Abstract
Non-coding RNAs of complex tertiary structure are involved in numerous aspects of the replication and processing of genetic information in many organisms; however, an understanding of the complex relationship between their structural dynamics and function is only slowly emerging. The Neurospora Varkud Satellite (VS) ribozyme provides a model system to address this relationship. First, it adopts a tertiary structure assembled from common elements, a kissing loop and two three-way junctions. Second, catalytic activity of the ribozyme is essential for replication of VS RNA in vivo and can be readily assayed in vitro. Here we exploit single molecule FRET to show that the VS ribozyme exhibits previously unobserved dynamic and heterogeneous hierarchical folding into an active structure. Readily reversible kissing loop formation combined with slow cleavage of the upstream substrate helix suggests a model whereby the structural dynamics of the VS ribozyme favor cleavage of the substrate downstream of the ribozyme core instead. This preference is expected to facilitate processing of the multimeric RNA replication intermediate into circular VS RNA, which is the predominant form observed in vivo.
Keywords: catalytic RNA, Neurospora, RNA dynamics, non-coding RNA folding, single molecule fluorescence resonance energy transfer
Introduction
During the past few years it has been recognized that non-coding RNAs play a much wider than anticipated role in many aspects of the replication, processing, modification and regulation of genetic information.1–3 Non-coding RNAs of complex tertiary structure whose structure-function relationships have been well studied include the self-cleaving ribozymes, in which catalytic activity depends directly on structure and can be measured without the need for reporter molecules or other macromolecular interactions.4–6 The VS ribozyme (Fig 1) is the catalytic motif embedded in circular VS RNA, which is found in mitochondria of geographically and genetically divergent species of the bread mold Neurospora.7,8 Similar to a retrotransposon, VS RNA is converted into a DNA plasmid through enzymatic action of a reverse transcriptase encoded on the coexisting V retroplasmid.9 The reverse transcriptase requires a circular monomeric VS RNA template to generate the VS plasmid, which in turn is transcribed by mitochondrial RNA polymerase into multimeric VS RNA copies. Self-cleavage and re-ligation by the embedded VS ribozyme to regenerate circular monomeric VS RNA is thus an important feature of satellite replication in the fungal host.
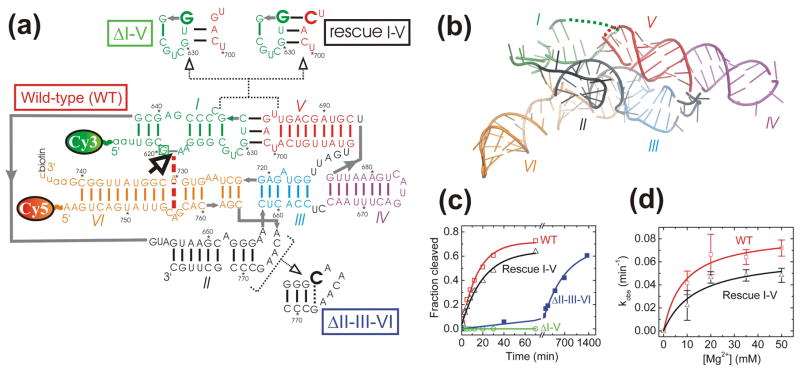
Fig. 1.
Structure and function of the well-studied G11 VS ribozyme that preserves the native VS RNA sequence. (a) Secondary structure depiction of the four VS ribozyme variants studied here, as based on two similar low-resolution structural models.18,20 Helices are color-coded, the bold open arrow next to the boxed G620 denotes the cleavage site, the red dashed line indicates a key interaction in the active site, gray lines denote direct backbone connectivities, and the Cy3, Cy5 and biotin labeling sites are indicated. Mutations in the ribozyme variants are shown next to heir names. All single molecule FRET experiments employed a 2′-O-methyl modification of G620 to render the substrate non-cleavable. (b) Tertiary structure model of the VS ribozyme (Zamel and Collins, unpublished data) using the same helix color code as in panel a. Some strand connectivities are incomplete in the model; the ones in the I-V kissing loop are indicated by dashed lines. (c) Cleavage time courses (data points) for each of the four ribozyme variants studied here, with single-exponential fits (lines) yielding the observed rate constants reported in the text. (d) Magnesium dependence of the observed cleavage rate constants of WT and Rescue I-V.
The VS ribozyme is the largest and most complex of the small pathogenic ribozymes and not coincidentally the least understood.4,5,10,11 Structural details at atomic resolution have been derived only in NMR studies of small segments such as stem-loops I, V and VI, which may well change conformation upon assembly in the ribozyme.12–16 The global architecture of the naturally occurring VS ribozyme sequence (Fig 1a) and its relation to catalytic activity have so far been studied by ensemble activity and fluorescence resonance energy transfer (FRET) assays, mutagenesis, native gel electrophoresis, hydroxyl radical footprinting, and UV-induced crosslinking.17–21 In combination, these studies have yielded two similar, if incomplete models of how the six helical elements self-assemble to juxtapose helices I and VI in the catalytic core (Fig. 1b).18,20 This model depicts the global architecture of the active VS ribozyme as built from the II-III-VI and III-IV-V three-way junctions and the I-V kissing loop interaction (Fig. 1a and b). While it is known that ribozymes commonly undergo dynamic structural changes along their reaction pathways,22–24 no study has yet addressed whether alternate global folds of the VS ribozyme exist, what their rates of interconversion may be, and how they may relate to biological function.
Single molecule FRET is a particularly suitable tool to identify even short-lived conformational isomers and report on their exchange kinetics and heterogeneities under equilibrium conditions, as recently demonstrated for the Tetrahymena23,25,26 and hairpin ribozymes.6,27–31 Here we present the global folding dynamics of single VS ribozyme molecules as observed in real-time by total internal reflection FRET microscopy.32 We combine the quantified population distributions and interconversion kinetics with ensemble activity assays and single-site as well as second-site revertant mutagenesis to show: (i) how the dynamics of the I-V kissing interaction affect catalytic activity; (ii) that a slow local conformational change must be traversed before the ribozyme undergoes self-cleavage; and (iii) that the II-III-VI junction acts as a structural scaffold for kissing loop formation. Our results lead to a kinetic model of the VS ribozyme reaction pathway whose relation to VS RNA replication we discuss in light of a recently designed class of fast-cleaving ribozymes.33
Results
Single wild-type VS ribozyme molecules display dynamic and heterogeneous three-state folding
Our design for single molecule FRET experiments takes advantage of the well-studied ribozyme variant G11, derived from the natural VS RNA sequence from four nucleotides upstream to 163 nucleotides downstream of the cleavage site.34 This sequence was only slightly adapted to optimize the ribozyme for FRET studies (Fig 1a). In particular, we labeled the 5′ end of G11 with the FRET donor Cy3 by an enzymatic ligation approach that also allowed us to introduce a chemistry-blocking 2′-O-methyl modification at G620 for structural studies. In addition, we opened the non-essential35 (data not shown) closing loop of helix VI to attach a 5′-Cy5 FRET acceptor and a 3′-biotin for surface immobilization to take advantage of prism-based total internal reflection excitation (Materials and Methods). Our ribozyme is thus designed to specifically report on distance changes between helices I and VI that are distal in primary sequence (Fig 1a), yet juxtaposed to form the catalytic core as shown in the proposed tertiary structure model (Fig. 1b).
First, we characterized the catalytic activity of our labeled wild-type (WT) ribozyme by standard ensemble assays (Fig. 1c). The cleavage time course is fit well with a single exponential, yielding an observed rate constant of 0.065 ± 0.006 min−1 (total cleavage extent of ~75%) under our standard conditions (40 mM Tris-HCl, pH 8.0, 50 mM KCl, and 35 mM MgCl2 at 25 °C, see Material and methods). The unmodified G11 ribozyme has a very similar cleavage rate constant (0.14 min−1 in the same buffer (up to ~95% cleavage extent), but at 37 °C instead of 25 °C,34 which likely accounts for these differences) suggesting that our WT modifications minimally interfere with function. The cleavage rate constant saturated at 35 mM Mg2+ (Mg1/2 = 9 mM when fit with a hyperbolic binding equation, Fig. 1d), which was used in all subsequent single molecule experiments.
