Abstract
Monitoring adverse events (AEs) is an important part of clinical research and a crucial target for data standards. The representation of adverse events themselves requires the use of controlled vocabularies with thousands of needed clinical concepts. Several data standards for adverse events currently exist, each with a strong user base. The structure and features of these current adverse event data standards (including terminologies and classifications) are different, so comparisons and evaluations are not straightforward, nor are strategies for their harmonization. Three different data standards - the Medical Dictionary for Regulatory Activities (MedDRA) and the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) terminologies, and Common Terminology Criteria for Adverse Events (CTCAE) classification - are explored as candidate representations for AEs. This paper describes the structural features of each coding system, their content and relationship to the Unified Medical Language System (UMLS), and unsettled issues for future interoperability of these standards.
Keywords: Biomedical research, Adverse effects, Informatics, Forms and Records Control, Terminology, Systematized Nomenclature of Medicine
1. INTRODUCTION
An adverse event (AE) is broadly defined as any clinical event, sign, or symptom that goes in an unwanted direction. 1 Adverse events can encompass physical findings, complaints, and laboratory results. 1 Per FDA (U.S. Food and Drug Administration) and ICH (International Conference on Harmonisation) definitions, adverse events also include worsening of pre-existing conditions, so comprehensive AE coding systems must encompass diseases, disorders, and conditions as well. The structure and features of contending adverse event data standards (including terminologies and classifications) are different, so comparisons between and evaluations of data standards are not straightforward, nor are activities for determining concept equivalence across them. This paper describes 3 candidate data standards for representing adverse events: the Medical Dictionary for Regulatory Activities (MedDRA) and the Systematic Nomenclature of Medicine Clinical Terms (SNOMED CT) terminologies, and the Common Terminology Criteria for Adverse Events (CTCAE) classification. These data standards are weighed against general standards criteria and terminology desiderata, and studied in relation to the Unified Medical Language System (UMLS). The UMLS is not designed to be a coding standard, but rather acts as a “Rosetta stone” for translating synonymous or quasi-synonymous concepts across different source vocabularies. We use the UMLS as a tool to examine overlap between the candidate standards. Our description of these heterogeneous coding systems and insights on interoperability strategies should provide a timely resource for current discussions of uniform representation for adverse events and clinical research data. 2
2. BACKGROUND
2.1 Adverse Events in Clinical Research
Adverse event data drives decisions to terminate a study, or revise protocols, procedures or informed consent documents, in the interest of protecting human subjects. The need to monitor adverse events is a fundamental part of clinical research 1 , and the rationale for standard coding of AEs is similar to that for other clinical research data – it enables researchers to record information in a consistent manner. Data standards facilitate the sharing of data, and shared AE data has patient safety implications. 3, 4 Currently, two data standards, MedDRA and CTCAE, are widely used for AE reporting. A third, SNOMED CT, is an emerging standard for clinical care data, and therefore has an implied future relationship with any AE reporting data standard. Various functions for uniform AE data representation - including standardized AE reporting systems, the extraction of AEs from existing clinical care data sources, adverse event data management and communication systems, and aggregate analysis - dictate the requirements for an ideal AE data standard, and influence future relationships between competing data standards.
2.2 Medical Dictionary for Regulatory Activities (MedDRA)
The current standard for the regulated reporting of drug-related adverse events to the FDA, and other international regulatory entities per the ICH, is MedDRA. MedDRA is a medical terminology used by the regulated biopharmaceutical industry for data related to pre-marketing to post-marketing data reporting activities. 5 The scope includes signs, symptoms, diseases, diagnoses, therapeutic indications, qualitative result concepts (e.g., present, absent, increased, decreased), surgical and medical procedures, or medical, social, and family history. Maintenance and Support Services Organization (MSSO) updates and maintains MedDRA and distributes licensed copies to users in industry and regulatory agencies. 6 This is a proprietary commercial terminology, but free or reduced user fees are available for qualified institutions.
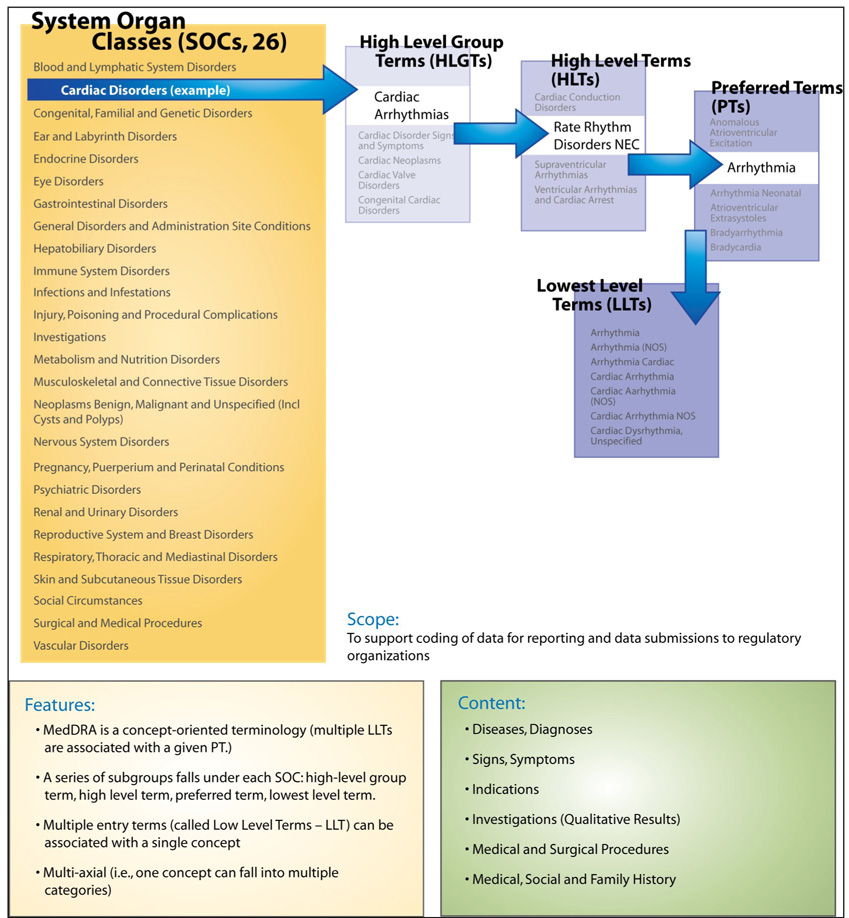
MedDRA is a multi-hierarchical terminology, with 26 top-level hierarchies called System Organ Classes (SOC) shown in Figure 1. Moving down the hierarchy for an SOC, High Level Group Terms (HLGTs) and High Level Terms (HLTs) organize MedDRA terms into “clinically relevant” groups. Preferred terms (PTs) are the “concepts” in MedDRA, each being a specific, unique and nonambiguous medical concept. One or more Lower Level Terms (LLTs) correspond to each PT; the LLTs are effectively entry terms that include synonyms, quasi-synonyms, lexical variants, and occasionally sub-elements. 6
Figure I.
