Abstract
The recent, rapid progress in our understanding of cytoplasmic RNA-mediated antiviral innate immune signaling was initiated by the discovery of retinoic acid-inducible gene I (RIG-I) as a sensor of viral RNA [1]. It is now widely recognized that RIG-I and related RNA helicases, melanoma differentiated-associated gene-5 (MDA5) and laboratory of genetics and physiology-2 (LGP2), can initiate and/or regulate RNA and virus -mediated type I IFN production and antiviral responses. As with other cytokine systems, production of type I IFN is a transient process, and can be hazardous to the host if unregulated, resulting in chronic cellular toxicity or inflammatory and autoimmune diseases [2-9]. In addition, the RIG-I-like receptor (RLR) system is a fundamental target for virus-encoded immune suppression, with many indirect and direct examples of interference described. In this article, we review the current understanding of endogenous negative regulation in RLR signaling and explore direct inhibition of RLR signaling by viruses as a host immune evasion strategy.
RLR signaling and its control
RIG-I and MDA5, two so-called RIG-I-like receptor (RLR) family proteins have been identified as cytoplasmic sensors of viral RNA [1, 10]. RIG-I and MDA5 belong to the DExD/H box RNA helicase family and also have two caspase activation and recruitment domains (CARD) N-terminal to the helicase region, implicated in relaying the signal downstream. Although similar, the two proteins differ in specificity of virus recognition as well as RNA binding specificity [11] as reviewed elsewhere [12]. For MDA5 neither biological substrate specificity nor exact RNA binding have been clearly specified experimentally, however MDA5 is thought to be the primary receptor for signaling initiated by cytoplasmic accumulation of the well-studied synthetic dsRNA analog, poly(I:C) [11, 13]. While the helicase region represents one surface for interactions with dsRNAs, for the prototype, RIG-I, substrate recognition and binding specificity has been linked to a domain C-terminal to the helicase region. This RIG-I region has the ability to recognize 5′ tri-phosphorylated ends of double-stranded (ds) or single stranded (ss) RNA [14, 15]. Structural analysis has determined that this regulatory domain contains an obligatory zinc binding module, which is conserved in both MDA5 and LGP2 sequence alignments [16]. A basic groove has been suggested as the 5′triphosphate binding site and this is borne out in site-directed mutagenesis experiments [14-17].
It is commonly observed that cytokine signal transduction is transient and must be tightly regulated to prevent unwanted or inappropriate cellular responses that may lead to differentiation or apoptosis. Several mechanisms are thought to underlie the tight regulation of the RIG-I signaling response to prevent unnecessary signaling under steady state conditions. It has been demonstrated that RIG-I can autonomously control its basal activity. The 5′-triphosphate binding C-terminal region of RIG-I was initially referred to as a repressor domain (RD) [18] because its presence in RIG-I was associated with low steady-state signaling in the absence of RNA ligands. Control of basal RIG-I activity by the RD is mediated by auto-inhibition, keeping the protein in an inactive “closed conformation” [18]. RNA ligand binding is thought to induce RIG-I structural alteration to expose the CARD to downstream signaling partners [19-22]. Although structural elements of the RIG-I C-terminus are preserved in MDA5, this autoregulation function does not appear to be conserved, as MDA5 exhibits high basal signaling activity [10, 18]. This dissimilarity suggests there are additional means to control MDA5 signal transduction in the absence of ligand stimulation. Currently available data suggest that while expressed modestly at steady state, MDA5 biosynthesis is highly responsive to dsRNA and interferon stimulation [18]. Accumulation of MDA5 may therefore drive signaling responses subsequent to the initial RIG-I response.
Negative regulators of RLRs
In addition to the autonomous inhibitory mechanisms mentioned above, several additional proteins have been recently described as supporting the attenuation or negative regulation of dsRNA signaling. Current experimental data suggest that there are several layers of direct and indirect feedback inhibition that individually or in combination give rise to the overall antiviral signaling profiles both generally and with tissue and cell-specific variations (Table I, Fig. 1).
Table I.
| Cellular Inhibitors of RLR signaling | ||||
|---|---|---|---|---|
| Inhibitors | Target(s) | Proposed mechanisms | Type of negative regulation | Refs. |
| LGP2 | dsRNA, RIG-I, IPS-1 containing complex | dsRNA sequestration, Competition with IKKi for IPS-1, Inhibition of RIG-I dimerization | Negative feedback (RLRs) | [18, 23-26] |
| A20 | Unknown | deubiquitination of the target protein(s)? | Negative feedback (RLRs, TLRs etc) | [28-30] |
| Pin1 | IRF-3 | phosphorylation-dependent isomerization | Negative feedback (RLRs, TLRs) | [41] |
| SIKE | TBK1 and IKKe | Sequestration of kinases | Inhibition of steady state (RLRs, TLRs) | [45] |
| Atg5-Atg12 | RIG-I, MDA5, IPS-1 | Sequestration of RLRs and IPS-1 | Inhibition of steady state (RLRs) | [54] |
| RNF125 | RIG-I, MDA5, IPS-1 | Proteosomal degradation of RLRs and IPS-1 | Negative feedback (RLRs) | [55] |
| DUBA | TRAF3 | deubiquitination of TRAF3 | Negative feedback (RLRs, TLRs etc) | [57] |
| NLRX1 | IPS-1 | competition with RLRs for IPS-1 | Inhibition of steady state (RLRs) | [63] |
| ISG15 | RIG-I | Sequestration of RIG-I | Negative feedback (RIG-I) | [66] |
| DAK | MDA5 | Sequestration of MDA5 | Inhibition of steady state (MDA5) | [67] |
| CYLD | RIG-I, TBK1 | deubiquitination of RIG-I | Inhibition of steady state and activated state | [61, 62] |
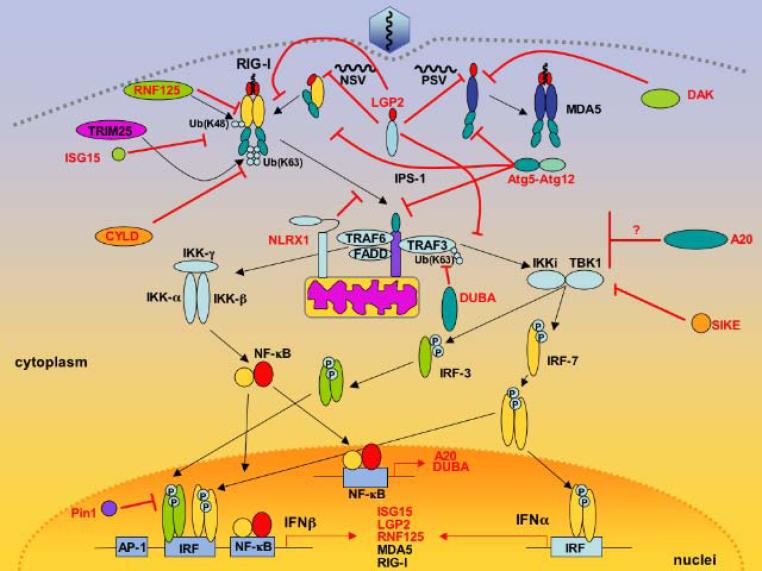
Figure 1.