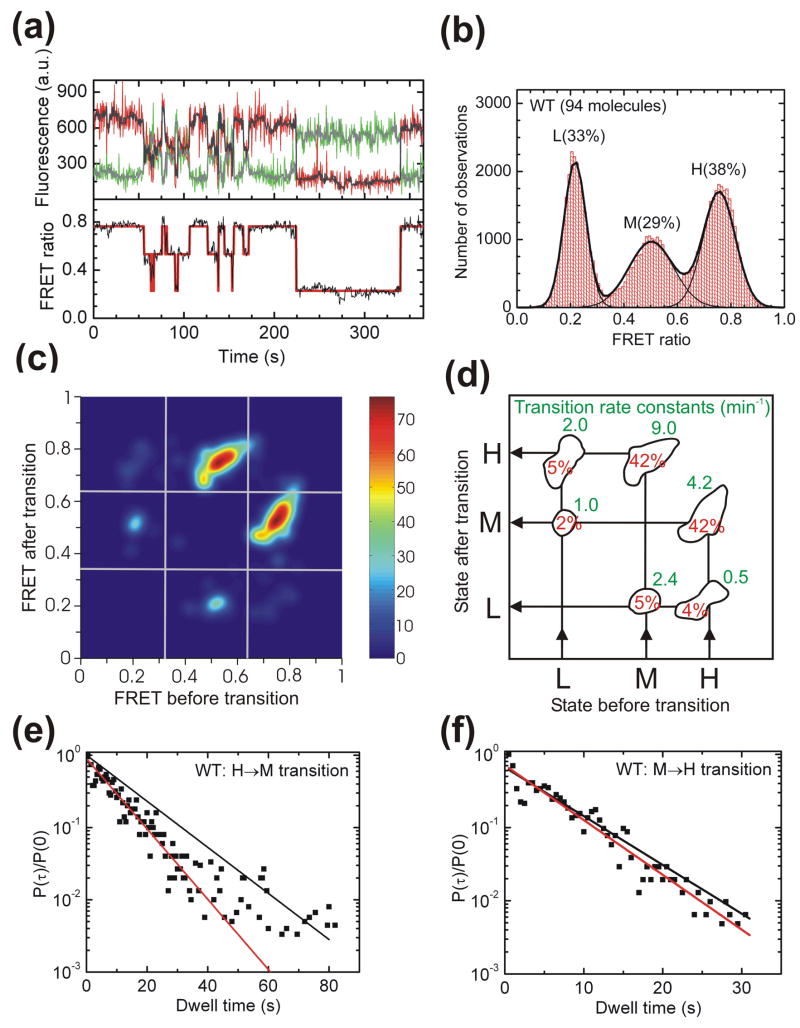
Immobilized single VS ribozyme molecules exhibit anticorrelated donor and acceptor fluorescence intensities as signature of global conformational changes (Fig. 2a). (For a discussion of our evidence that fluorophore blinking is insignificant under our low-illumination conditions, please refer to Materials and methods.) Three distinct FRET states and thus distinguishable RNA folds are observed for WT. The particular ribozyme molecule in Fig. 2a alternatively populates a high (H) FRET state with a FRET ratio (defined as IA/(IA+ID), where ID and IA are the donor and acceptor fluorescence intensities, respectively) of ~0.76, a mid (M) FRET state at ~0.50, and a low (L) FRET state at ~0.22. All analyzed time traces end with single-step donor photobleaching, confirming a single molecule as the source of the signal. A histogram of the FRET ratios of 94 WT molecules is well described by a triple Gaussian that yields the relative population distributions of the three FRET states (Fig. 2b). Donor-acceptor distances of 35 Å, 55 Å, and 96 Å (each ± 5 Å) were estimated for the H, M, and L states, respectively, after corrections for donor leakage into the acceptor channel, fluorophore photobleaching, and differential detector efficiencies for the donor and acceptor emissions (Materials and Methods).36,37
Fig. 2.
Single molecule FRET analysis of the WT VS ribozyme. (a) Representative FRET time trace of a WT molecule. The raw Cy3 donor and Cy5 acceptor fluorescence signals are green and red, respectively. Superimposed in gray are the data after applying a non-linear filter as described.38 The FRET ratio (black trace below) is calculated from the filtered data as IA/(IA + ID) and reveals three distinct states in Hidden Markov modeling (HMM, red line). (b) An aggregate FRET histogram of 94 WT molecules shows Gaussian distributions for three states with FRET values of ~0.76 (H), ~0.50 (M) and ~0.22 (L) and the indicated abundances. (c) Transition density contour map from HMM analysis54 of all transitions for 77 well defined WT molecules. (d) Relative contributions and mean rate constants of the transitions illustrated in panel C. (e, f) The normalized probability densities40 of all dwell times preceding an H→M (E) and M→H (F) transition are fit with single exponential decay functions (red lines) that are compared to decay functions (black lines) using the rate constant from the HMM analysis in panel D.
Shot noise in the raw data was reduced by employing a non-linear filter as described.38 Hidden Markov modeling (HMM) was then used to determine for each molecule the most probable path traversed between the three FRET states (Fig 2a).39 The transition density contour map in Fig. 2c,d summarizes all six possible observed transitions between initial and final FRET states for the analyzed WT molecules. Single-exponential transition rate constants for the H to the M state of 4.2 min−1 and for the M to the H state of 9.0 min−1 were derived from the hidden Markov fitting procedure39 (see Materials and methods) (Fig. 2d). Together, these two conformational changes account for 84% of all observed transitions (Fig. 2d). Despite the overall predominance of these transitions, 9 out of the 94 molecules included in Fig. 2b,c,d never sample the H state. These molecules are instead characterized by long dwell times in the M and L states (Supplementary Fig. S1). This small subset of molecules and the generally low transition rate constants for leaving the L state (Fig. 2d) lead to the observation of 33% L state in the aggregate histogram of Fig. 2b. These findings provide evidence for static heterogeneity at the single molecule level, as previously noticed for other RNAs such as the hairpin ribozyme,27–30 and suggest that the L state is a rarer structural state of the VS ribozyme.
Markov processes as assumed in HMM consist of a combination of state-to-state transitions with single-exponential decay kinetics,39 which may not always reflect the full complexity of single molecule behavior. To further characterize the nature of the kinetics of the H→M and M→H transitions, we therefore plotted the normalized probability densities40 of the corresponding dwell times obtained from the HMM path analysis for all WT molecules that accessed the H state at least once (Fig. 2e,f). A single-exponential decay fit yields a rate constant of 6.5 min−1 for the H→M transition, which is significantly faster than the previously described, averaged Markov rate constant of 4.2 min−1 (compare red and black lines in Fig. 2e). By contrast, both analysis methods yield similar rate constants for the M→H transition (10.1 min−1 for the single exponential decay fit and 9.0 min−1 for the averaged Markov value in Fig. 2f). The discrepancy observed for the H→M transition is due to a higher occurrence of long dwell times than is predicted for a single-exponential distribution (fitting to a double exponential failed to converge), providing evidence for heterogeneity also in the kinetic behavior of the H→M transition. Since no sub-populations of distinct kinetic behavior can be discerned in this case we cannot distinguish static (never-changing) and dynamic (time-dependent) heterogeneity.
What is the structural nature of the H state and what role, if any, does it play in function? The tertiary structure model (Fig. 1b) contains the previously established essential I-V kissing loop interaction which has been shown to help dock the cleavage site in helix I with the residues in helix VI that are involved in catalysis.18,20,35,41,42 Our measured distance between the FRET probes of (35 ± 5) Å for the H state agrees well with the distance of 38 Å between the C1′ carbons of G620 and A743 in the model (please note that while A743 is indeed where our Cy5 acceptor is attached, the sequence upstream of G620, to which the Cy3 donor is attached, is absent from the incomplete model, Figs. 1a and 1b). This agreement supports the hypothesis that the observed high FRET state reports on the catalytically active docked structure.