MedDRA Upper-level Categories – called System Organ Class (SOC); (from NCI Terminology Browser:[http://nciterms.nci.nih.gov/NCIBrowser/Startup.do]
2.3 Systematized Nomenclature for Medicine Clinical Terms (SNOMED CT)
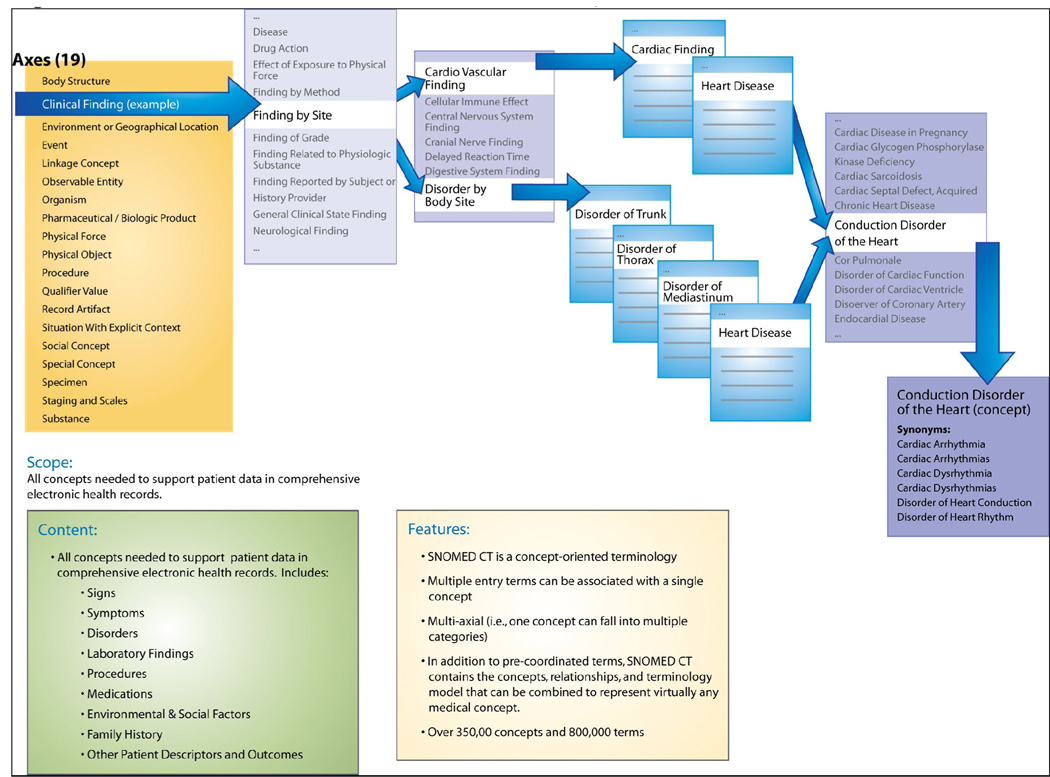
SNOMED CT is a large, formalized, comprehensive medical terminology intended to represent anything of clinical relevance in electronic medical records. 7, 8 Its scope includes: clinical findings, anatomy, test names and results, organisms, specimens, pharmaceutical and biologic products, medical and surgical procedures, social, medical, and family history, clinical and environmental events and other content areas. SNOMED CT is a proprietary and commercial terminology (College of American Pathologists), but in 2003 the National Library of Medicine negotiated a government license that allows access to core SNOMED CT content for United States users for at least 5 years. As of 2007, the ownership of SNOMED CT was officially transferred to the International Health Terminology Standards Development Organisation (IHTSDO), of which the United States is one of the founding member countries. This transfer was designed to increase global access, development, and use of SNOMED CT; however,the organization is new and increased global access to SNOMED CT has not yet been realized. The current right to use SNOMED CT within the United States remains unchanged.
In addition to the nearly 800,000 pre-coordinated terms, SNOMED CT has the terminology model, concepts, and relationships to construct almost any desired concept. SNOMED CT is a multi-hierarchical terminology, meaning that concepts can have more than one parent concept, usually from within the same hierarchy (or axis), although there is a very small number of cases of cross-axial hierarchical relationships. (Figure II). Additionally, the sophisticated terminology model of SNOMED CT allows concepts to be associated through a host of relationships – other than hierarchical – with other concepts (e.g., laterality, course, causative agent).
Figure II.