RLR signaling pathway and its negative regulatory molecules. Positive signals and molecules are indicated as black arrows and black letters, respectively. Negative signals and negative regulators are indicated as red lines and red letters. For references, see text. Abbreviations; NSV: Negative stranded virus; PSV: Positive stranded virus; RIG-I: retinoic acid inducible gene-I; MDA5: melanoma differentiated-associated gene-5; LGP2: laboratory of genetics and physiology-2; RNF125: ring-finger protein 125: TRIM25: tripartite motif 25; SIKE: Suppressor of IKKε; DAK: Dihydroxyacetone kinase; FADD: Fas-Associated protein with Death Domain; DUBA: Deubiquitinating enzyme A; TRAF: tumor necrosis factor receptor-associated factor; Ub: Ubiquitin; CYLD: cylindromatosis; ATG: Autophagy-related gene; NLR: nucleotide-binding domain and leucine-rich repeat.
LGP2
RIG-I was identified as a positive regulator of dsRNA signaling by a reporter gene based cDNA library screen. The identified reporter-activating clone encoded a truncated RIG-I encompassing only the N-terminal CARD regions, and it was found that sole expression of the CARD region of either RIG-I or MDA5 can constitutively and positively activate downstream signaling [1, 23]. In contrast, a CARD-deficient C-terminal fragment, referred to as RIG-IC, Δ-CARD or C-RIG, can function as a dominant negative inhibitor. Domain structure similarity between the RIG-IC fragment and the CARD-deficient third member of the RLR family, LGP2, suggested that LGP2 may be a natural negative regulator of dsRNA signaling [18, 23-25]. Indeed, expression of LGP2 negatively regulates Sendai virus and Newcastle disease virus (NDV)- mediated IFNβ gene activation, and RNAi mediated knock-down of LGP2 can enhance antiviral gene expression. Moreover, it was observed that LGP2 mRNA and protein are inducible by dsRNA or IFN treatments as well as virus infection, consistent with LGP2 functioning as a negative feedback regulator. LGP2-deficient mice were created by homologous recombination, and the absence of LGP2 resulted in a complex phenotype [26]. Experimental data support the role of LGP2 as a negative regulator for the negative strand RNA virus, vesicular stomatitis virus (VSV), as the knockout mice exhibited greater resistance to VSV infection.
Several mechanisms may account for the LGP2 inhibitory effects. As a homolog of RIG-IC, one attractive model is that LGP2 can sequester RNA ligands from recognition by RIG-I or MDA5 [23, 25]. Alternatively, it was found that LGP2 can suppress IFNβ promoter activity induced by non-RNA activators, including expression of the constitutively active RIG-I N-terminal CARD fragment or the signaling adaptor molecule IPS-1/MAVS/CARDIF/VISA [19-22] from plasmid vectors. These RNA-independent stimuli are not suppressed by the artificial dominant negative RIG-IC fragment, which implicates additional suppression activity of LGP2. Mechanistic experiments found that LGP2 co-immunoprecipitates with the adaptor protein, IPS-1/MAVS/CARDIF/VISA, and exhibits competitive binding with the downstream kinase, IKKi/IKKε for a common site in IPS-1[24]. LGP2 has also been demonstrated to associate with RIG-I to inhibit its auto-oligomerizaton via the LGP2 C-terminal region comparable to the RIG-I RD [18]. In this model, dimerization of RIG-I by 5′-triphosphorylated ssRNA, proposed to be an active form of RIG-I, [16] is replaced by a RIG-I:LGP2 hetero-oligomer.
Unexpected in view of the evidence in favor of LGP2 as a negative regulator, the LGP2-deficient mice were found to be more susceptible to infections with encephalomyocarditis virus (EMCV), indicating a role for LGP2 as a positive regulator of antiviral signaling by this picornavirus. Resistance to EMCV has been demonstrated to be controlled by MDA5 rather then RIG-I signaling, and therefore the greater susceptibility to this virus may be interpreted to indicate that LGP2 contributes to positive regulation of MDA5 mediated antiviral signaling [26]. Further mechanistic studies will be needed to clarify the functions of LGP2 in both RIG-I and MDA5 –mediated antiviral signal transduction.
A20
A20 was originally identified as a TNF-inducible gene in human umbilical vein endothelial cells [27] and was found to be induced by a wide range of stimuli including NF-κB. The anti-apoptotic protein has been known as a negative regulator of NF-κB and was recently shown to block RIG-I-mediated signaling to the IRF-3, IRF-7 and NF-κB pathways. Expression of A20 blocks the constitutively active RIG-I N-terminus from signaling IRF-3 phosphorylation, dimerization, and DNA binding [28] as well as Sendai virus, NDV and dsRNA mediated IFNβ promoter activity [29, 30]. Experiments indicate that A20 intervenes downstream of the mitochondrial adaptor protein IPS-1 but upstream of the kinases (TBK1 and IKKi) that phosphorylate IRF-3. Therefore, it seems likely that A20 could also inhibit MDA5-mediated signaling, but this remains to be demonstrated formally.
A20 is a unique protein that possesses a N-terminal de-ubiquitination domain, called ovarian tumor (OTU) domain, and a C-terminal ubiquitin ligase domain. Inhibition of NF-κB signaling by A20 relies on cooperation of these two ubiquitin-related domains [31], but only the C-terminal ubiquitin ligase domain is essential for RIG-I signaling interference [28], suggesting that the A20 target protein is most likely modified by K48 ubiquitin leading to proteosomal degradation [29, 30]. A20 deficient mice develop severe multi-organ inflammation [32] and A20-deficient fibroblast displayed prolonged NF-κB activity and are unable to terminate tumor necrosis factor (TNF)-induced NF-κB activation. Macrophages derived from A20 deficient mice are incapable of terminating TLR-induced NF-κB activation [32, 33], indicating that A20 functions as a negative feedback regulator for several branches of the immune systems.
Pin1
It has been well established that upon virus infection, IRF-3 can be phosphorylated at two C-terminal phospho-acceptor clusters (Ser385-Ser386 and Ser396-Ser398-Ser402-Thr404-Thr405) [34-37]. These modifications are critical for IRF-3 activation [38-40]. An additional phosphorylation site, Ser339, was also found to be involved in IRF-3 regulation [41]. While the specific kinase(s) for this phosphorylation site are unknown, accumulation of phosphorylation occurs with delayed kinetics relative to activating stimuli. Due to a neighboring proline at position 340, phosphorylation of Ser 339 produces a binding site for the WW domain of the peptidyl-prolyl isomerase, Pin1. This interaction possibly subjects IRF3 to cis-trans isomerization around the Ser-Pro peptide bond. The Ser339-Pro motif is conserved evolutionarily among human, mouse, rat, and, chicken, suggesting that Pin1 activity is fundamental to IRF-3 regulation.