Mutation of the kissing loop further links the H state to the active docked structure
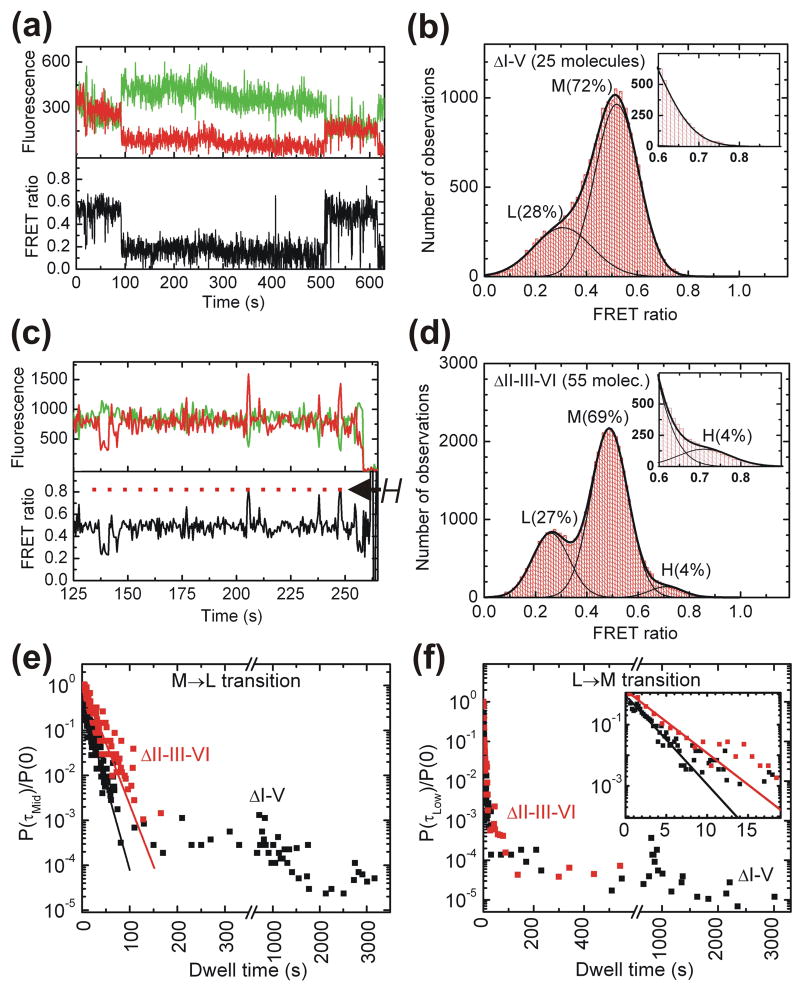
To ask whether the I-V kissing loop interaction is present in our H state, we designed a ΔI-V ribozyme with a C632→G mutation (Fig. 1a) that is predicted to weaken the interaction by eliminating one of three base pairs. This mutant was previously shown to lack self-cleavage activity41 and accordingly our version yields no detectable product (Fig. 1c). Significantly, the folding behavior of ΔI-V is markedly different from that of WT (Fig. 3a). ΔI-V molecules spend long periods of time in global folds characterized by FRET ratios of ~0.48 (M) and ~0.27 (L) (the M state is typically interrupted by brief excursions into the L state, Fig. 3a). A FRET histogram obtained from 25 molecules shows that the H state was not detected (Fig. 3b). Due to the long lifetime of the L state (the molecule in Fig. 3a, for example, dwells in it for ~8 min), its dwell times were ill-defined due to either premature donor photobleaching or the fact that the molecule already inhabits the L state at the beginning of the measurement. Separate measurements were therefore conducted over one hour where the laser was intermittently shuttered for an overall exposure time of only 9 min to limit photobleaching. The normalized probability densities of all observed M→L and L→M transition dwell times for ΔI-V (Fig. 3e and f) show a clear distinction between short- and long-lived M and L states. The short lived dwell times were fit to single exponential decay functions yielding rate constants of 5.0 min−1 for M→L and 40 min−1 for L→M. The average values for the long dwell times are 17 min and 14 min for the M and the L state, respectively, which must be viewed as lower bounds given the natural limitation of our observation window.
Fig. 3.
Single molecule FRET analysis of the ΔI-V and ΔII-III-VI variants. (a) Representative FRET time trace of a ΔI-V molecule. The raw Cy3 donor and Cy5 acceptor fluorescence signals are green and red, respectively. Compared to WT, the H state is not observed in ΔI-V and is replaced by long lived L and M states. The M state shows brief excursions to the L state. (b) An aggregate FRET histogram of the raw data for the 25 ΔI-V molecules is well described by Gaussian distributions for only two states with FRET ratios of ~0.48 (M) and ~0.27 (L), but no H state. (c) Representative FRET time trace of a ΔII-III-VI molecule, showing brief excursions into the H state, as well as an early excursion into the L state. (d) An aggregate FRET histogram of the raw data for 55 ΔII-III-VI molecules reports a ~0.69 (H) state that is populated ~4% of the time in addition to the ~0.49 (M) and ~0.25 (L) states (see inset). Please note that not all molecules visit all three states due to molecular heterogeneity so that these aggregate histograms do not reflect equilibrium distributions. (e, f) The short dwell times of the normalized dwell time probability densities40 preceding an M→L (E) and L→M (F) transition in the ΔI-V and ΔII-III-VI variants are fit with single exponential decay functions (lines), revealing discrepancies from homogeneous kinetics. Please note that the signal-to-noise ratio in the M state due to shot noise did not allow for reliable HMM analysis of the short-lived H state of the ΔII-III-VI ribozyme. Visual inspection of the traces yielded approximate transition rate constants of ~3 min−1 for the M→H transition and ~40 min−1 for the H→M transition.
The failure to observe the H state upon weakening of the kissing loop interaction suggests that adoption of the H state requires an intact kissing loop. The accompanying loss of cleavage activity links the H state with biological function of the VS ribozyme. Consistent with these observations, previous hydroxyl radical footprinting studies have observed a decrease in solvent protection of the riboses of several nucleotides involved in the kissing loop interaction when all three interacting bases of either loop I or V were mutated.18 An NMR study of stem-loop V predicts that the three kissing base pairs can form a short A-type helix,15 which we find to be disrupted by the single G-G mismatch in ΔI-V (Fig. 1a). The time resolution of our data collection (500 ms) allows the possibility that stems I and V may transiently (for <1 s) come into close contact, but their interaction is not sufficiently stabilized to be observed by either single molecule FRET or catalytic activity.
The II-III-VI junction plays the role of an architectural scaffold
Correct juxtaposition of helices II, III, and VI has been implicated in playing an important role in VS ribozyme function. In particular, an A656C mutation at this junction has been shown to substantially reduce the cleavage rate constant in a trans-cleaving variant of the VS ribozyme, presumably by forming a structurally detrimental Watson-Crick base pair with G768.19 To further explore the structural basis for this large reduction in activity we introduced the same mutation, generating ribozyme ΔII-III-VI (Fig 1a), and assayed its cleavage activity (Fig. 1c). The ΔII-III-VI mutant cleaves with a substantially reduced rate constant of 0.002 min−1, as expected,19 further validating the relevance of our experimental design.
Fig. 3c shows a representative section of a time trace of a single ΔII-III-VI molecule. It predominantly resides in the M state but also shows brief excursions into a high-FRET state similar to the H state of WT. These transient excursions are also evident from the aggregate FRET histogram, which includes a 4% contribution of the H state (Fig. 3d). By contrast, the H state is never observed to be accessed by the ΔI-V ribozyme (Fig. 3b).