SNOMED Structure and Upper-level Categories, called Axes
Concepts in SNOMED CT are defined through relationships to other concepts. These defining relationships consist of at least one hierarchical (“is a”) relationship and other non-hierarchical relationships. For example, the concept “Cardiomegaly” is fully defined by one hierarchical relationship (is a “Structural disorder of heart”) and two non-hierarchical relationships (finding site of “Heart structure” and associated morphology of “Hypertrophy”).9 A concept is considered “fully defined” if its definition is sufficient to distinguish it from all its parent or sibling concepts, but “primitive” if it is just partially defined by its relationships. Of the 307,220 SNOMED CT concepts, only 46,567 (15.2%) are fully defined. Formal definitions in SNOMED CT are represented in the Description Logic formalism and can be validated computationally. 10
The U.S.-based Consolidated Health Informatics Initiative (CHI) made an official recommendation in 2004 that SNOMED CT be a standard terminology for Anatomy, Diagnoses and Problem Lists, Laboratory result contents, Non-laboratory Interventions and Procedures, and Nursing. 11 As SNOMED CT is indeed the pending standard for clinical data in the health care delivery domain, and because the vision of the National Health Information Infrastructure includes interoperability between clinical care data and research data 12, 13, SNOMED CT should be considered as a candidate standard for adverse event data. Regardless, SNOMED CT will have a relationship with other adverse event coding systems as use cases and applications that bring together clinical research and healthcare information systems emerge. Additionally, the relationship between SNOMED CT and clinical research has become more tangible since the FDA recently named a subset of SNOMED CT concepts as a standard for drug indications in the Structured Product Label. 14
2.4 Common Terminology Criteria for Adverse Events (CTCAE)
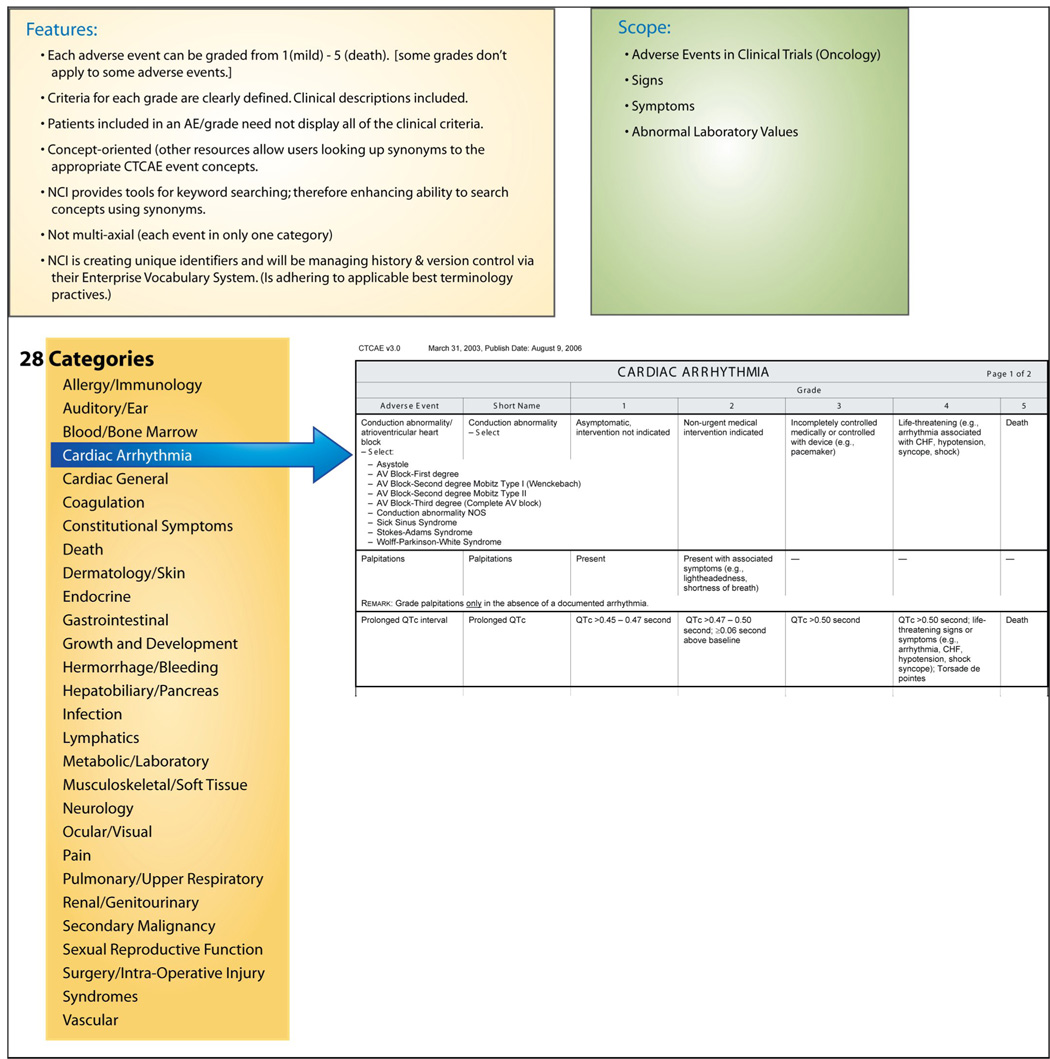
The Common Terminology Criteria for Averse Events (CTCAE) is an established (20 years) classification system for adverse event data collection and reporting in oncology research. The CTCAE was developed and is maintained by the National Cancer Institute (NCI), and its use is required for all NCI-funded studies. CTCAE is the dominant standard for reporting AEs in clinical trials in the cancer domain. 15 In the CTCAE, adverse events are grouped into one of 28 categories – broad classifications of AEs based on anatomy (e.g., “Dermatology/Skin”) or pathophysiology (“Hemorrhage/Bleeding”). Embedded in the CTCAE is a severity grading scale (1=mild, 2=moderate, 3=severe, 4=Life-threatening/ disabling, 5=death) for each event. For any given adverse event, specific definitional criteria are listed by grade. (Figure III) These clinical criteria are not required – there are often several clinical criteria that constitute the description for each AE and grade, yet only one need be present for inclusion in that grade. There are recent extensions to the CTCAE to represent radiation therapy AEs. Although the CTCAE is a classification and not a terminology, it is managed by NCI using well-accepted “good terminology practices” (e.g., concept permanence, non-semantic identifiers). Access to the classification, training, and supporting tools from the NCI is free to all users.
Figure III.
High-Level Categories and Structure for CTCAE.
2.5 Unified Medical Language System
The UMLS is a multi-purpose resource that includes concepts and terms from over 100 different source vocabularies, and establishes linkages across these source vocabularies with its own concept structure and semantic type categorization of concepts in the biomedical domain. 16−18 The UMLS, developed by the National Library of Medicine in 1986, is composed of three main knowledge components: Metathesaurus, Semantic Network and SPECIALIST Lexicon. The Semantic Network is designed to categorize concepts (using semantic types) in the UMLS Metathesaurus and organize relationships among the concept categories (using semantic relationships). Semantic types and relationships correspond to nodes and links in graphical representations of the Semantic Network. The SPECIALIST Lexicon is a natural language processing system that can assist in navigating UMLS concepts by using syntactic, grammatical, and spelling variations.
The UMLS tools support the mapping of multiple data standards to a common set of concepts via the Metathesaurus 19. The Metathesaurus contains over 1 million biomedical concepts from over 100 controlled vocabularies and classification systems, including MedDRA, SNOMED CT, and the CTCAE. The purpose of the Metathesaurus is to provide a common representation for information exchange between health-related information systems using various coding systems and vocabularies. The UMLS provides mappings across terms from different terminologies by integrating terms (from multiple source terminologies) with the same meanings into the same UMLS concept in the Metathesaurus. Using the UMLS, synonymy between two terms (from one or different source vocabularies) is asserted if they belong to a single UMLS concept. We use the UMLS concept as a lingua franca to determine synonymy and, as a result content overlap, across the three AE coding systems described above.