For other Pin 1 substrates, this type of isomerization has been shown to regulate the stability and localization of the substrate protein, with profound effects on transcriptional activation, cell cycle progression, and cell death. For example, Pin1 association with the tumor suppressor p53 generates conformational changes that enhances transactivation activity [42, 43]. For IRF-3, Pin1 association was demonstrated to facilitate ubiquitin-mediated proteosomal degradation. Pin 1 deficient mouse embryonic fibroblasts (MEFs) are more hostile to replication of Newcastle disease virus (NDV) and encephalomyocarditis virus (EMCV), consistent with a negative regulation phenotype. As Pin1 inhibits at the level of IRF-3, it controls TLR3, RIG-I, and MDA5-mediated signaling by eliminating active IRF-3 pools characterized by Ser339 phosphorylation. Interestingly, Pin1 also has been reported as a positive regulator of the NF-κB component p65, which is also involved in IFNβ promoter activation [44]. Further work will be needed to determine the exact mechanisms involved in Pin1 regulation of antiviral signaling.
SIKE
Suppressor of IKKε, SIKE, has been identified as an IKKε-associated protein by yeast two-hybrid screening [45]. Further characterization of SIKE revealed that it also can associates with TBK1 and that SIKE expression can efficiently inhibit IKKε, TBK1, TRIF, TLR3 and RIG-I-mediated IFNβ and interferon stimulated response element (ISRE) promoter activation. In this study an interaction between the kinases IKKε and TBK1 with TRIF, IRF-3 and RIG-I was observed. This interaction was disrupted by expression of SIKE. This effect is specific to components of virus- and TLR-3-triggered IRF-3 activation pathway but does not disrupt components of the NF-κB pathway. Furthermore, gel filtration experiments have demonstrated that SIKE exists in a higher order complex associating with TBK1 as well as unidentified cellular protein. VSV infection or poly(I)•poly(C) treatment appear to dissociate SIKE from TBK1 as it converts these complexes to the size of monomeric and dimeric SIKE that are free from TBK1. RNAi-based knock down of SIKE without VSV infection or poly(I)•poly(C) treatment leads to increased induction of IFNβ and ISRE promoter but not NF-κB promoter activity. Accordingly, SIKE knock down decreases VSV growth while overexpression of SIKE increases VSV growth in HEK293 cells. Endogenous protein levels of SIKE protein remain unchanged during virus infection or TLR3 activation. This study suggests that SIKE functions as a suppressor of TBK1 and IKKε by sequestering TBK1 and IKKε into an inactive complex to avoid unnecessary activation of those kinases.
Atg5-Atg12 conjugate
Autophagy is a cellular process that mediates a nonspecific bulk degradation/recycling pathway that is essential for turnover of stable macromolecules, maintenance of an amino acid pool upon starvation, and the cellular response to a variety of hormonal stimuli [46]. In addition, the autophagy pathway is crucial for defense against infections with intracellular bacteria [47-49], plays a key role in recognizing signatures of viral infection through TLR7 in plasmacytoid dendritic cells, and represents a critical effecter mechanism to restrict viral replication [50, 51]. However, autophagosomes have also been exploited by certain viruses as a niche for viral replication, where its membrane scaffold structure serves as a place for RNA replication [52, 53]. A recent study has demonstrated that RLR signaling is negatively regulated by some autophagic proteins [54]. The small ubiquitin-like Atg12 becomes conjugated to its substrate, Atg5, to produce the Atg5-Atg12 conjugate, a key regulator of the autophagic process that associates directly with the CARD domains of RIG-I, MDA5 and IPS-1. It is postulated that Atg5-Atg12 conjugate can bridge RIG-I or MDA5 to IPS-1. The presence of the Atg5-Atg12 conjugate in the RIG-I-IPS-1 complex might lead to an inactive status. MEFs from Atg5 deficient mice produced more IFNβ and IP10 mRNA after VSV infection or cytoplasmic delivery of poly(I)•poly(C) compared to wild type (WT) MEFs. Accordingly, absence of Atg5 leads to diminished VSV growth. In contrast, Herpes simplex virus-1 (HSV-1) –mediated induction of IFNβ and IP10 mRNA shows an opposite effect. Authors have shown that an Atg5 point mutant, which can not be conjugated to Atg12, does not inhibit IPS-1 dependent IFNα, IFNβ, and NF-κB promoter activity. In addition, Atg7 deficient MEFs, in which Atg5-Atg12 conjugation is defective, produce higher amount of type I IFNs in response to transfection of poly(I)•poly(C). Collectively, this study has indicated that Atg5-Atg12 conjugate, but not monomeric Atg5 or Atg12, acts as a suppressor of RLR signaling.
RNF125
It has recently become clear that RIG-I is regulated by ubiquitin conjugation mediated by a RING finger family protein, RNF125, an E3-ubiquitin ligase that specifies their proteosomal degradation [55]. RNF125 was identified as an UbcH8 interacting protein by yeast two-hybrid screening based on the prediction that UbcH8, which is an E2 enzyme for both ubiquitin and ISG15, might associate with the E3 ligase for RIG-I. The N-terminal region of RNF125 associates with RIG-I via the CARD and RNA helicase domain but not with the RD, and expression of RNF125 increases RIG-I ubiquitination and destabilization. Conversely, RNAi-mediated knock down of RNF125 decreases the level of ubiquitin-modified RIG-I resulting and stabilizes RIG-I. Both MDA5 and IPS-1 were also shown to be ubiquitination targets of RNF125. As a result, virus or dsRNA -mediated IFNβ production is inhibited by expression of RNF125 and enhanced by RNF125 knock-down. As RIG-I abundance decreases with the onset of the RNF125 accumulation, RNF125 also has the properties of a negative feedback regulator of RIG-I. While the exact locus for RNF125-mediated RIG-I ubiquitination is not known, its functional consequence differs from ubiquitination of RIG-I on K172 [56]. This residue of RIG-I, located in the CARD region, is a target for efficient TRIM25-induced ubiquitination that is essential for antiviral signaling. The activating ubiquitin modification is K63-linked, a type not typically associated with proteasome mediated degradation. It will be interesting to explore the potential interplay between these independent ubiquitin based regulatory mechanisms.
DUBA
An siRNA based screening for OTU deubiqitinating (DUB) family members identified DUBA, as a negatively regulator of antiviral signaling [57]. The data indicate that reduction of DUBA by siRNA knock down augments TLR3, 4, 7, RIG-I, and MDA5-mediated IFN induction. Experiments demonstrate that increased expression of DUBA had the opposite effect, indicating that DUBA acts as a negative regulator. Knockdown of DUBA did not alter NOD2, TLR2, TLR3, or TNF-dependent NF-κB reporter activity, but did increase RIG-I and MDA5-mediated responses, suggesting a more general role for IFN activation and a limited role on NF-κB activation. A biochemical approach has identified TNF receptor-associated factor 3 (TRAF3) as a DUBA interacting protein. TRAF3 is a critical adaptor molecule required for induction of type I IFNs in response to viral infection and TLR activation [58, 59]. TRAF3 directly associates with IPS-1 to induce IFNs in RLR signaling by linking TBK1/IKKε kinases [60]. Expression of DUBA cleaves Lys63-linked ubiquitin chains on TRAF3 and depletion of DUBA increases ubiquitinated TRAF3. Expression of DUBA decreases interaction between TRAF3 and TBK1, suggesting that ubiquitination of TRAF3 controls the activity of the kinase-containing complex. As DUBA protein is also increased in macrophages by LPS stimulation, it may also function as part of a negative feedback circuit.