In conjunction with our earlier observation that the H state reports on formation of the kissing loop, our single molecule studies of the A656C mutant provide evidence that a correct architecture of the II-III-VI junction is also a prerequisite for kissing loop formation. This finding implies that the A656C mutation induces local changes in the II-III-VI junction that have a cascading, distal effect preventing loops I and V from docking. Previous steady-state FRET measurements on the isolated II-III-VI junction have posited a change in the tilt angle between helix II and the III/VI helical stack as a consequence of the local change in sequence.19 Such a rearrangement in three-way junction architecture may then disfavor kissing loop formation as observed here.
Disruption of the II-III-VI junction also has a profound effect on the transition kinetics between the M and L states. While many single ΔI-V molecules with an unaltered II-III-VI junction exhibit very long dwell times in the M state (~17 min, see above and Fig. 3e, black symbols), the normalized probability density plot for the M→L transition of ΔII-III-VI instead is well described by a single rate constant of 3.6 min−1 with few, if any molecules exhibiting long dwell times in the M state (Fig. 3e, red symbols). The normalized probability distribution of L→M transition dwell times for ΔII-III-VI, by contrast, remains multi-exponential including relatively long dwell times (Fig. 3f). Fitting the dwell times of the rapid L→M transition yields a rate constant of 26 min−1 (Fig. 3f), while the average of all longer dwell times is 3.2 min, somewhat shorter than the 14 min observed for ΔI-V.
Taken together, these observations are consistent with the notion that a properly folded II-III-VI junction, in the absence of the I-V kissing loop interaction, is associated with a very stable M state that forms an architectural scaffold for the long-range kissing loop interaction to build on. This stable scaffold is disrupted in the A656C mutant, consistent with the general lack of long M dwell times for ΔII-III-VI in Fig. 3e. By contrast, the long fluorophore distance in the L state (96 Å) together with the fact that both the ΔI-V and ΔII-III-VI ribozymes similarly show long L dwell times leads us to propose that L states generally do not have a properly folded II-III-VI junction, thus unraveling the central architectural scaffold of the VS ribozyme in a way that dramatically increases the fluorophore distance. This model thus associates the L state with a misfolded II-III-VI junction.
Second-site reversion mutagenesis rescues both H state and ribozyme activity
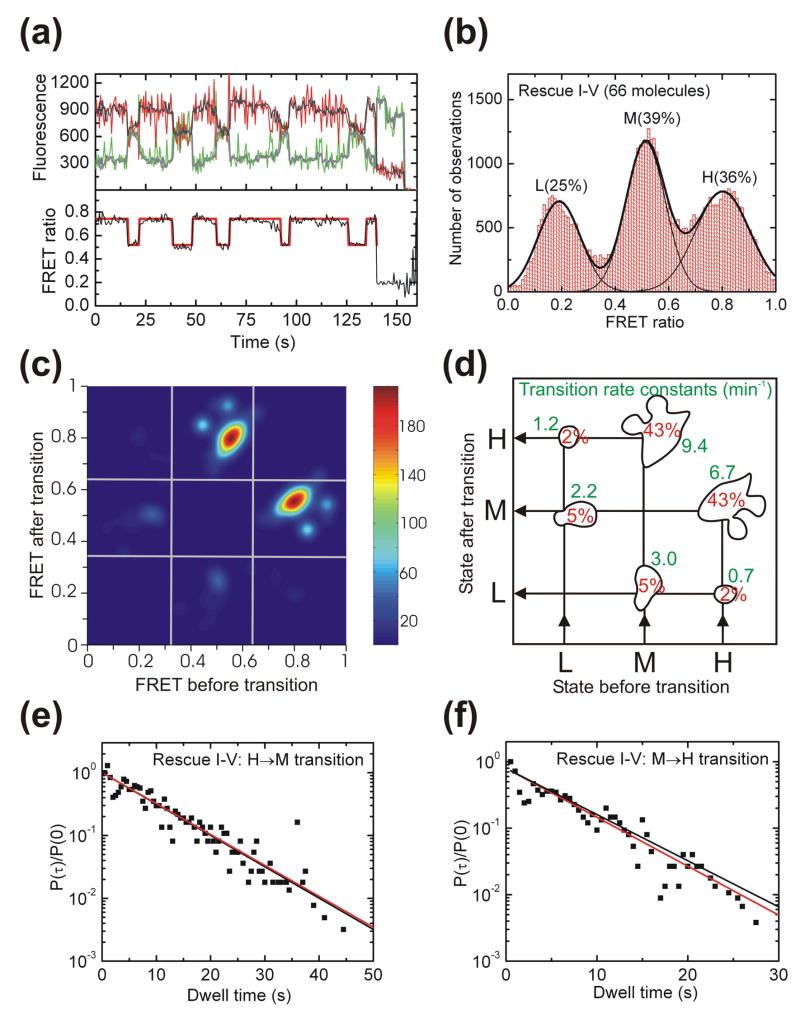
Our ΔI-V ribozyme completely loses its H state, presumably because of an inability to form a stable kissing loop interaction. If so, recovery of this interaction should restore the structural dynamics observed for WT. To test this hypothesis we designed a Rescue I-V ribozyme with a second-site reversion mutation that flips the WT C:G to a G:C base pair (Fig. 1a). The representative time trace in Fig. 4a shows that the H state is indeed recovered, further providing evidence that the H state reports on the formation of the catalytically essential kissing loop interaction. Rescue I-V molecules spend 36% of their time in the H state (Fig. 4b) versus 38% for WT (Fig. 2b), suggesting that kissing loop rescue is nearly complete. This notion is further supported by the very similar transition density contour maps of Rescue I-V and WT (Fig. 4c and d versus Fig. 2c and d).
Fig. 4.
Single molecule FRET analysis of the Rescue I-V ribozyme. (a) Representative FRET time trace of a Rescue I-V molecule. The raw Cy3 donor and Cy5 acceptor fluorescence signals are green and red, respectively. Superimposed in gray are the data after applying a non-linear filter as described.38 The FRET ratio (black trace below) is calculated from the filtered data as IA/(IA + ID) and reveals three distinct states in Hidden Markov modeling (HMM, red line). (b) An aggregate FRET histogram of 66 Rescue I-V molecules shows Gaussian distributions for three states with FRET values of ~0.80 (H), ~0.51 (M) and ~0.19 (L) and the indicated abundances. (c) Transition density contour map from HMM analysis54 of all transitions for 62 well defined Rescue I-V molecules. (d) Relative contributions and mean rate constants of the transitions illustrated in panel C. (e, f) The normalized probability densities40 of all dwell times preceding an H→M (E) and M→H (F) transition are fit with single exponential decay functions (red lines) that are compared to decay functions (black lines) using the rate constant from the HMM analysis in panel D.
The transition density contour map of Rescue I-V is also similar to that of WT with the predominant transitions occurring between the H and M states. Comparison of the averaged transition rate constants from the contour map with those derived from analysis of the normalized dwell time probability densities shows good agreement (Fig. 4e and f). This relative homogeneity of the transition kinetics contrasts with the significant kinetic heterogeneity (non-single-exponential behavior) observed for the H→M transition of WT (Fig. 2e), thus illustrating how the simple reversal of a G:C basepair leads to subtle differences in dynamic behavior detectable by single molecule probing.