3. COMPARISON HIGHLIGHTS
3.1. Coverage and Content Overlap
As shown in Figure I & Figure II, the intended scope of both MedDRA and SNOMED CT extends beyond adverse events; only CTCAE explicitly limits its coverage to adverse events. In order to get a quantitative sense of the coverage and overlap of the three vocabularies, we present some counts of the source terms and their corresponding UMLS concepts (based on the 2007AA release of UMLS). SNOMED CT is by far the largest vocabulary, with 307,220 preferred terms (every SNOMED CT concept has exactly one preferred term). MedDRA is significantly smaller, and contains 17,505 preferred terms - the level of granularity most commonly used in AE reporting. The CTCAE has 1,060 preferred terms (ignoring the hierarchical terms and terms beginning with grading information). A crude assessment of coverage by the number of preferred terms shows that, interestingly, SNOMED CT is about 17 times bigger than MedDRA which is in turn about 17 times bigger than CTCAE. Due to its broad scope of coverage, not all SNOMED CT concepts are relevant to AE reporting. The “Clinical Finding” axis is the most relevant axis. There are 107,743 preferred terms in the findings axis, which is about 6 times greater in size than MedDRA.
The number of UMLS concepts containing preferred terms from SNOMED CT, MedDRA and CTCAE is 284,859, 17,197 and 1,060 respectively. The numbers for SNOMED CT and MedDRA are smaller than the number of preferred terms because one UMLS concept can contain more than one preferred term from the same terminology. Overlap is defined by the presence of preferred terms from different terminologies within the same UMLS concept. It is found that 22% of CTCAE overlaps with MedDRA and 51% of MedDRA overlaps with SNOMED CT. As expected, most of the overlap (79%) between MedDRA and SNOMED CT occurs within the “Clinical Finding” axis of SNOMED CT.
3.2 Structural Comparisons
Hierarchical relationships are present in all 3 coding systems. In general, the number of hierarchical relationships can be a measure of the depth and granularity of a terminology or classification. Hierarchical relationships may be conceptualized as “navigational pathways” – either cognitive steps that users would take to browse the coding system, or relationships that facilitate the retrieval and aggregate analysis of stored data. Because MedDRA’s hierarchies are rigidly defined by the terminology structure rather than by its content, there are never more than 4 levels of “depth” for a given MedDRA term. SNOMED CT, in contrast, has no restrictions on the depth of a tree (e.g., the deepest hierarchy in SNOMED CT has 30 levels). To illustrate, the concept “Arrhythmia” in MedDRA has 2 different “paths” upward to the SOC of “Cardiac Disorders”, but each of those paths can only contain 3 MedDRA concepts (an HLT, and HLGT, and the SOC). [Example: 1.) ‘Arrhythmia’ (pt) → ‘Supraventricular arrhythmias’ → (hlt) → ‘Cardiac arrhythmias’ (hlgt) → ‘Cardiac Disorders’ (soc) 2.) ‘Arrhythmia’ (pt) → ‘Ventricular arrhythmias and cardiac arrest’ (hlt) → ‘Cardiac arrhythmias’ (hlgt) → ‘Cardiac Disorders’ (soc).]
The same concept in SNOMED CT can also have multiple pathways upward to the SNOMED CT high-level “Clinical Finding” concept, but there are no restrictions on the depth of each path. In the “palpitations” example, there are 4 different conceptual pathways to the high-level concept “Clinical Finding” in SNOMED CT. One such path upward includes the following concepts [note: read → as “is-a”): ‘Palpitations’ → ‘Finding related to awareness of heartbeat’ → ‘Cardiac finding’ → ‘Cardiovascular finding’ → ‘Finding by site’ → ‘Clinical Finding’. Another path is ‘Palpitations’ → ‘Mediastinal finding’ → ‘Finding of region of thorax’ → ‘Finding of trunk structure’ → ‘Finding of body region’ → ‘Finding by site’ → ‘Clinical Finding’. In contrast, the structure of the CTCAE classification only has two levels of depth for a given adverse event concept – e.g., ‘Palpitations’ → ‘Cardiac Arrhythmias’.
The MedDRA terminology is structured to assume that the LLTs are clinically equivalent to their associate Preferred Term. MedDRA’s use of preferred terms associated with multiple LLT entry terms approximates a concept-orientation, a fundamental characteristic of sound terminologies, although the relationship (whether synonymous or quasi-synonymous) of LLTs to PTs is not explicitly stated. 20 While synonymy is context-dependent, there are clearly examples of LLTs (e.g., “Junctional bradycardia”, “Junctional tachycardia” and “Reciprocating tachycardia”) that are not synonymous with each other or to their associated PT (e.g., “Nodal arrhythmia”).
SNOMED CT allows polyhierarchy within axes (the 19 broadest groupings of concepts), meaning that a given concept can have any number of parent concepts from the same axis. The polyhierarchy feature is desirable for large terminologies, because it allows multiple strategies for terminology navigation and data integration, and can enable the automated placement of new concepts (to facilitate terminology growth and maintenance). While not a true polyhierarchy, a given PT in MedDRA can be associated with multiple HLTs (e.g., the PT “Gastric Ulcer Haemorrhage” is associated with both the HLT “Gastric Ulcers and Perforation” - part of the “Gastrointestinal Disorders” SOC, and also with the HLT “Gastrointestinal Haemorrhages - part of the ”Vascular Disorders” SOC.) To maintain that coded data is not counted twice, however, the HLTs and HLGTs are each only associated with exactly one SOC. In that regard, Bosquet et al (2005) observed that MedDRA behaves like a classification, and the lack of polyhierarchy inside of SOCs can lead to inconsistencies and difficulties querying complete groups of related concepts.
The MedDRA structure is based on a rigid level of imposed term specificity rather than by features of the underlying concepts. The SOC → HLGT → HLT → PT structure causes very large groups (in terms of numbers of concepts) at the SOC and HLGT levels and smaller groups at the HLT level that might have limited utility for analyses of aggregated data. 21 The counts for the different MedDRA term types are: SOC 26, HLGT 332, HLT 1682, PT 17,504. Aggregate analysis by SOC and HLGT levels, for example, can include a great many MedDRA concepts for a given SOC or HLGT grouping, yet analysis at the HLT or PT level can yield very granular groups, each containing relatively few AE concepts, with limited power for analysis.
Combining medically related AE terms can facilitate detection of safety issues by increasing the power of the analysis through larger numbers, a strategy of particular interest for examining AE data encoded with highly specific MedDRA concepts. 21 However, it has been cautioned that generating results at the HLT level is not valuable, as a given HLT can contain many heterogenous concepts (e.g., high blood pressure and low blood pressure). 21 Consequently, there can be a risk that events of varying clinical importance can be grouped together, and results can be skewed by high numbers of relatively unimportant events.