CYLD
Another OTU DUB family protein, the tumor suppressor, CYLD, has been identified as a negative regulator of RIG-I by two groups [61, 62]. RNA interference targeting CYLD results in an enhancement in Sendai virus-triggered IFNβ secretion and experiments using CYLD-deficient MEFs and dendritic cells (DCs) show constitutive activation of TBK1/IKKε kinases, which is associated with hyper-induction of type I IFNs by VSV infection. The data indicate that CYLD targets RIG-I as well as TBK1 for deubiquitination that leads to inactivation of the signaling. CYLD associates with the CARD domain of RIG-I and removes K63-linked ubiquitin from the RIG-I CARDs that are conjugated by the E3 ubiquitin ligase, TRIM25. Immunoprecipitation experiments show that CYLD coprecipitates not only with RIG-I but also with TBK1 and IKKε. Interestingly, TBK1 or IKKε but not RIG-I specifically precipitates two species of CYLD in size, suggesting phosphorylation of CYLD by these kinases.
Loss of CYLD in DCs causes accumulation of ubiquitination of RIG-I indicating that RIG-I is constantly in the cycle of ubiquitination-deubiquitination with deubiquitination being a dominant event in steady state. Furthermore, the accumulation of ubiquitinated RIG-I in CYLD-deficient cells is associated with constitutive activation of TBK1/IKKε kinases. CYLD protein level is reduced in the presence of TNF and viral infection. CYLD is most likely a negative regulator that inhibits RIG-I in both, steady state and activated state, to prevent unnecessary signaling event. However, the precise mechanism that controls TRIM25-mediated ubiquitination and CYLD-mediated deubiquitination remains to be elucidated.
NLRX1
A member of nucleotide-binding domain and leucine-rich repeat containing (NLR) protein family, NLRX1 has been identified as a negative regulator of RLR signaling [63]. Combination of microscopy based and biochemical approaches has revealed that NLRX1 is a ubiquitously expressed protein that resides at the outer mitochondrial membrane where IPS-1 is located. The amino terminus of NLRX1 contains a mitochondrial-targeting sequence making it the first NLR shown to have this feature. Intriguingly, association between endogenous NLRX1 and IPS-1 is observed and the interaction is mediated by the CARD-like domain of IPS-1 and a putative nucleotide-binding domain (NBD) of NLRX1. Expression of NLRX1 results in the potent inhibition of RIG-I, MDA5, and IPS-1 -mediated IFNβ promoter activity, but does not alter TLR3-mediated signaling. Deletion of the leucine-rich repeat (LRR) abolishes the IPS-1 inhibitory activity, though it is dispensable for interaction with IPS-1. Depletion of NLRX1 by siRNA enhances IPS-1-induced IFNα, IFNβ, IL-6, RANTES, TNFα mRNA as well as virus-induced secretion of IFNβ. Mechanistic studies suggests that NLRX1 competes with RIG-I for IPS-1 interaction, implying that association between RIG-I and IPS-1 through CARD-CARD interaction is disrupted by NLRX1 in the mitochondrial compartment. Together, these results support a role of NLRX1 as a negative regulator of RLR signaling. One distinction between this negative regulator and others described here is that NLRX1 protein and mRNA levels do not change when cells are activated by expression of positive factors such as RIG-I, MDA5, IPS-1, and IRF-3/7 or even with IFNβ treatment. Therefore, NLRX1 seems to act as a inhibitor of steady state antiviral signaling rather than working as a negative feedback inhibitor.
Interestingly, NLRX1 has also been reported to be a positive regulator of the immune system that potently triggers the generation of reactive oxygen species (ROS). NLRX1 synergistically potentiates ROS production induced by TNFα, Shigella infection and double-stranded RNA treatment, resulting in amplified NF-κB-dependent and JUN amino-terminal kinases-dependent signaling [64]. Clearly this protein has several functions in immune regulation, and further research will reveal their relative contributions to overall antiviral responses.
ISG15
The ubiquitin-like IFN stimulated gene 15 (ISG15) protein has been reported to modify RIG-I [65] and can negatively regulate RIG-I signaling activity [66]. When ISG15-specific E1 (UBE1L), E2 (UbcM8), and ISG15 are expressed together in cells, robust ISG15 modification of RIG-I is observed. Expression of the ISG15 system decreases the basal level of ISRE and IFNβ promoter activity as well as NDV-mediated IFNβ promoter activity and delays cytoplasmic poly(I)•poly(C)- mediated IFNβ promoter activity. Interestingly, the basal level of RIG-I and ISG proteins but not mRNA are much higher in UBEL1-deficient murine fibroblasts than in WT cells. Accordingly, UBEL1 -/- cells produce much higher IFNβ mRNA than WT cells in response to NDV infection. As ISG15 modification system is only present in IFN stimulated cells, it is tempting to speculate that ubiquitin and ISG15 may coordinately regulate the fate of RIG-I or compete for control of RIG-I activity.
Dihydroxyacetone kinase (DAK)
Only one MDA5 specific negative regulator has been reported. The protein kinase, DAK, was found to interact with MDA5 in a yeast two hybrid screen [67]. DAK is a member of the evolutionarily conserved family of dihydroxyacetone kinases. In bacteria, DAK phosphorylates dihydroxyacetone to produce DHA phosphate, an obligatory precursor for the biogenesis of glyceryl ether phospholipids. Mammalian DAK displays dual activities as flavin adenine dinucleotide (FAD)- adenosine monophosphate (AMP) lyase and ATP-dependent Pha kinase [68, 69]. However, the physiological functions of DAK in higher eukaryotes are unknown. In immunoprecipitation experiments, DAK associates with MDA5 but not RIG-I, and the CARD domain-containing fragment of MDA5 is sufficient for the association. Expression of DAK inhibits MDA5-mediated IFNβ and ISRE reporter gene activity, but not RIG-I-, IPS-1 or TLR3 activity. DAK does not inhibit MDA5-mediated NF-κB activation in reporter gene assay, suggesting that DAK selectively inhibits MDA5 mediated IRF-3 activation. The observed interaction between endogenous MDA5 and DAK is decreased when the cells are infected with Sendai virus, suggesting that the association is disrupted upon virus infection, which is similar to SIKE's dissociation from TBK1 during virus infection. Therefore DAK might keep MDA5 inactive under steady state conditions, although the detailed mechanism of DAK regulation of MDA5 remains to be elucidated.