Rescue of the kissing loop interaction and reemergence of the H state coincide with substantial recovery of catalytic activity in Rescue I-V (Fig. 1c). The Mg2+ dependence curve of Rescue I-V is predicted to saturate at 0.065 min−1 with a Mg2+ half-titration point of 14 mM, just slightly below and above the WT values of 0.084 min−1 and 9 mM, respectively (Fig. 1d). These findings provide additional strong evidence that link the H state observed in single molecule FRET experiments to on-pathway folding of the kissing loop interaction and to catalytic activity. Consequently, the fact that a lower estimate of 10% of WT molecules never sample the active H state but instead dwell in the M and L states (Supplementary Fig. 1) at least partially explains why a significant fraction of ribozymes remain inactive in ensemble cleavage assays (Fig. 1c), a common observation for the VS ribozyme independent of fluorophore labeling,42,43 as well as for ribozymes in general.27,29
Alternative connectivity between the ribozyme and cleavage site reveals a stably docked downstream cleavage site
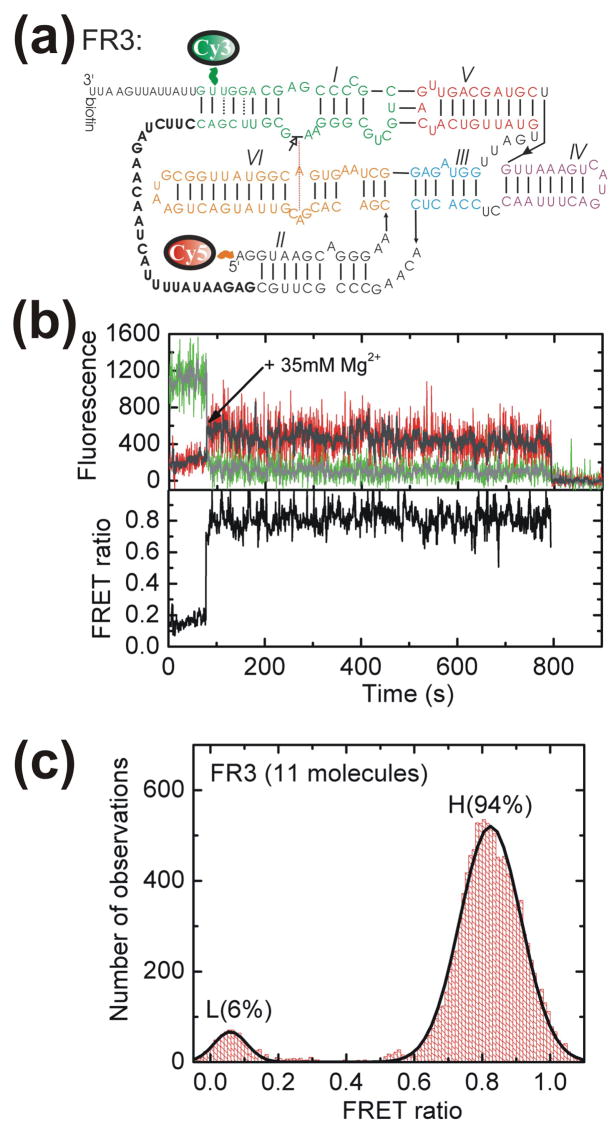
Because natural VS RNA is generated as a head-to-tail multimer, a given ribozyme core is flanked by an upstream cleavage site (as in our G11-based WT ribozyme, see Fig. 1) and another cleavage site several hundred nucleotides downstream.44 Bulk solution kinetic analysis has previously shown that the downstream cleavage site is cleaved much faster than the upstream site.33,45 The correlation between slow cleavage and unstable docking of the upstream site in G11-derived ribozymes raises the possibility that fast cleavage of the downstream site may at least in part be due to the greater stability of its docking. To test this hypothesis we used single-molecule FRET to examine the folding of a ribozyme derived from the fast-cleaving VS ribozyme RS1933, designated FR3, that contains a tether placing the substrate 26 nucleotides downstream of the ribozyme core (Fig. 5a). Addition of 35 mM Mg2+ (the same concentration as in our G11-based experiments) induces a conformational change in the FR3 RNA from a low to a high FRET state that, as the (lower bound) ~11 minute H state dwell in Fig. 5b shows, is very stable and non-dynamic. Such stable folding is consistently observed for FR3 molecules, leading to a strong (94%) bias towards the high FRET, presumably docked, state with only short excursions to low FRET states as illustrated by the aggregate FRET histogram in Fig. 5c. This FR3 behavior starkly contrasts with our observations on WT, where frequent changes among FRET states are observed (Fig. 2).
Fig. 5.
Docking of the alternatively connected, catalytically active FR3 ribozyme. (a) Secondary structure of the FR3 ribozyme showing its alternative connectivity (bold) to the downstream stem-loop I substrate. Fluorophore labeling sites were chosen similar to those of WT in Fig. 2. (b) Example trace of a FR3 molecule. The raw Cy3 (donor) and Cy5 (acceptor) intensities are shown along with the non-linear averaged data (Haran, 2004) in gray. The FRET ratio calculated as IA/(IA + ID) for the averaged data is shown in black. (c) An aggregate FRET ratio histogram for data points after addition of 35 mM Mg2+ is well described by a gaussian distribution of two states.
Discussion
The structural dynamics of tertiary structured RNA and their links to biological function are still ill-understood, despite recent advances in our appreciation of the breadth of non-coding RNA function.1–3 Here we have exploited single molecule FRET to dissect the global structural dynamics of the VS ribozyme. This complex non-coding RNA contains combinations of tertiary structure elements commonly found in large RNAs, such as three-way junctions and a kissing loop, and is endowed with catalytic activity that reports on the biologically relevant fold.
Our findings support the notion that complex RNAs often fold into their active states by traversing discrete structural intermediates, similar to recent observations on other complex ribozymes such as group I23,25 and group II introns.46 Each newly formed intermediate acquires additional tertiary interactions whose formation is contingent upon the proper folding of the previous state. In the case of the VS ribozyme, the correctly folded II-III-VI junction serves as the structural scaffold upon which the kissing loop interaction builds. In turn, the kissing loop serves as a scaffold to juxtapose the internal loops of helices I and VI for cleavage of the former. We also observe evidence for static (never-changing) and possibly dynamic (time-dependent) heterogeneity between individual molecules of the VS ribozyme, suggestive of a deeply furrowed folding free energy landscape with independent pathways separated by barriers too high for crossing by thermal motion, consistent with observations for other ribozymes such as group I introns23,25,26 and the hairpin ribozyme.27–31
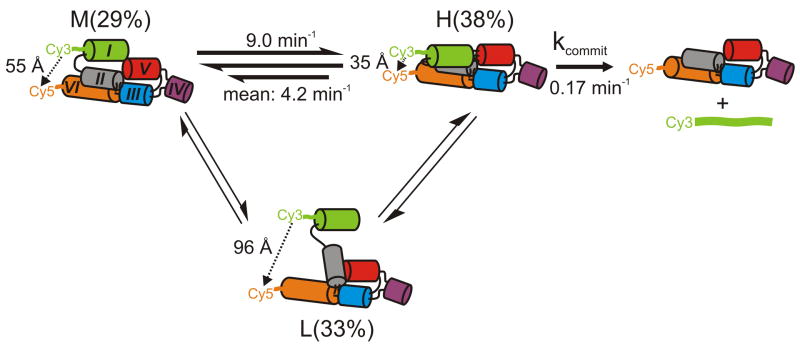
The minimal mechanistic model in Fig. 6 summarizes the structural dynamics of the full-length G11 VS ribozyme as directly observed and quantified here. We observe for WT altogether three states of distinct FRET values and thus fluorophore distances that are likely to lie on the folding pathway towards catalysis: L, M, and H states, which are about equally populated in the time and molecule average, although transitions into the long-lived L state are relatively rare (thin arrows in Fig. 6). Our data support a model wherein population of the H state, attainable from both the L and M states, is necessary but not sufficient for catalysis. This on-pathway H state requires a properly folded II-III-VI junction, whose disruption in ΔII-III-VI greatly reduces both the probability of kissing loop formation (compare Figs. 2b and 3c) and catalytic activity (Fig 1c). The correctly folded II-III-VI junction thus serves as a structural scaffold for the formation the kissing loop interaction which we observe as the H state.
Fig. 6.
Structural and kinetic model of the reaction pathway of the WT VS ribozyme. The helix colors match those in Fig. 1b. The reported H to M transition rate constant is the mean value obtained from the HMM analysis; the dual arrows represent heterogeneity (either static, i.e., never-changing or possibly dynamic, i.e., time-dependent in nature) in this transition as illustrated in Fig. 2e. The fluorophore distances and relative abundances of the H, M and L states were obtained from single molecule FRET, while kcommit was modeled. L states, while often long-lived, are only infrequently accessed by a sub-population of molecules (static heterogeneity, thinner arrows).