Although the structure of MedDRA inherently limits the number of hierarchical relationships, a series of specialized, manually-created Special Search Queries (SSCs) gathers concepts that are functionally related (e.g., the “Pain” SSC is a collection of several hundred concepts from different parts of MedDRA - such as “Migraine” (PT) from the “Vascular Disorders” and “Nervous System Disorders” SOCs, and “Myalgia” part of the “Musculoskeletal and Connective Tissue Disorders” SOC). 22 These queries support the aggregation and analysis of MedDRA-coded data, since the SSCs essentially offer the same functionality as true polyhierarchical coding schemes like SNOMED CT. The down-side is that these lists must be manually maintained – i.e., as new terms are added to MedDRA, all relevant existing queries must be manually updated to include the new terms, since the MedDRA structure does not provide the foundation for any sort of automated placement of new concepts. 23
The design of SNOMED CT includes a model for post-coordination – the creation of new concepts by combining existing concepts. Additionally, associative – or non-hierarchical - relationships in SNOMED CT provide a way for decomposing concepts into canonical representations (e.g., for identifying equivalence [or differences] between terms)‥ There are none of these “associative” relationships in MedDRA, since the model does not claim to support them. There are altogether 1 million associative (i.e. non-hierarchical and non-mapping) relationships in SNOMED CT. More relationships might indicate potential for effective coding (via browsing/navigating terminology), automated terminology maintenance (via automated concept placement and quality control), increased retrieval, and knowledge discovery (via data mining). However, there is no definitive argument that more relationships make a given terminology superior; only the context of intended use can determine ideal terminology for a given application or domain.
In contrast to MedDRA and SNOMED CT terminologies, the CTCAE is a classification system. Classifications, typically used for data aggregation, are generally considered “output” rather than “input” systems because they do not have the granularity required for the primary documentation of clinical care. Because the clinical criteria for each AE and grade are not required, the clinical specificity of CTCAE data is not precise (i.e., records with a given CTCAE event/grade code represent cases with one, some, or all of the specified clinical criteria, but the real event severity characteristics are uncertain.) However, the lack of a globally accessible standard AE terminology, and associated electronic tools to facilitate efficient and rapid use of terminology at the point of research data collection, make classifications such as CTCAE an appealing option from the perspective of work flow integration, ease of use, and user acceptance.
3.3. Comparison of general features of MedDRA, SNOMED CT, and CTCAE
Of the three AE coding system, each has pros and cons based upon its structure, content, access, and available tools and support. In addition to historical, political and popularity issues, examination of the structural differences between different terminologies (MedDRA and SNOMED CT) and between terminologies and classification systems (CTCAE) can offer insight into the issues that might arise in evaluating or integrating various data standards. It is important to note that standards have pros and cons that are relative to their intended purpose, and that a specified purpose should also dictate the relative importance of the features that we selected for Table I.
Table I.
Comparison of Selected Features
| Criterion | MedDRA (ratings in grey borrowed from Bousquet et al.) [22] | SNOMED CT | CTCAE, v.3 |
|---|---|---|---|
| Selected Terminology Desiderata[19] | |||
| Vocabulary content – comprehensive for AE domain; should include signs, symptoms, lab values, diseases and disorders. | E | E | F |
| Concept orientation | E | E | F |
| Concept orientation | E | E | E |
| Concept permanence | E | E | E |
| Non-semantic concept identifiers | E | E | E |
| Polyhierarchy | P | E | N/A |
| Formal definitions | N | E | N |
| Rejection of ‘not elsewhere classified terms’ | N | N | N |
| Multiple granularities | F | E | N/A |
| Multiple consistent views | P | E | N |
| Context representation | P | F | P |
| Graceful evolution | E | E | E |
| Recognized redundancy | N | F/P | N |
| Coding system type | terminology | terminology | classification |
| Post-coordination | no | yes | N/A |
| Incorporated in UMLS | yes | yes | yes |
| Designed specifically for representing AEs | yes | no | yes |
| Scope | AEs, PE findings, Medical History | Clinical findings | Toxicities |
| Global acceptance for AE reporting (i.e., ICH) | yes | no | no |
| FDA acceptance | yes | Yes for SPL; no for AE | no |
| Mappings to other possible standards | yes | yes | some |
| Free or low-cost use – US Users | no | yes | yes |
| Free or low-cost use – International Users | no | some | yes |
| Already in-use for Pharmaceutical Companies | yes | no | some |
| “Human readable” concept definitions | no | no | yes |
| Established rules for assigning codes | yes | some | yes |
| Training & Guidance Resources exist | E | P | E |
| Training & Guidance Resources are affordable/accessible | fee | fee | free |
| Previous/Demonstrated Organizational Experience in Adverse Events | yes | no | yes |
| Explicit Severity Grading | no | no | yes |
E=Excellent, F=Fair, P=Poor, N=None; N/A = Not applicable
As a classification system, the CTCAE does not attempt to offer the knowledge content of SNOMED CT, but its simplicity may make it easier to use. The CTCAE has a long history of use in clinical trials and fairly comprehensive coverage. There are adverse events for some conditions outside of oncology that may not be covered by CTCAE (e.g., treatments for neurological disorders such as Prader-Willi Syndrome might have psychosocial or behavioral effects.) Also, pregnancy-related outcomes and effects of confidentiality breeches are not included. Further, by definition, worsening of existing conditions can constitute AEs, and the CTCAE, which was created to classify drug toxicities, is not comprehensive in this regard. However, the CTCAE is the only candidate AE data standard that includes standardized severity criteria. This has practical implications for clinical research workflow and adverse event management, as serious (~more severe) events have different reporting requirements and implications on protocol continuation. 24
Standards criteria for sound terminologies have been widely cited and are the foundation of standards efforts such as caBIG, CDISC, and others. 25, 26 Evaluation criteria for classifications fundamentally differ from those for formal terminologies. Classifications have a requirement to count instances once and only once for a given group or class, and are generally considered unsuitable for primary clinical data collection. Therefore, each of the AE coding systems we present was designed for different purposes and has different expectations. In attempt to compare the “apples and oranges” of these heterogeneous coding systems, we present a common list of features as a basis to compare the MedDRA, SNOMED CT and CTCAE coding systems. The information in Table 1 is not presented to evaluate any of these coding systems as the features we include in our summary have not been weighed for importance for the AE or clinical research domain. The “best” data standard might not be the one that rates “E” on everything in the table (though none do). The ideal standard would certainly have coverage enough to support the domain, as well as have a structure, supporting tools, training, experience and user base to be used consistently and reliably for the data observed. Additionally, the content representations should be specific enough to support the immediate intended use-cases (i.e., intended purpose) for the encoded data.