Viral negative regulators of the RLR signaling pathway
The importance of the IFN system as a primary antiviral defense has made it a strong selective pressure for virus evolution. Though effective, the RLR signaling pathway offers many potential targets for virus evasion, interference, and antagonism, including the initiating ligand (cytoplasmic non-self RNA); the RNA sensors, RIG-I and MDA5; signaling adaptor molecules such as IPS-1 and TRAFs; kinases involved in signal propagation and transcription factor activation (e.g., TBK1, IKKs); and the transcription factors that regulate antiviral gene expression (IRF-3, IRF-7, NF-κB) [70]. The recent literature is rich with descriptions of viral interference with antiviral signaling [71-73], although for many viral proteins the exact cellular target or mode of interference awaits further description. One outstanding example, for which the mechanism of inhibition on both the TLR3 and the RLR pathways has been characterized in great detail, is the hepatitis C virus protein complex NS3/4A. The NS3/4A protease function was known to be essential for proper posttranslational cleavage of the hepatitis C virus polyprotein when it was also identified as responsible for inhibition of IRF3 phosphorylation [74]. Sequence alignments of known NS3/4A specific cleavage sites in components of TLR3 signaling revealed a potential cleavage site in TRIF/TICAM-1 (Toll-IL-1 receptor domain-containing adaptor inducing IFNβ), an adaptor molecule in the TLR3 signaling pathway. In vitro cleavage assays confirmed the predicted cleavage of TRIF into two fragments, both incapable of IFNβ induction [75]. Shortly after the identification of TRIF as an NS3/4A substrate, NS3/4A was also shown to specifically interfere with the RLR system. This inhibition could be restored by IKKε expression, indicating a target upstream of the kinases [76]. With the identification of IPS-1 (Cardif/Visa/MAVS) [19-22] as the adaptor molecule in the RLR pathways, two groups have reported specific cleavage of IPS-1 by NS3/4A, explaining the mechanism of RLR inhibition by HCV [20, 77]. The importance and effectiveness of NS3/4A mediated immune response interference was underlined by the fact that it is not unique to HCV, but homologous proteases with IPS-1 cleavage activity were found in the related Flavivirus, GB Virus B [78], as well as the Picornavirus hepatitis A Virus [79, 80].
As with HCV, several viruses or their encoded proteins have been identified as inhibitors of RLR-induced signaling (Table II). However, in many cases the precise point of interference has yet to be clearly delineated. A common technique employed for determination of interference with the RLR mediated antiviral response takes advantage of the signaling activity of specific RNA ligands or ectopically expressed RIG-I N-terminal CARD region, full length MDA5, IPS-1 or downstream serine kinases as means to dissect the target for viral interference. Inhibition on the level of kinases or transcription factors will also affect TLR signaling as RLR and TLR pathways converge in later steps of their signaling cascade. Here, we focus instead on examples of viral strategies that are well described in their abilities to specifically circumvent the RLR response by RNA processing or direct interference with MDA5 or RIG-I.
Table II.
| Viral Inhibitors of RLR signaling | ||||
|---|---|---|---|---|
| Virus | Inhibitors | Target(s) | Proposed mechanisms | Refs. |
| Human cytomegalovirus | TRS1 | viral RNA | dsRNA sequestration, PAMP masking | [104-106] |
| Murine cytomegalovirus | m142/m1 43 | viral RNA | dsRNA sequestration, PAMP masking | [107, 108] |
| Vaccinia virus | E3L | viral RNA | dsRNA sequestration, PAMP masking | [109, 110] |
| Reovirus | σ3 | viral RNA | dsRNA sequestration, PAMP masking | [111, 112] |
| Rotavirus | NSP3 | viral RNA | dsRNA sequestration, PAMP masking | [113] |
| Rotavirus | NSP1 | IRF-3 | IRF-3 degradation | [114] |
| Herpes simplex virus | Us11 | viral RNA | dsRNA sequestration, PAMP masking | [115, 116] |
| Herpes simplex virus | ICP0 | IRF-3 | IRF-3 sequestration/degradation | [117-119] |
| Human herpesvirus 6 | IE1 | IRF3 | Inhibition of IRF3 dimerization/IRF3 degradation | [120] |
| Human herpesvirus 8 | vIRF | IRF3 promotor binding site | Virus encoded IRFs imitate non functional IRF | [121] |
| Human papillomavirus | E6 | IRF-3 | IRF-3 sequestration | [122] |
| Human papillomavirus | E7 | IκB kinase complex | Inhibition of kinase complexes | [123] |
| Poxvirus | N1L | IκB kinase complex | Inhibition of kinase complexes | [124] |
| Hepatitis C virus | NS2 | Unknown | Unknown | [125] |
| Hepatitis C virus | NS3/4A | IPS-1 | IPS-1 cleavage | [20, 74, 76, 77] |
| GB Virus B | NS3/4A | IPS-1 | IPS-1 cleavage | [78] |
| Hepatitis A virus | NS3/4A | IPS-1 | IPS-1 cleavage | [79, 80] |
| West Nile | Unknown | Unknown | Unknown | [126] |
| Porcine reproductive and respiratory syndrome virus | Unknown | Unknown | Unknown | [127] |
| Mengovirus | Leader | Unknown | Inhibition of IRF-3 dimerization | [128] |
| Pestivirus | Npro | IRF-3 | Proteasomal IRF-3 degradation | [129, 130] |
| Hantavirus | G1 | TBK1 | Inhibition of TBK1 | [131] |
| Paramyxovirus | V | MDA5 | MDA5 Complex inactivation | [10, 85] |
| Paramyxovirus | C | Unknown | Unknown | [132] |
| Human metapneumovirus | G | RIG-I | RIG-I complex inactivation | [102] |
| Human, bovine respiratory syncytial virus | NS1 | Unknown | Unknown | [133, 134] |
| Human, bovine respiratory syncytial virus | NS2 | Unknown | Unknown | [133, 134] |
| Rabiesvirus | P | TBK-1 | Inhibition of TBK1 | [135] |
| Thogotovirus | ML | IRF-3 | Inhibition of IRF-3 dimerization | [136] |
| Borna disease virus | P | TBK-1 | Inhibition of TBK1 | [137] |
| Ebolavirus | Vp35 | viral RNA, TBK1 | dsRNA sequestration, PAMP masking, interference with kinase activity | [138, 139] |
| Influenza A,B | NS1 | viral RNA, RIG-I, IPS-1 | dsRNA sequestration, PAMP masking, RIG-I/IPS-1 complex inactivation | [15,93-96,140] |
| Human immunodeficiency virus 1 | TAT | viral RNA | dsRNA sequestration, PAMP masking | [141] |
RNA processing as a RIG-I evasion strategy
The specific ligand for RIG-I signaling was identified as 5′-triphosphate containing ssRNA or dsRNA [14-17]. As most viruses do not cap their genomic RNA or mRNA, 5′triphosphate ends are often detected as non-self RNA in virus infected cells. Many viral proteins have RNA binding affinity, and in some cases the same proteins act as antiviral antagonists [72]. Although it is often difficult to clearly define the specificity of multifunctional proteins, it has been suggested that RNA binding underlies the immune evasion properties of many viral IFN antagonists (Table II).