Our mechanistic model allows us to further evaluate the catalytic relevance of the H state. All observed transition rate constants between the three FRET states are at least an order of magnitude faster than the cleavage rate constant of 0.065 min−1 under the same conditions; the M→H and mean H→M conformational transitions are 138- and 65-fold faster, respectively (Fig. 6). This rate difference indicates that the downstream pathway towards phosphodiester cleavage and product release minimally perturbs the equilibrium distribution between the L, M, and H states. We therefore define kcommit as the rate constant that describes the probability of cleavage and product dissociation from the H state (5′-product binds only weakly and is thus assumed to be irreversibly released under our conditions (J. Olive and R.A.C, unpublished)). Straightforward analytical modeling of the reaction scheme in Fig. 6 predicts that the rate constant observed in our cleavage assays is simply the product of kcommit and the fraction of time the RNA spends in the H state as reported in our FRET histograms. This simplest possible model assumes that the 2′-O-methyl modification does not disrupt the ground state interaction formed by the natural 2′-OH. kcommit is then calculated to be (0.065 min−1)/(0.38) = 0.17 min−1 for WT, (0.002 min−1)/(0.04) = 0.05 min−1 for ΔII-III-VI, and (0.05 min−1)/(0.36) = 0.14 min−1 for Rescue I-V. The reasonable agreement among the three kcommit values as calculated from our experimental results lends further support to the minimal reaction scheme in Fig. 6. It also suggests that WT and mutants must traverse similar local energy barriers for catalysis to occur after formation of the catalytically competent H FRET state.
The slow rate constant kcommit associated with cleavage of the G11 VS ribozyme (Fig. 6) indicates that there is a significant additional barrier(s) that must be overcome before catalysis can occur. What is the nature of this barrier: conformational or chemical? In our G11 based ribozyme substrate stem-loop I is connected via its 3′ end to the 5′ end of helix II. However, this is only one of the relative arrangements of stem-loop I with respect to the ribozyme core found in nature: Because the VS RNA replication intermediate is naturally a head-to-tail multimer, another copy of stem-loop I is located downstream of the ribozyme core. Indeed, in vitro assays of multimeric VS RNA show that the downstream site is cleaved preferentially.45 A family of alternative ribozymes (RS19 and its derivatives such as FR3) connects the 5′ end of stem-loop I to the 3′ terminus of helix II through a long (>25-nucleotide) linker, mimicking the natural topology of multimeric VS RNA, and results in an observed cleavage rate several hundred-fold faster than that of G11.33 This finding suggests that reaction chemistry can be very rapid in certain VS ribozyme configurations so that the well-studied G11 ribozyme may be rate-limited by a local conformational change that FRET between the termini of helices I and VI cannot detect. We speculate that the alternative connectivity of the family of RS19 constructs affords the substrate stem-loop I additional conformational flexibility that optimizes the catalytic core conformation concomitantly with kissing loop formation, leading to rapid cleavage.
The discovery of VS variants with greatly enhanced cleavage rates compared to G11 led to the hypothesis that the downstream substrate is preferentially cleaved,33 which was confirmed when multimeric VS RNA was shown to preferentially cleave downstream.45 These findings, however, shed little light on how the 881-nucleotide VS RNA selects against the upstream site. Our single molecule characterization of the G11 based WT in comparison with the RS19-based FR3 offers new insight into this question. The readily reversible formation of the I-V kissing-loop formation in WT (Fig. 2) stands in sharp contrast to the much more stably formed high FRET conformer observed for FR3 (Fig. 5). A difference in docking stability alone, however, is not sufficient to account for the previously reported several hundred-fold difference in the cleavage rate constant, as evident from observation of a slow, rate-limiting kcommit.33 We hypothesize that these fast ribozymes also alleviate our proposed local conformational barrier, perhaps via their alternate connectivity and/or increased linker length.
Inside the mitochondria of the fungus Neurospora, one expects selective pressure on the replication cycle of VS RNA to minimize the population of vulnerable linear monomer. By contrast, the circular monomeric VS RNA is exonuclease resistant while still serving as a suitable substrate for the accompanying reverse transcriptase.9 Multimeric VS RNA may have adapted to such selective pressures by fine-tuning its ribozyme motif to minimize the lifetime of the exonuclease vulnerable 2′,3′-cyclic phosphate and 5′-OH termini required for the ligation into the circular VS RNA. Such self-protection of the RNA requires: (i) slow cleavage of the proximal stem-loop I substrate upstream of the ribozyme motif; (ii) dynamic docking and undocking of the proximal (upstream) stem-loop substrate with the catalytic core, as demonstrated here, to allow for dynamic exchange with the distal (downstream) stem-loop I until that is cleaved; and (iii) dissociation of the cleaved distal (downstream) stem-loop I product. This leaves a 5′-OH in the immediate vicinity of the corresponding proximal (upstream 2′,3′-cyclic phosphate) product that was generated by its distal VS ribozyme partner. Our work here demonstrates that the upstream substrate would be readily exchangeable for the more stably docking downstream substrate, as prerequisite for the above model.
Materials and methods
Preparation of topWT, topΔI-V, toprescue 1-V, and topΔII-III-VI
QuickChange site-directed mutagenesis (Stratagene) on a plasmid encoding G1134 was used to introduce a transcription initiation site at G622 (2 nucleotides downstream of the cleavage site) and a termination site at the capping loop of helix VI through insertion of an EcoRI site. Based on this plasmid, defined as the WT sequence, further site-directed mutagenesis was performed to generate plasmids topΔI-V, toprescue 1-V, and topΔII-III-VI and confirmed by DNA sequencing.
Transcription on EcoRI linearized plasmids was carried out with 20 mM GMP, 1 mM GTP, 4 mM ATP, UTP, CTP, 40 mM Tris-HCl, pH 8.0, 25 mM MgCl2, 2 mM Spermidine, 5 mM DTT, 0.01% TritonX-100, 1 unit inorganic pyrophosphatase, and 0.2 mg T7 RNA Polymerase in a 2-ml reaction at 37 °C for 6 h. This yielded ~95% 5′-monophosphate initiated transcripts. The 3′ ends of these transcripts were biotinylated after periodate oxidation as described.47
Each of the 5′-monophosphorylated transcripts was ligated to ligS1 as follows. 3.7 μM ligS1, 2.45 μM splintS1 and 1.85 μM transcript (ratios taken from ref. 48) in 200 μl of 50 mM Tris-HCl, pH 7.5, 10 mM DTT, 1 mM ATP was heated to 90 °C for 2 min, then incubated for 10 min at room temperature. MgCl2 was then added to 10 mM and recombinant T4 DNA ligase to 6 μM (final concentrations) and the reaction incubated at 30 °C for 6 h. The ligation reactions were desalted by gel filtration on NAP-10 columns (Pharmacia) and ethanol precipitated before purification on denaturing, 8 M urea, 10% (w/v) PAGE. Ligated RNA (yield typically ~40%) was detected based on its Cy3 fluorescence, cut out, eluted into 1 ml H2O, 1 mM EDTA overnight at 4°C, and ethanol precipitated. Typical total yields of ligated RNA were 10–40 pmol per reaction. Cleavable versions of topWT, topΔI-V, toprescue 1-V, and topΔII-III-VI were prepared in the same way except that a ligS1 RNA variant was used without the 2′-O-methyl at G620, and the 3′ ends of the transcripts were not biotinylated.
Preparation of synthetic oligonucleotides
RNA strands LigS1 (5′-Cy3-AAUUGCmGAA-3′) and b1 (5′-NH2-AAGUCAGUAUUGCAGCACAGCACAAGCC CGCUUGC-3′), where mG and NH2 represent a 2′-O-methylated G and a 5′-Amino Modifier C6 (Glen Research), respectively, were purchased from the HHMI Keck Foundation Biotechnology Resource Center at Yale University and deprotected as suggested by the manufacturer (http://info.med.yale.edu/wmkeck). The RNA was purified by denaturing, 8 M urea, 20% (w/v) polyacrylamide gel electrophoresis (PAGE) and C8-reverse-phase HPLC chromatography as described.49 Cy5 was attached postsynthetically to the 5′ end of b1 via the succinimidyl ester of Cy5, as described49 to generate strand Cy5b1. Oligodeoxynucleotide splintS1 (5′-CGGGGCGACGACG CCCTTCGCAATT-3′) was purchased from Invitrogen.