4. DISCUSSION
4.1 Evaluation issues
The evaluation of data standards includes assessing content coverage and the structural and maintenance features that affect its use for a given purpose. 20, 27 The characteristics and features described in the previous section are not meant to be exhaustive nor authoritative. We emphasize again that no data standard can be evaluated outside the context of its intended purpose. The table of attributes does not consider the relative importance of each feature relative to specified data standards goals and objectives. In order to meaningfully evaluate heterogeneous coding system for adverse events, the clinical research community as a whole should add to and weigh these criteria. Meanwhile, our summary can be used to objectively look at the problem of multiple data standards and develop strategies for their interoperation. While the “best” terminologies might rate well in all of the structural criteria, in a given context the needs might not justify the time and resource commitments that are inevitable with big terminology implementation. We remind the reader that implementations of large terminologies such as SNOMED CT or GALEN are incomplete in most organizations, can take years to implement, and the quality or consistency of use of such standards are undetermined. However, if specific use-cases determine that complex terminologies such as SNOMED CT are warranted, then this might be a worthy goal. Further work should examine the performance of these three data standards in terms of real applications and planned data analyses.
In addition to the features presented in Table I, other criteria that are important to the implementation and usefulness of the standards should be considered. A weighing of certain structural features, combined with quantitative estimates for burden and costs of implementation, could shed light on ideal data standards and strategies of this area. Other criteria related to the access, use, and support of each terminology will also be relevant in terms of increased likelihood of adoption. Finally, measures of reliability in coding, ease in coding, efficiency of term retrieval, and data mining potential might illuminate best terminologies for primary or secondary coding. Results from this type of inquiry might generate a measure of complexity and act as a surrogate for predicting the burden of implementation. The intended purpose of the standards should dictate the evaluation criteria and rating for various competing standards.
4.2. Possibilities for the Future
Because each of the AE data standards we discuss has positive features and a strong user base in distinct areas of clinical research and care delivery activities, the continuation of their use – at least in the short-term - is probable. However, the relationship between them needs to be defined. Long-term activities of relevant stakeholders might include the harmonization of heterogeneous coding systems. Strategies include either dividing content domains among data standards so that they can be pieced together with no content overlap, or, when there is content overlap, creating data representations and terminological structures that can “fold” into each other and co-exist. In the long-term, purpose-driven cooperation of relevant stakeholders could possibly spawn a co-evolution of the three heterogeneous data standards, resulting in assimilation of the three coding schemes into one. Short-term possibilities for future relationships between heterogeneous standards include the mapping of concepts across terminologies, so that data native in one coding standard can be “translated” to another coding system to support other applications for secondary uses of the data.
4.3. Mappings
The need for mappings between biomedical terminologies usually arises when data encoded in one terminology is reused for a secondary purpose that requires a different system of encoding. Mapping is the most common work-around strategy to solve problems of data interoperability in the absence of a single terminology standard. However, the mapping approach has genuine shortcomings and limitations and should not obviate the quest for a single unified terminology standard.
The process for creating cross-terminology mappings itself is time-consuming and labor-intensive. Mapping to and from gigantic terminologies like SNOMED CT will take many person-years of work. Automated mapping tools have been developed, but their relatively low levels of recall and precision dictate their use as assistants to human editors rather than a substitute for human resources. Another problem with the use of mappings is the poorly-defined nature of the mapping relationship. Mapping is the process of finding a concept in a target terminology that “best matches” a particular concept in the source terminology. What “best matching” means can range from exact synonymy to mere “relatedness”. In most situations, mappings are created from more finely granular terminologies (like SNOMED CT) to less granular terminologies or classifications such as ICD-9. As a result, many of the non-synonymous mappings are from a narrower to a broader concept. There are potential issues in using the non-synonymous mappings for term translation, among them are the problems of information loss and ambiguity. Information loss occurs when a more specific term is translated into a less specific term (e.g. “Recent total retinal detachment” in SNOMED CT can only be mapped to “Retinal detachment” in MedDRA). Ambiguity occurs when there are multiple eligible mapping for a particular source concept (e.g. “Hearing loss” in SNOMED CT can be mapped to “Deafness”, “Hearing impaired” or “Hypoacusis” in MedDRA).
Another issue of mappings is that their use is almost always context specific. One set of mappings created for a specific use case may not be usable in another. The creation of a set of mappings is only the first step. Ongoing maintenance and updating of mappings is another major effort. Theoretically, all mappings should be reviewed whenever the terminologies on either end of the mappings are updated. This is no easy undertaking for terminologies like SNOMED CT and MedDRA which are quite large and have relatively frequent updates. How best to handle versioning in mappings is still a largely unresolved issue.
There are also more specific problems in mapping between the three terminologies under discussion. As mentioned before, CTCAE does not cover diseases which are not adverse events and this will limit the extent to which the other two terminologies can be mapped to it. In addition, the grading information in CTCAE is not easily matched in the other terminologies in which severity grading is often absent or defined differently. Currently, a set of mappings from CTCAE to MedDRA is available from the Cancer Therapy Evaluation Program (CTEP) website of NCI.28 This is jointly maintained by CTEP and MedDRA and regularly updated. The mappings only cover the base terms of CTCAE and not the grading information. Even though it is felt that a standardized mapping of grading information is important, the best approach of achieving this has not been decided upon.29
Practical needs for mappings between MedDRA and SNOMED CT may require them to be available sooner rather than later. If the time it will take to create full mappings between the two vocabularies is considered too long, it may be worthwhile to identify most commonly used subsets of terms to be mapped first. One way to identify the subsets is by looking at the frequency distribution of the terms that are stored in real-life databases. In addition, the Problem List Subset released by the Veterans Health Administration and Kaiser Permanente for the purpose of FDA’s Structured Product Labeling could potentially identify priority SNOMED CT terms to be mapped.
Regardless of the methodology employed to create the mappings, the UMLS will be an invaluable resource to utilize. As mentioned earlier, there is significant overlap between MedDRA and SNOMED CT within the concept structure of the UMLS. This means that half of MedDRA’s PTs already have SNOMED CT mappings within the UMLS. Even better is the fact that these are all synonymous mappings (i.e., context independent) and can be used regardless of the directionality of the intended mapping (i.e. they are valid either as SNOMED CT to MedDRA mappings or vice versa).