More specific than RNA sequestration from RLR detection, permanent genomic RNA processing to mask 5′end modifications was recently implicated as an IFN evasion mechanism [81, 82]. Genomic RNA of representative negative-stranded RNA virus (NSV) families was tested for IFN induction. In agreement with other cases [14, 15] a strong IFN response was found to be dependent on the 5′triphosphate. Interestingly, genomic RNA from members of Bunyaviridae, specifically Hantaan virus (HTNV) and Crimean-Congo hemorrhagic fever virus (CCHFV), as well as Bornaviridae, Borna disease virus (BDV), did not induce an IFN response. In all three cases, the evasion of virus-induced RIG-I signaling correlates with the presence of 5′monophosphate genomic RNA [82, 83]. The exact mechanisms for removal of 5′triphosphate residues are largely unknown. Alternate mechanism have been proposed including a prime- and realign replication mechanism for HTNV [83] and programmed genome trimming for BDV [84]. In both models the 5′monophosphates suggest an endonuclease or pyrophosphatase activity.
Paramyxovirus V proteins target MDA5
To date the only viral protein known to interfere specifically with MDA5 is the paramyxovirus V protein. A wide variety of paramyxoviruses target MDA5 but not RIG-I via their V protein and can specifically block MDA5 mediated IFN induction [10, 85]. MDA5 was isolated in a screen for immunoprecipitated host cell proteins that interact with the V protein of parainfluenza virus 5 (PIV5) [10]. It was demonstrated that the C-terminal domain, a highly conserved hallmark of paramyxovirus V proteins, is necessary and sufficient for the inhibition of and interaction with MDA5. The inhibition of MDA5 is a general property of V protein C-termini and yeast-two hybrid results are suggestive of a direct protein:protein interaction. Nonetheless, the exact mechanism of interference is yet to be explained, in part due to the lack of a clear understanding of the role of MDA5 in antiviral signaling. Interestingly, despite a 30% sequence identity between MDA5 and RIG-I, no interaction was detected between RIG-I and a V protein. This strong preference for MDA5 seems in contradiction to the finding that MDA5 deficient mice exhibit normal resistance to paramyxovirus infection while RIG-I deficient mice are more susceptible [11, 86]. It can be speculated that additional MDA5 functions besides IFN induction, such as the described proapoptotic activity [87], might play a role in the strong evolutionary selection for the V protein to target MDA5.
Influenza A Virus NS1 proteins target RIG-I
The nonstructural protein 1 (NS1) of influenza A is well known as an antagonist of antiviral host responses [88]. NS1 has been demonstrated to inactivate several immune effectors, including the downregulation of cellular mRNA processing [89, 90], interaction and blocking of PKR [91, 92] as well as RNA sequestration from 2′5′-OAS activation [93]. IFN production is highly elevated in cells infected with NS1 deficient influenza A strains compared to cells infected with wildtype virus [94], and overexpression of NS1 can block IFN production by other viruses or PRR ligands [94, 95]. RIG-I and not MDA5 is responsible for influenza A detection [11, 96], specifically the 5′triphosphate of influenza A genomic RNA is recognized by RIG-I [15]. Several publications demonstrated the inhibition of RIG-I mediated signaling by NS1 [15, 96-98]. While initially dsRNA sequestration by NS1 was suggested as the main mechanism for prevention of PRR signaling, recently an interaction between RIG-I and NS1 was demonstrated [15, 96]. Interestingly, constitutive RIG-I CARD induced signaling was also efficiently blocked by NS1 [96]. While direct interference with RIG-I may explain many actions of NS1, some findings suggest a more complicated mode of interference. For example, antiviral signaling induced by ectopic expression of IPS-1 is also inhibited by NS1 and both RIG-I and NS1 co-fractionate with IPS-1 in an insoluble cell fraction. It is unknown whether the interaction between RIG-I and NS1 is direct, but coimmunoprecipitation assays suggest that the interaction may be influenced by RNA as NS1 RNA binding mutants bound poorly to RIG-I [15]. Nonetheless NS1 mutants that are defective for RNA binding retain some ability to block IFNβ promoter transcription [99, 100].
Human Metapneumovirus Glycoprotein G targets RIG-I
The newly discovered human metapneumovirus (hMPV) belongs to the family of Paramyxoviridae and was identified as a leading cause of respiratory tract infection among children [101]. Recombinant hMPV lacking the G gene (rhMPV-ΔG) replicated efficiently in vitro but was highly attenuated in a hamster model system [102]. The role of the gene product, glycoprotein G, a transmembrane protein, is largely unknown, however recent findings demonstrated a role in hMPV induced innate immune response. In cell culture, infection with rhMPV-ΔG lead to higher levels of type I IFNs and other cytokines compared to infection with recombinant wildtype virus [103]. Coexpression studies of RIG-I and MDA5 with the hMPV G protein revealed that the G protein can specifically block RIG-I mediated IFNβ induction. In addition, RIG-I and the G protein were shown to co-precipitate in co-expression studies as well as in hMPV infected cells [103]. As with the Influenza NS1 protein it is unknown whether the RIG-I G protein interaction is direct and what constitutes the precise mechanism of interference.