Preparation of FR3 RNA
FR3 is derived from the RS19 family of VS ribozymes,33 in which stem-loop I, which contains the site of cleavage, is attached via a linker of arbitrary sequence to the 3′ end of the ribozyme core (Fig. 5a). The sequence of stem-loop I was altered to introduce a 5-amino-allyl uridine at a position predicted from molecular modeling (not shown) to be on the exterior face of helix Ia for Cy3 labeling. Synthesis of FR3 was accomplished by VS ribozyme-mediated ligation using a strategy similar to that used previously to incorporate a site-specific 4-thio-uridine nucleotide into stem-loop I.21 The ribozyme portion was obtained by in vitro transcription by T7 RNA polymerase of a linearized plasmid template, followed by self-cleavage and gel-purification of the upstream cleavage product which ends at G620 and contains a 2′3′ cyclic phosphate terminus; the transcription mixture included 4 mM ApG dinucleotide in which the adenosine contained an amino group at the end of a six-carbon linker attached to the 5′ phosphate (Dharmacon, Inc) to allow for subsequent labeling of the 5′ end of the RNA with Cy5 mono-reactive dye (GE Healthcare). A second RNA beginning at position 621 (with a 5′ hydroxyl) was chemically-synthesized (Dharmacon, Inc.) and contains a 5-amino-allyl-uridine for subsequent labeling with Cy3 mono-reactive dye (GE Healthcare) at the position indicated in Fig. 5a and a 3′ terminal biotin. Incubation of these two Cy-labeled RNAs in the presence of 200 mM Mg2+ results in ligation to form the full-length FR3 RNA, which was gel-purified and ethanol-precipitated.
Single-Molecule FRET measurements
A 10-μl solution of 100 nM top strand and 200 nM Cy5b1 in 40 mM Tris-HCl, pH 8.0, 50 mM KCl was heated to 70 °C for 2 min and cooled at room temperature for 10 min to anneal the VS ribozyme strands. The annealed ribozyme was diluted to 200 pM in 1× standard buffer50 (40 mM Tris-HCl, pH 8.0, 50 mM KCl and 35 mM MgCl2) and flowed into a microfluidic channel on a quartz slide, coated first with biotinylated BSA, then streptavidin as described,27,51 to generate a surface density of ~0.1 molecules per μm2 for optical resolution. Similar surface tethering has been extensively used and characterized as non-perturbing for the hairpin ribozyme.27–29,52 After 2 min, the slide was washed with several volumes of 1× buffer. In parallel, 200 μl oxygen scavenger solution (OSS) was prepared that contained ~750 μg/ml glucose oxidase, 90 μg/ml catalase, 10% (w/v) glucose, and 1% (v/v) β-mercaptoethanol in 1× standard buffer. This solution was then flowed into the microfluidic channel or stored under nitrogen for later use. The donor and acceptor fluorescence signals of optically resolved single molecules were simultaneously monitored in real-time by prism-based total internal reflection fluorescence video microscopy as described,27–29. There was little evidence of β-mercaptoethanol induced blinking of the Cy5 under our illumination conditions of 9 W/cm2; we observed few excursions to a low-FRET state, which may suggest acceptor blinking, and none of the traces were analyzed after they had reached zero signal, as would indicate donor blinking. Our illumination conditions lie well below the excitation intensities used in experiments that characterized β-mercaptoethanol induced blinking of Cy5.53 The optimal I-CCD frame rate was found to be two frames per second (fps), and all time traces were taken at 2 fps resolution unless otherwise stated.
Single Molecule Data Analysis
The aggregate FRET histograms were over all reported molecules contain background corrected FRET ratios only for molecules where both Cy3 and Cy5 were confirmed to be fluorescent. This criterion introduces a possible bias into the calculation of 4% H state for the ΔII-III-VI ribozyme, as the FRET ratio of the L state is indistinguishable from molecules with photobleached Cy5 (data not shown), making the terminal state ambiguous for traces that end in low FRET. In addition, observed dwell times in terminal states are biased towards smaller values due to premature Cy3 photobleaching and our limited observation window (see also below). The average time ΔII-III-VI molecules spend in their longer lived M and L states as based on confirmed dwell times is ~30 s each. To correct for the above effects, ambiguous terminal states were assigned to be of this length. The H state was still found to be 4% abundant, suggesting that our histogram analysis is robust against possible bias.
For dwell time analysis, noise in time traces was reduced using a previously described non-linear filter with m = 3 and p = 20 as parameters.38 Hidden Markov modeling (HMM) with two to five FRET states was then used to determine the most likely traversed path through each well defined trace as described,39 with each result undergoing visual inspection to ensure the best possible fit; a two□ or three-state description sufficed for all traces. The HMM algorithm assigns a single-exponentially distributed probability to each transition. All individual probabilities for each type of transition (H→M, M→H etc.) were averaged to yield the rate constants reported with their respective significant figures in Figures 2d and 4d. The raw dwell times that arose from the HMM analysis were also used to calculate normalized probability densities as described,40 and fit using Marquardt-Levenberg nonlinear least-squares regression in Origin 7.5. These density plots present an independent test for the single-exponential distribution of dwell times that is assumed in the HMM analysis. All rate constants reported here from single molecule FRET analysis were corrected for the bias towards smaller values due to premature Cy3 photobleaching and our limited observation window by subtracting 0.2 min−1 as described.29
Distance estimates
To obtain fluorophore distance estimates R for the various FRET states the apparent FRET efficiency was calculated from , 36 where Φ and η signify the fluorophore quantum yields and detector channel efficiencies, respectively, and the donor and acceptor intensities ICy3 and ICy5, respectively, were corrected for leakage of 20% of donor photons from the donor into the acceptor channel. R was then calculated from Eapp = c [1+ (R/R0)6]−1, where c = 0.69 and R0 = 54 Å were used as described.37
Ensemble cleavage assays
Cleavable versions of topWT, topΔI-V, toprescue I-V, and topΔII-III-VI were 3′ end labeled with [32P]-pCp using T4 RNA ligase. Trace amounts of 3′ end labeled top strand were combined with 500 nM Cy5b1 in 40 μl of 40 mM Tris-HCl, pH 8.0, 50 mM KCl, heated to 70 °C for 2 min, equilibrated in a 25 °C bath for 10 min, and a 4-μl aliquot taken for a zero time point. The reaction was initiated by adding 4 μl of the appropriate stock solution of MgCl2. At defined times 4-μl aliquots were taken and quenched by the addition of 10 μl 80% (v/v) formamide, 0.025% (w/v) xylene cyanol, 0.025% (w/v) bromophenol blue and 50 mM EDTA. The cleaved and uncleaved products were separated by denaturing, 8 M urea, 10% (w/v) PAGE. The gel was exposed to a PhosphorImager screen overnight and quantified using ImageQuant software (Molecular Dynamics). The fraction cleaved versus time was fit with the single exponential first-order rate equation y(t) = y0 + A (1−e(−kobst)) to determine the observed cleavage rate (kobs) and extent of cleavage (A). The Mg2+-dependence of the cleavage rate constant was fit with a hyperbolic, non-cooperative binding equation of the form:
where kmax is the cleavage rate under saturating Mg2+ conditions and Mg1/2 is the magnesium half-titration point.
Supplementary Material
Acknowledgments
The authors wish to thank Taekjip Ha for HMM analysis software, Liz McDowell for software that converts HMM data into transition density plots, and John Hsieh for a batch of recombinant T4 DNA ligase. This work was supported by grant 43875-AC4 from the American Chemical Society Petroleum Research Fund to N.G.W. and a grant from the Canadian Institutes for Health Research to R.A.C..