4.4 Context and scope of adverse event data collection
Obviously, adverse events are habitually collected in a regulated research environment, but often the context of AE data collection is to support the local management of AEs. The data needs to support various tasks, and should be delineated and considered for determining idea standards and strategies for interoperability. Additionally, local IRBs have standards and requirements that might vary from those at FDA or ICH, and the data requirements of these other stakeholders should be considered. Does the decision-making and handling of adverse events depend upon constructs such as seriousness and severity, and if so should these constructs be explicitly included in the terminology? Given the industry investment and FDA/ICH endorsement at this time, it seems likely that MedDRA may continue to be a standard for FDA reporting. If so, there will be future needs for mappings between SNOMED CT and MedDRA, but it is uncertain which direction, scope, and priority these mappings should take.
Since SNOMED CT is the likely standard for the collection of clinical data in EMR, the “collect once, use many” paradigm would dictate mapping SNOMED CT (as the source clinical data collection) to specialized terminologies and classifications that would support adverse event reporting as a secondary data use. It is a daunting task to create mappings for every SNOMED CT concept (even if we only take the “Clinical findings” hierarchy) to MedDRA. One approach is to take the synonymy in UMLS as a start (which already covers a substantial proportion) and expand on it gradually. The frequency of occurrence of SNOMED CT and MedDRA terms in actual AE data can help to identify the high-priority terms that need to be mapped first.
The ultimate vision is a reuse of clinical data to support clinical research – “collect once, use many”. 12 The requirements for supporting use cases will imply very different requirements in terms of directionality and coverage and will impact any strategy for achieving standardized data in this domain. The clinical research community should define clearly what the intended purposes of uniform AE reporting standards are. Additionally, they must also clearly define whether real opportunities for data sharing between clinical care and research exist. Within that context, a clear understanding of the standards requirements for both structural and content features is critical.
The clinical regulated research community should decide, based upon workflow, whether AE reporting can justify primary or secondary data. Primary AE data currently are used for regulatory reporting and local institutional reporting and communication of AE handling across research organizations or networks. The use of clinical data, such as that from an EMR, as source data to an AE detection system, for example, would represent a secondary use of the data. In these cases, the AE representation could some day be derived from the clinical data that will likely be represented in SNOMED CT. This goal would imply a need for mappings in the direction of SNOMED CT to MedDRA. The importance of connecting the health care and research domains for this and other use cases should lead to the development of tools that allow data to be interoperable in the appropriate direction and maintain the level of precision needed for the secondary uses.
Data analysis issues might justify the need for mappings in the MedDRA – SNOMED CT direction, if data mining on MedDRA encoded data is determined as a priority by the field. The higher number of defined relationships in SNOMED CT enable both data mining and automated placement of new terms, both of which would strengthen MedDRA. It has been suggested that the mapping of MedDRA to a third-generation system, such as SNOMED CT, might facilitate the automated maintenance and curation of MedDRA. 23 This would allow the current MedDRA structure to be kept (presumably for initial data collection and reporting) to ensure that end users have a common view of the data and computational properties to MedDRA. Others have suggested the addition of knowledge (i.e., concept properties), borrowed from existing robust terminology such as SNOMED CT, could be added to existing clinical research terminologies (such as MedDRA and WHO-ART) to facilitate growth and maintenance and automatic concept classification 30 , also implying a mapping in the MedDRA – SNOMED CT direction.
Practically, mapping is also limited by the content and granularity of the component terminologies. Since, for example, CTCAE doesn’t cover diseases or pregnancy-related outcomes, those items could not be mapped to CTCAE. Similarly, excess SNOMED CT content that does not exist in MedDRA cannot be mapped. Further, the granularity in SNOMED CT would be lost in any mapping to a less expressive terminology or classification. Because the CTCAE grading information is not part of MedDRA, SNOMED CT, or UMLS Semantic Network, many of the severity semantics of the CTCAE are lost in conventional mappings. And the mapping of post-coordinated SNOMED CT concepts is not feasible to do in advance.
Having multiple standards does complicate data sharing and requires resources to achieve interoperability. An ultimate goal would be to have the same standard, or harmonious standards, across the different user communities. Perhaps CTCAE and MedDRA can co-evolve so that future versions can be more compatible to enable more meaningful transformation of encoded data (as opposed to the retrospective mapping approach)? This is not an unreasonable suggestion as both vocabularies are serving similar purposes and have some structural similarities (limited hierarchical levels based upon term structure versus concept semantics). Realization of future co-evolution of competing terminologies will require regular communication between the developers of the two coding systems.
5. CONCLUSION
Whether coding systems become standard often depends on factors other than their intrinsic quality. As the three vocabularies all have long history and substantial user base, it is likely that they will all survive and continue to be used. At this point, we think that discussion should not focus on evaluation topics but rather business processes and how mappings (specifying the directionality) can support research process.
The structure and features of current adverse event coding systems are different, so comparisons and evaluations are not straightforward, nor are strategies for their harmonization. Because each has a strong user base, it is likely that efforts for data standardization within the AE domain will entail mapping. Various uses for uniformly represented adverse event data, including initial reporting, extracting AEs from other clinical data sources, adverse event data management and communication systems, and aggregate analysis dictate the requirements for ideal data standards and influence future relationships between these competing data standards. The clinical research community as a whole should focus discussion around ideal flows of adverse event information across healthcare and research settings, and identify and prioritize the requirements for supporting coding systems. This dialogue should include future standards, cooperation, and harmonization of competing terminologies.
Acknowledgments
Grant support:
The project described was supported by Grant Number RR019259 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents aresolely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. This work was also supported in part through cooperative agreements by The National Institute of Child Health and Human Development (NICHD), National Institutes of Health (NIH); the Juvenile Diabetes Research Foundation (JDRF); Mead Johnson Inc; The Canadian Institutes of Health Research; the European Foundation for the Study of Diabetes (EFSD) and the Commission of the European Communities, specific RTD programme “Quality of Life and Management of Living Resources”, proposal number QLK1-2002-00372. It does not reflect its views and in no way anticipates the Commission’s future policy in this area.
The authors also thank the Office of Rare Diseases for their support of the RDCRN. The authors wish to thank Dr. Olivier Bodenreider of the NLM for his initial direction on this work, and Jennifer Lloyd of USF for her assistance with figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Rachel L. Richesson, University of South Florida College of Medicine, Tampa, FL, USA.
Kin Wah Fung, National Library of Medicine, Bethesda, MD, USA.
Jeffrey P. Krischer, University of South Florida College of Medicine, Tampa, FL, USA.