As the newly discovered cytoplasmic RLR pathways have proven their importance for innate immune responses, we expect rapid progress in the discovery of additional viral strategies to interfere with RLR signaling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13:543–51. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- Gresser I, Aguet M, Morel-Maroger L, Woodrow D, Puvion-Dutilleul F, Guillon JC, Maury C. Electrophoretically pure mouse interferon inhibits growth, induces liver and kidney lesions, and kills suckling mice. Am J Pathol. 1981;102:396–402. [PMC free article] [PubMed] [Google Scholar]
- Gresser I, Tovey MG, Maury C, Chouroulinkov I. Lethality of interferon preparations for newborn mice. Nature. 1975;258:76–8. doi: 10.1038/258076a0. [DOI] [PubMed] [Google Scholar]
- Heylbroeck C, Balachandran S, Servant MJ, DeLuca C, Barber GN, Lin R, Hiscott J. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J Virol. 2000;74:3781–3792. doi: 10.1128/jvi.74.8.3781-3792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Takaoka A, Taniguchi T. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Park JM, Zhang K, Luo JL, Maeda S, Kaufman RJ, Eckmann L, Guiney DG, Karin M. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature. 2004;428:341–345. doi: 10.1038/nature02405. [DOI] [PubMed] [Google Scholar]
- Peters K, Chattopadhyay S, Sen GC. IRF-3 activation by sendai virus infection is required for cellular apoptosis and avoidance of persistence. J Virol. 2008;82:3500–3508. doi: 10.1128/JVI.02536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A, Honda K, Taniguchi T. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem Biophys Res Commun. 2003;306:860–866. doi: 10.1016/s0006-291x(03)01049-0. [DOI] [PubMed] [Google Scholar]
- Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A. 2004;101:17264–9. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem. 2007;282:15315–8. doi: 10.1074/jbc.R700007200. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–64. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Cui S, Eisenächer K, Kirchhofer A, Brzózka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr., Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–7. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–40. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr., Akira S, Yonehara S, Kato A, Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–8. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80:12332–42. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–8. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Opipari AWJ, Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem. 1990;265:14705–14708. [PubMed] [Google Scholar]
- Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Yamamoto M, Miyagishi M, Taira K, Nakanishi M, Fujita T, Akira S, Yamamoto N, Yamaoka S. A20 is a negative regulator of IFN regulatory factor 3 signaling. J Immunol. 2005;174:1507–1512. doi: 10.4049/jimmunol.174.3.1507. [DOI] [PubMed] [Google Scholar]
- Wang YY, Li L, Han KJ, Zhai Z, Shu HB. A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-kappaB and ISRE and IFN-beta promoter. FEBS Lett. 2004;576:86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. Deubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–6. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641–50. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A. 2004;101:233–8. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–51. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–96. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–74. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Yoneyama M, Ito T, Takahashi K, Inagaki F, Fujita T. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J Biol Chem. 2004;279:9698–702. doi: 10.1074/jbc.M310616200. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature. 2002;419:853–857. doi: 10.1038/nature01120. [DOI] [PubMed] [Google Scholar]
- Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, Gu L, Tang X, Lu KP, Xiao ZX. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature. 2002;419:849–853. doi: 10.1038/nature01116. [DOI] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Lee HK, Iwasaki A. Autophagy and antiviral immunity. Curr Opin Immunol. 2008;20:23–9. doi: 10.1016/j.coi.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O'Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, Zhang Z, Arnott D, Dixit VM. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–32. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- Hacker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Hacker G, Mann M, Karin M. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–7. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–11. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. Embo J. 2006;25:3257–63. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu X, Lee AJ, Jin W, Chang M, Wright A, Imaizumi T, Sun SC. Regulation of IKK-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008 doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CS, O'Donnell M, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, Xavier R, Ting AT. Tumor suppressor CYLD is a negative regulator of RIG-I-mediated anti-viral response. EMBO rep. 2008 doi: 10.1038/embor.2008.136. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, Lich JD, Heise MT, Chen Z, Ting JP. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–7. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin SE. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci U S A. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Hwang SY, Imaizumi T, Yoo JY. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F, Li S, Tian Y, Zhang M, Xu LG, Zhang Y, Wang RP, Chen D, Zhai Z, Zhong B, Tien P, Shu HB. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci U S A. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bächler C, Flükiger-Brühwiler K, Schneider P, Bähler P, Erni B. From ATP as substrate to ADP as coenzyme: functional evolution of the nucleotide binding subunit of dihydroxyacetone kinases. J Biol Chem. 2005;280:18321–18325. doi: 10.1074/jbc.M500279200. [DOI] [PubMed] [Google Scholar]
- Cabezas A, Costas MJ, Pinto RM, Couto A, Cameselle JC. Identification of human and rat FAD-AMP lyase (cyclic FMN forming) as ATP-dependent dihydroxyacetone kinases. Biochem Biophys Res Commun. 2005;338:1682–1689. doi: 10.1016/j.bbrc.2005.10.142. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–24. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–30. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem Pharmacol. 2008;75:589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- Foy E, Li K, Wang C, Sumpter R, Jr., Ikeda M, Lemon SM, Gale M., Jr. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–8. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr., Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–7. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman A, Grandvaux N, Lin R, Ottone C, Akira S, Yoneyama M, Fujita T, Hiscott J, Meurs EF. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. J Virol. 2005;79:3969–78. doi: 10.1128/JVI.79.7.3969-3978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–22. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Benureau Y, Rijnbrand R, Yi J, Wang T, Warter L, Lanford RE, Weinman SA, Lemon SM, Martin A, Li K. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J Virol. 2007;81:964–76. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, Grotheer D, Berk I, Schlemminger S, Vallbracht A, Dotzauer A. Hepatitis A virus suppresses RIG-I-mediated IRF-3 activation to block induction of beta interferon. J Virol. 2005;79:10968–77. doi: 10.1128/JVI.79.17.10968-10977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, Lemon SM. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A. 2007;104:7253–8. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider U, Martin A, Schwemmle M, Staeheli P. Genome trimming by Borna disease viruses: viral replication control or escape from cellular surveillance? Cell Mol Life Sci. 2007;64:1038–42. doi: 10.1007/s00018-007-6545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, Mirazimi A, Weber F. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol. 1995;69:5754–62. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider U, Schwemmle M, Staeheli P. Genome trimming: a unique strategy for replication control employed by Borna disease virus. Proc Natl Acad Sci U S A. 2005;102:3441–6. doi: 10.1073/pnas.0405965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–45. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DC, Gopalkrishnan RV, Lin L, Randolph A, Valerie K, Pestka S, Fisher PB. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene. 2004;23:1789–800. doi: 10.1038/sj.onc.1207300. [DOI] [PubMed] [Google Scholar]
- Krug RM, Yuan W, Noah DL, Latham AG. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–9. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Chen Z, Li Y, Krug RM. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. Embo J. 1999;18:2273–83. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- Hatada E, Saito S, Fukuda R. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J Virol. 1999;73:2425–33. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SL, Katze MG. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J Interferon Cytokine Res. 1998;18:757–66. doi: 10.1089/jir.1998.18.757. [DOI] [PubMed] [Google Scholar]
- Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006;103:7100–5. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–96. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–73. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr., Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–24. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, Fujita T, Katz JM, Donis RO, Sambhara S. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol. 2007;36:263–9. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, Hippenstiel S, Suttorp N, Wolff T. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9:930–8. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–66. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber B, Schneider J, Wolff T. Double-stranded RNA binding of influenza B virus nonstructural NS1 protein inhibits protein kinase R but is not essential to antagonize production of alpha/beta interferon. J Virol. 2006;80:11667–77. doi: 10.1128/JVI.01142-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–24. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S, Skiadopoulos MH, Yang L, Lamirande EW, Tran KC, Murphy BR, Collins PL, Buchholz UJ. Recombinant human Metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J Virol. 2004;78:12877–87. doi: 10.1128/JVI.78.23.12877-12887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Liu T, Shan Y, Li K, Garofalo RP, Casola A. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog. 2008;4:e1000077. doi: 10.1371/journal.ppat.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassady KA. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J Virol. 2005;79:8707–15. doi: 10.1128/JVI.79.14.8707-8715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ, Hakki M, De Niro KL, Geballe AP. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J Virol. 2004;78:197–205. doi: 10.1128/JVI.78.1.197-205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki M, Geballe AP. Double-stranded RNA binding by human cytomegalovirus pTRS1. J Virol. 2005;79:7311–8. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ, Hanson LK, Brown CE, Janzen DM, Geballe AP. Double-stranded RNA binding by a heterodimeric complex of murine cytomegalovirus m142 and m143 proteins. J Virol. 2006;80:10173–80. doi: 10.1128/JVI.00905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valchanova RS, Picard-Maureau M, Budt M, Brune W. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J Virol. 2006;80:10181–90. doi: 10.1128/JVI.00908-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EJ, Marie I, Prakash A, Garcia-Sastre A, Levy DE. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by Vaccinia virus E3L protein. J Biol Chem. 2001;276:8951–7. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Condit RC, Vijaysri S, Jacobs B, Williams BR, Silverman RH. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J Virol. 2002;76:5251–9. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E, Denzler KL, Tartaglia J, Perkus ME, Paoletti E, Jacobs BL. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olland AM, Jane-Valbuena J, Schiff LA, Nibert ML, Harrison SC. Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 A resolution. Embo J. 2001;20:979–89. doi: 10.1093/emboj/20.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Pettiford S, Jiang B, Jacobs BL. Products of the porcine group C rotavirus NSP3 gene bind specifically to double-stranded RNA and inhibit activation of the interferon-induced protein kinase PKR. J Virol. 1994;68:3821–9. doi: 10.1128/jvi.68.6.3821-3829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JW, Ewen J, Ettayebi K, Hardy ME. Zinc-binding domain of rotavirus NSP1 is required for proteasome-dependent degradation of IRF3 and autoregulatory NSP1 stability. J Gen Virol. 2007;88:613–20. doi: 10.1099/vir.0.82255-0. [DOI] [PubMed] [Google Scholar]
- Poppers J, Mulvey M, Khoo D, Mohr I. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J Virol. 2000;74:11215–21. doi: 10.1128/jvi.74.23.11215-11221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Mohr I. Inhibition of cellular 2′-5′ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J Virol. 2007;81:3455–64. doi: 10.1128/JVI.02520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson G, Zhang Y, Jones C. The Bovine herpesvirus 1 gene encoding infected cell protein 0 (bICP0) can inhibit interferon-dependent transcription in the absence of other viral genes. J Gen Virol. 2005;86:2697–702. doi: 10.1099/vir.0.81109-0. [DOI] [PubMed] [Google Scholar]
- Melroe GT, DeLuca NA, Knipe DM. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol. 2004;78:8411–20. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saira K, Zhou Y, Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J Virol. 2007;81:3077–86. doi: 10.1128/JVI.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska J, Gravel A, Fink K, Grandvaux N, Flamand L. Inhibition of transcription of the beta interferon gene by the human herpesvirus 6 immediate-early 1 protein. J Virol. 2007;81:5737–48. doi: 10.1128/JVI.02443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ, Jr., Barber GN, Hiscott J. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene. 2001;20:800–11. doi: 10.1038/sj.onc.1204163. [DOI] [PubMed] [Google Scholar]
- Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–72. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitkovsky D, Hehner SP, Hofmann TG, Moller A, Schmitz ML. The human papillomavirus oncoprotein E7 attenuates NF-kappa B activation by targeting the Ikappa B kinase complex. J Biol Chem. 2002;277:25576–82. doi: 10.1074/jbc.M201884200. [DOI] [PubMed] [Google Scholar]
- DiPerna G, Stack J, Bowie AG, Boyd A, Kotwal G, Zhang Z, Arvikar S, Latz E, Fitzgerald KA, Marshall WL. Poxvirus protein N1L targets the I-kappaB kinase complex, inhibits signaling to NF-kappaB by the tumor necrosis factor superfamily of receptors, and inhibits NF-kappaB and IRF3 signaling by toll-like receptors. J Biol Chem. 2004;279:36570–8. doi: 10.1074/jbc.M400567200. [DOI] [PubMed] [Google Scholar]
- Kaukinen P, Sillanpaa M, Kotenko S, Lin R, Hiscott J, Melen K, Julkunen I. Hepatitis C virus NS2 and NS3/4A proteins are potent inhibitors of host cell cytokine/chemokine gene expression. Virol J. 2006;3:66. doi: 10.1186/1743-422X-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Gale M., Jr. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80:2913–23. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Xiao S, Jiang Y, Jin H, Wang D, Liu M, Chen H, Fang L. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-beta production by interfering with the RIG-I signaling pathway. Mol Immunol. 2008;45:2839–46. doi: 10.1016/j.molimm.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hato SV, Ricour C, Schulte BM, Lanke KH, de Bruijni M, Zoll J, Melchers WJ, Michiels T, van Kuppeveld FJ. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell Microbiol. 2007;9:2921–30. doi: 10.1111/j.1462-5822.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, Goodbourn S. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol. 2006;80:11723–32. doi: 10.1128/JVI.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca SA, Herbert RJ, Crooke H, Drew TW, Wileman TE, Powell PP. Loss of interferon regulatory factor 3 in cells infected with classical swine fever virus involves the N-terminal protease, Npro. J Virol. 2005;79:7239–47. doi: 10.1128/JVI.79.11.7239-7247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alff PJ, Gavrilovskaya IN, Gorbunova E, Endriss K, Chong Y, Geimonen E, Sen N, Reich NC, Mackow ER. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J Virol. 2006;80:9676–86. doi: 10.1128/JVI.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahle L, Marq JB, Brini A, Hausmann S, Kolakofsky D, Garcin D. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J Virol. 2007;81:12227–37. doi: 10.1128/JVI.01300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert B, Marozin S, Conzelmann KK. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J Virol. 2003;77:8661–8. doi: 10.1128/JVI.77.16.8661-8668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann KM, Tran KC, Collins PL. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J Virol. 2005;79:5353–62. doi: 10.1128/JVI.79.9.5353-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozka K, Finke S, Conzelmann KK. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J Virol. 2005;79:7673–81. doi: 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings S, Martinez-Sobrido L, Garcia-Sastre A, Weber F, Kochs G. Thogoto virus ML protein suppresses IRF3 function. Virology. 2005;331:63–72. doi: 10.1016/j.virol.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Unterstab G, Ludwig S, Anton A, Planz O, Dauber B, Krappmann D, Heins G, Ehrhardt C, Wolff T. Viral targeting of the interferon-{beta}-inducing Traf family member-associated NF-{kappa}B activator (TANK)-binding kinase-1. Proc Natl Acad Sci U S A. 2005;102:13640–5. doi: 10.1073/pnas.0502883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas WB, Loo YM, Gale M, Jr., Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–78. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Towner JS, Nichol ST. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–84. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Dauber B, Heins G, Wolff T. The influenza B virus nonstructural NS1 protein is essential for efficient viral growth and antagonizes beta interferon induction. J Virol. 2004;78:1865–72. doi: 10.1128/JVI.78.4.1865-1872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra RK, McMillan NA, Desai S, McSwiggen J, Hovanessian AG, Sen G, Williams BR, Silverman RH. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–7. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]