica-Worms (Washington University) for providing the YWHA cDNA.
Footnotes
Supported by National Institutes of Health grant HD38520 to S.V.
REFERENCES
- 1.Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Fu HA, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Yaffe MB. How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 4.van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signaling, and apoptosis. Bioessays. 2001;23:936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- 5.Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- 6.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meek SEM, Lane WS, Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 8.Rubio MP, Peggie M, Wong BHC, Morrice N, MacKintosh C. 14-3-3s regulate fructose-2,6-bisphosphate levels by binding to PKB-phosphorylated cardiac fructose-2,6-bisphosphate kinase/phosphatase. EMBO J. 2003;22:3514–3523. doi: 10.1093/emboj/cdg363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 2005;re10 doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- 10.Lau JMC, Wu CL, Muslin AJ. Differential role of 14-3-3 family members in Xenopus development. Dev Dyn. 2006;235:1761–1776. doi: 10.1002/dvdy.20816. [DOI] [PubMed] [Google Scholar]
- 11.Darling DL, Yingling J, Wynshaw-Boris A. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr Top Dev Biol. 2005;68:281–315. doi: 10.1016/S0070-2153(05)68010-6. [DOI] [PubMed] [Google Scholar]
- 12.Uchida S, Kubo A, Kizu R, Nakagama H, Matsunaga T, Ishizaka Y, Yamashita K. Amino acids C-terminal to the 14-3-3 binding motif in CDC25B affect the efficiency of 14-3-3 binding. J Biochem. 2006;139:761–769. doi: 10.1093/jb/mvj079. [DOI] [PubMed] [Google Scholar]
- 13.Margolis SS, Walsh S, Weiser DC, Yoshida M, Shenolikar S, Kornbluth S. PP1 control of M phase entry exerted through 14-3-3-regulated Cdc25 dephosphorylation. EMBO J. 2003;22:5734–5745. doi: 10.1093/emboj/cdg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Winkler K, Yoshida M, Kornbluth S. Maintenance of G(2) arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J. 1999;18:2174–2183. doi: 10.1093/emboj/18.8.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermeking H, Benzinger A. 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol. 2006;16:183–192. doi: 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC, Hao ZL, Jayes FC, Bush LA, Shetty J, Shore AN, Reddi PP, et al. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol. 2003;256:73–88. doi: 10.1016/s0012-1606(02)00126-4. [DOI] [PubMed] [Google Scholar]
- 17.Vossenaar ER, Zendman AJW, van Venrooij WJ, Pruijn GJM. PAD, a growing family of citrullinating enzymes: genes, features, and involvement in disease. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 18.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chemical Biology. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 20.Raijmakers R, Zendman AJW, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes F, van Veelen PA, Drijfhout JW, Pruijn GJM. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol. 2007;367:1118–1129. doi: 10.1016/j.jmb.2007.01.054. [DOI] [PubMed] [Google Scholar]
- 21.Chavanas S, Mechin MC, Takahara H, Kawada A, Nachat R, Serre G, Simon M. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved noncoding segments and a new human gene, PADI6. Gene. 2004;330:19–27. doi: 10.1016/j.gene.2003.12.038. [DOI] [PubMed] [Google Scholar]
- 22.Zhang JY, Dai JL, Zhao EP, Lin Y, Zeng L, Chen JZ, Zheng H, Wang Y, Li X, Ying K, Xie Y, Mao YM. cDNA cloning, gene organization, and expression analysis of human peptidylarginine deiminase type VI. Acta Biochim Pol. 2004;51:1051–1058. [PubMed] [Google Scholar]
- 23.Esposito G, Vitale AM, Leijten FPJ, Strik AM, Koonen-Reemst A, Yurttas P, Robben T, Coonrod S, Gossen JA. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007;273:25–31. doi: 10.1016/j.mce.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Kline D, Kline JT. Repetitive calcium transients and the role of calcium in exocytosis and cell-cycle activation in the mouse egg. Dev Biol. 1992;149:80–89. doi: 10.1016/0012-1606(92)90265-i. [DOI] [PubMed] [Google Scholar]