REFERENCES
- 1.Friedman LM, Furberg CD, DeMets DL. Fundamentals of Clinical Trials. Third ed. New York: Springer-Verlag; 1998. Assessing and Reporting Adverse Events; pp. 170–184. [Google Scholar]
- 2.Richesson RL, Krischer JP. Data Standards in Clinical Research: Gaps, Overlaps, Challenges and Future Directions. Journal of the American Medical Informatics Association. 2007;14(6):687–696. doi: 10.1197/jamia.M2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Souza T, Kush R, Evans JP. Global clinical data interchange standards are here! Drug Discovery Today. 2007 February;12(3–4):174–181. doi: 10.1016/j.drudis.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Levin R. Data Standards for Regulated Clinical Trials: FDA Perspective. [Accessed July 21, 2006];CDISC. Available at http://www.cdisc.org/pdf/2004_06_14_cdisc.pdf.
- 5.Brown EG, Wood L, Wood S. The Medical Dictionary for Regulatory Activities (MedDRA) Drug Safety. 1999 Feb;20(2):109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 6.MSSO. MedDRA Introductory Guide V9.1: Maintenance and Support Services Organization (MSSO) 2006. [Google Scholar]
- 7.Spackman KA, Campbell KE, Cote RA. SNOMED RT: A Reference Terminology for Health Care. Paper presented at: American Medical Informatics Association Fall Symposium; 1997. [1997]. [PMC free article] [PubMed] [Google Scholar]
- 8.Spackman KA. SNOMED CT milestones: endorsements are added to already-impressive standards credentials. Healthc Inform. 2004 Sep;21(9):54–56. [PubMed] [Google Scholar]
- 9.CAP. 2006. Jan, [Accessed January 8, 2007]. SNOMED Clinical Terms® Guide Abstract logical models and representational forms CMWG revision, version 5. [Google Scholar]
- 10.Spackman KA, Campbell KE. Compositional concept representation using SNOMED: towards further convergence of clinical terminologies. Proc AMIA Symp. 1998:740–744. [PMC free article] [PubMed] [Google Scholar]
- 11.CHI. CHI Executive Summaries: Consolidated Health Informatics. 2004 http://www.whitehouse.gov/omb/egov/documents/CHIExecSummaries.doc.
- 12.Tang PC. Position Paper. AMIA Advocates National Health Information System in Fight Against National Health Threats. Journal of the American Medical Informatics Association. 2002;9(2):123–124. doi: 10.1197/jamia.M1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Healthcare Terminologies and Classifications: An Action Agenda for the United States. AMIA; 2006. American Medical Informatics Association and American Health Information Management Association Terminology and Classification Policy Task Force. http://www.amia.org/inside/initiatives/docs/terminologiesandclassifications.pdf. [Google Scholar]
- 14.FDA. FDA News. FDA Advances Federal E-Health Effort. U.S. Food and Drug Administration; 2006
- 15.Trotti A. The need for adverse effects reporting standards in oncology clinical trials. Journal of Clinical Oncology. 2004 January 1;22(1):19–22. doi: 10.1200/JCO.2004.10.911. [DOI] [PubMed] [Google Scholar]
- 16.Bodenreider O, Burgun A, Botti G, Fieschi M, Le Beux P, Kohler F. Evaluation of the Unified Medical Language System as a medical knowledge source. Journal of the American Medical Informatics Association. 1998;5(1):76–87. doi: 10.1136/jamia.1998.0050076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NLM. U.S. National Library of Medicine 8600. Bethesda, MD 20894: Rockville Pike; 2000. [Last updated: 19 April 2000. Accessed 3-2001]. Fact Sheet Unified Medical Language System. [Google Scholar]
- 18.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.NLM. [Accessed February 6, 2003];National Library of Medicine; Fact Sheet. UMLS ® Metathesaurus. 2003 1-13-03. Available http://www.nlm.nih.gov/pubs/factsheets/umlsmeta.html.
- 20.Cimino JJ. Desiderata for Controlled Medical Vocabularies in the Twenty-First Century. Methods of Information in Medicine. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 21.Almenoff J, Tonning JM, Gould AL, et al. Perspectives on the use of data mining in pharmacovigilance. Drug Safety. 2005;28(11):981–1007. doi: 10.2165/00002018-200528110-00002. [DOI] [PubMed] [Google Scholar]
- 22.Mozzicato P. Find More Like This. Standardised MedDRA Queries: Their Role in Signal Detection. Drug Safety. 2007;30(7):617–619. doi: 10.2165/00002018-200730070-00009. [DOI] [PubMed] [Google Scholar]
- 23.Bousquet C, Lagier G, Lillo-Le Lou A, Le Beller C, Venot A, Jaulent MC. Appraisal of the MedDRA Conceptual Structure for Describing and Grouping Adverse Drug Reactions. Drug Safety. 2005;28(1):19–34. doi: 10.2165/00002018-200528010-00002. [DOI] [PubMed] [Google Scholar]
- 24.Segal ES, Valette C, Oster L, et al. Risk management strategies in the postmarketing period : safety experience with the US and European bosentan surveillance programmes. Drug Safety. 2005;28(11):971–980. doi: 10.2165/00002018-200528110-00001. [DOI] [PubMed] [Google Scholar]
- 25.NCI. [Accessed May 25, 2006];National Cancer Institute; caBIG. Cancer Biomedical Informatics Grid. Data Standards. Available at: https://cabig.nci.nih.gov/workspaces/VCDE/Data_Standards/index_html/
- 26.Haber MW, Kisler BW, Lenzen M, Wright LW. Controlled Terminology for Clinical Research: A Collaboration Between CDISC and NCI Enterprise Vocabulary Services. Drug Information Journal. 2007;41:405–412. [Google Scholar]
- 27.Cimino JJ. Review paper: coding systems in health care. Methods Inf Med. Dec. 1996;35(4–5):273–284. [PubMed] [Google Scholar]
- 28.NCI. [Accessed January 22, 2008];National Cancer Institute Cancer Therapuetics Evaluation Program (CTEP); List of Codes and Values. 2007 June 4; Available at http://ctep.cancer.gov/guidelines/codes.html.
- 29.MSSO. Summary of MedDRA MSSO’s Blue Ribbon Panel Meeting on CTCAE-MedDRA Mapping. Northrop Grumman Corporation, MedDRA Maintenance and Support Services Organization (MSSO); 2006. Apr 6, [Accessed January 22, 2008]. Available at: http://www.meddramsso.com/MSSOWeb/docs/8466-100%20Summary%20BRP%20Apr2006.doc. [Google Scholar]
- 30.Alecu I, Bousquet C, Jaulent MC. Mapping of the WHO-ART terminology on SNOMED-CT to improve grouping of related adverse drug reactions. Paper presented at: Stud Health Technol Inform; 2006. [PubMed] [Google Scholar]