Abbreviations
- FRET
fluorescence resonance energy transfer
- WT
wild-type
References
- 1.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–1253. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- 4.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nature Rev Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 5.Doudna JA, Lorsch JR. Ribozyme catalysis: not different, just worse. Nat Struct Mol Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- 6.Ditzler MA, Aleman EA, Rueda D, Walter NG. Focus on function: Single molecule RNA enzymology. Biopolymers. 2007;87:302–316. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- 7.Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 8.Collins RA, Saville BJ. Independent transfer of mitochondrial chromosomes and plasmids during unstable vegetative fusion in Neurospora. Nature. 1990;345:177–179. doi: 10.1038/345177a0. [DOI] [PubMed] [Google Scholar]
- 9.Kennell JC, Saville BJ, Mohr S, Kuiper MT, Sabourin JR, Collins RA, Lambowitz AM. The VS catalytic RNA replicates by reverse transcription as a satellite of a retroplasmid. Genes Dev. 1995;9:294–303. doi: 10.1101/gad.9.3.294. [DOI] [PubMed] [Google Scholar]
- 10.Collins RA. The Neurospora Varkud satellite ribozyme. Biochem Soc Trans. 2002;30:1122–1126. doi: 10.1042/bst0301122. [DOI] [PubMed] [Google Scholar]
- 11.Lilley DM. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michiels PJ, Schouten CH, Hilbers CW, Heus HA. Structure of the ribozyme substrate hairpin of Neurospora VS RNA: a close look at the cleavage site. RNA. 2000;6:1821–1832. doi: 10.1017/s1355838200001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann B, Mitchell GT, Gendron P, Major F, Andersen AA, Collins RA, Legault P. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc Natl Acad Sci USA. 2003;100:7003–7008. doi: 10.1073/pnas.0832440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flinders J, Dieckmann T. The solution structure of the VS ribozyme active site loop reveals a dynamic “hot-spot”. J Mol Biol. 2004;341:935–949. doi: 10.1016/j.jmb.2004.06.084. [DOI] [PubMed] [Google Scholar]
- 15.Campbell DO, Legault P. Nuclear magnetic resonance structure of the Varkud satellite ribozyme stem-loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry. 2005;44:4157–4170. doi: 10.1021/bi047963l. [DOI] [PubMed] [Google Scholar]
- 16.Campbell DO, Bouchard P, Desjardins G, Legault P. NMR structure of varkud satellite ribozyme stem-loop V in the presence of magnesium ions and localization of metal-binding sites. Biochemistry. 2006;45:10591–10605. doi: 10.1021/bi0607150. [DOI] [PubMed] [Google Scholar]
- 17.Beattie TL, Olive JE, Collins RA. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc Natl Acad Sci USA. 1995;92:4686–4690. doi: 10.1073/pnas.92.10.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiley SL, Collins RA. Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J. 2001;20:5461–5469. doi: 10.1093/emboj/20.19.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafontaine DA, Norman DG, Lilley DM. Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J. 2001;20:1415–1424. doi: 10.1093/emboj/20.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafontaine DA, Norman DG, Lilley DM. The global structure of the VS ribozyme. EMBO J. 2002;21:2461–2471. doi: 10.1093/emboj/21.10.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiley SL, Sood VD, Fan J, Collins RA. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J. 2002;21:4691–4698. doi: 10.1093/emboj/cdf462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter NG, Hampel KJ, Brown KM, Burke JM. Tertiary structure formation in the hairpin ribozyme monitored by fluorescence resonance energy transfer. EMBO J. 1998;17:2378–2391. doi: 10.1093/emboj/17.8.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhuang XW, Bartley LE, Babcock HP, Russell R, Ha TJ, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 24.Pereira MJ, Harris DA, Rueda D, Walter NG. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: a conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 25.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J Mol Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 28.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang XW, Walter NG. Single-molecule enzymology of RNA: Essential functional groups impact catalysis from a distance. Proc Natl Acad Sci USA. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DM, Ha T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci USA. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nahas MK, Wilson TJ, Hohng S, Jarvie K, Lilley DM, Ha T. Observation of internal cleavage and ligation reactions of a ribozyme. Nat Struct Mol Biol. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- 32.Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Methods Enzymol. 2003;361:1–33. doi: 10.1016/s0076-6879(03)61003-7. [DOI] [PubMed] [Google Scholar]
- 33.Zamel R, Poon A, Jaikaran D, Andersen A, Olive J, De Abreu D, Collins RA. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc Natl Acad Sci USA. 2004;101:1467–1472. doi: 10.1073/pnas.0305753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo HC, De Abreu DM, Tillier ER, Saville BJ, Olive JE, Collins RA. Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J Mol Biol. 1993;232:351–361. doi: 10.1006/jmbi.1993.1395. [DOI] [PubMed] [Google Scholar]
- 35.Lafontaine DA, Wilson TJ, Zhao ZY, Lilley DM. Functional group requirements in the probable active site of the VS ribozyme. J Mol Biol. 2002;323:23–34. doi: 10.1016/s0022-2836(02)00910-5. [DOI] [PubMed] [Google Scholar]
- 36.Cosa G, Harbron EJ, Zeng Y, Liu HW, O’Connor DB, Eta-Hosokawa C, Musier-Forsyth K, Barbara PF. Secondary structure and secondary structure dynamics of DNA hairpins complexed with HIV-1 NC protein. Biophys J. 2004;87:2759–2767. doi: 10.1529/biophysj.104.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabanayagam CR, Eid JS, Meller A. Using fluorescence resonance energy transfer to measure distances along individual DNA molecules: corrections due to nonideal transfer. J Chem Phys. 2005;122:061103. doi: 10.1063/1.1854120. [DOI] [PubMed] [Google Scholar]
- 38.Haran G. Noise reduction in single-molecule fluorescence trajectories of folding proteins. Chem Phys. 2004;307:137–145. [Google Scholar]
- 39.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc Natl Acad Sci USA. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rastogi T, Beattie TL, Olive JE, Collins RA. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 1996;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson TJ, McLeod AC, Lilley DM. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins RA, Olive JE. Reaction conditions and kinetics of self-cleavage of a ribozyme derived from Neurospora VS RNA. Biochemistry. 1993;32:2795–2799. doi: 10.1021/bi00062a009. [DOI] [PubMed] [Google Scholar]
- 44.Saville BJ, Collins RA. RNA-mediated ligation of self-cleavage products of a Neurospora mitochondrial plasmid transcript. Proc Natl Acad Sci USA. 1991;88:8826–8830. doi: 10.1073/pnas.88.19.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon AH, Olive JE, McLaren M, Collins RA. Identification of separate structural features that affect rate and cation concentration dependence of self-cleavage by the Neurospora VS ribozyme. Biochemistry. 2006;45:13394–13400. doi: 10.1021/bi060769+. [DOI] [PubMed] [Google Scholar]
- 46.Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biochem Sci. 2007;32:138–145. doi: 10.1016/j.tibs.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Lambert MN, Vocker E, Blumberg S, Redemann S, Gajraj A, Meiners JC, Walter NG. Mg2+-induced compaction of single RNA molecules monitored by tethered particle microscopy. Biophys J. 2006;90:3672–3685. doi: 10.1529/biophysj.105.067793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ke A, Doudna JA. Crystallization of RNA and RNA-protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 49.Walter NG. Probing RNA structural dynamics and function by fluorescence resonance energy transfer (FRET) Curr Protocols Nucleic Acid Chem. 2002;11.10:11.10.11–11.10.23. doi: 10.1002/0471142700.nc1110s11. [DOI] [PubMed] [Google Scholar]
- 50.Andersen AA, Collins RA. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc Natl Acad Sci USA. 2001;98:7730–7735. doi: 10.1073/pnas.141039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 52.Liu S, Bokinsky G, Walter NG, Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting”. Proc Natl Acad Sci USA. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasnik I, McKinney SA, Ha T. Nonblinking and longlasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 54.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.