- 25.Mehlmann LM, Kline D. Regulation of intracellular calcium in the mouse egg—calcium release in response to sperm or inositol trisphosphate is enhanced after meiotic maturation. Biol Reprod. 1994;51:1088–1098. doi: 10.1095/biolreprod51.6.1088. [DOI] [PubMed] [Google Scholar]
- 26.Liu MY, Cai SL, Espejo A, Bedford MT, Walker CL. 14-3-3 interacts with the tumor suppressor tuberin at Akt phosphorylation site(s) Cancer Res. 2002;62:6475–6480. [PubMed] [Google Scholar]
- 27.Shen YH, Godlewski J, Bronisz A, Zhu J, Comb MJ, Avruch J, Tzivion G. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol Biol Cell. 2003;14:4721–4733. doi: 10.1091/mbc.E02-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zang MW, Waelde CA, Xiang XQ, Rana A, Wen R, Luo ZJ. Microtubule integrity regulates Pak leading to ras-independent activation of Raf-1— insights into mechanisms of Raf-1 activation. J Biol Chem. 2001;276:25157–25165. doi: 10.1074/jbc.M100152200. [DOI] [PubMed] [Google Scholar]
- 29.Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, Dalal SN, DeCaprio JA, Greenberg ME, Yaffe MB. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foschi M, Franchi F, Han JH, La Villa G, Sorokin A. Endothelin-1 induces serine phosphorylation of the adaptor protein p66(Shc) and its association with 14-3-3 protein in glomerular mesangial cells. J Biol Chem. 2001;276:26640–26647. doi: 10.1074/jbc.M102008200. [DOI] [PubMed] [Google Scholar]
- 31.Diao JY, Khine AA, Sarangi F, Hsu E, Iorio C, Tibbles LA, Woodgett JR, Penninger J, Richardson CD. X protein of hepatitis B virus inhibits Fasmediated apoptosis and is associated with up-regulation of the SAPK/JNK pathway. J Biol Chem. 2001;276:8328–8340. doi: 10.1074/jbc.M006026200. [DOI] [PubMed] [Google Scholar]
- 32.Berggard T, Linse S, James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 33.Angrand PO, Segura I, Volkel P, Ghidelli S, Terry R, Brajenovic M, Vintersten K, Klein R, Superti-Furga G, Drewes G, Kuster B, Bouwmeester T, Acker-Palmer A. Transgenic mouse proteomics identifies new 14-3-3-associated proteins involved in cytoskeletal rearrangements and cell signaling. Mol Cell Proteomics. 2006;5:2211–2227. doi: 10.1074/mcp.M600147-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 36.Haraguchi S, Naito K, Sato E. MAP kinase cascade, but not ERKs, activated during early cleavage of mouse embryos. Mol Reprod Dev. 1998;51:148–155. doi: 10.1002/(SICI)1098-2795(199810)51:2<148::AID-MRD4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 37.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 38.Tzivion G, Avruch J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 39.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 40.Zhang SH, Kobayashi R, Graves PR, PiwnicaWorms H, Tonks NK. Serine phosphorylation-dependent association of the band 4.1-related proteintyrosine phosphatase PTPH1 with 14-3-3 beta protein. J Biol Chem. 1997;272:27281–27287. doi: 10.1074/jbc.272.43.27281. [DOI] [PubMed] [Google Scholar]
- 41.Vossenaar ER, Radstake TRD, van der Heijden A, van Mansum MAM, Dieteren C, de Rooij DJ, Barrera P, Zendman AJW, van Venrooij WJ. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann Rheum Dis. 2004;63:373–381. doi: 10.1136/ard.2003.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca2+-induced activation of human PAD4. Nat Struct Mol Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 43.Kalous J, Solc P, Baran V, Kubelka M, Schultz RM, Motlik J. PKB/AKT is involved in resumption of meiosis in mouse oocytes. Biol Cell. 2006;98:111–123. doi: 10.1042/BC20050020. [DOI] [PubMed] [Google Scholar]
- 44.Gavin AC, Schorderet-Slatkine S. Ribosomal S6 kinase p90(rsk) and mRNA cap-binding protein eIF4E phosphorylations correlate with MAP kinase activation during meiotic reinitiation of mouse oocytes. Mol Reprod Dev. 1997;46:383–391. doi: 10.1002/(SICI)1098-2795(199703)46:3<383::AID-MRD18>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]


