Abstract
Scrub typhus (‘Tsutsugamushi’ disease in Japanese) is a mite-borne infectious disease. The causative agent is Orientia tsutsugamushi, an obligate intracellular bacterium belonging to the family Rickettsiaceae of the subdivision alpha-Proteobacteria. In this study, we determined the complete genome sequence of O. tsutsugamushi strain Ikeda, which comprises a single chromosome of 2 008 987 bp and contains 1967 protein coding sequences (CDSs). The chromosome is much larger than those of other members of Rickettsiaceae, and 46.7% of the sequence was occupied by repetitive sequences derived from an integrative and conjugative element, 10 types of transposable elements, and seven types of short repeats of unknown origins. The massive amplification and degradation of these elements have generated a huge number of repeated genes (1196 CDSs, categorized into 85 families), many of which are pseudogenes (766 CDSs), and also induced intensive genome shuffling. By comparing the gene content with those of other family members of Rickettsiacea, we identified the core gene set of the family Rickettsiaceae and found that, while much more extensive gene loss has taken place among the housekeeping genes of Orientia than those of Rickettsia, O. tsutsugamushi has acquired a large number of foreign genes. The O. tsutsugamushi genome sequence is thus a prominent example of the high plasticity of bacterial genomes, and provides the genetic basis for a better understanding of the biology of O. tsutsugamushi and the pathogenesis of ‘Tsutsugamushi’ disease.
Key words: Orientia tsutsugamushi, genome sequencing, obligate intracellular bacterium, repetitive sequence, IS element, integrative and conjugative element, gene amplification, genome reduction
1. Introduction
Orientia tsutsugamushi, a causative agent of scrub typhus or ‘Tsutsugamushi’ disease, is a Gram-negative bacterium belonging to the order Rickettsiales from the alpha-subdivision of Proteobacteria. The bacterium had been classified into the genus Rickettsia, but has recently been transferred to the genus Orientia, a genus newly created in the family Rickettsiaceae, based on the differences in the 16S ribosomal RNA (rRNA) sequence (Supplementary Fig. S1) and in several morphological and biochemical features.1
Like other members of the order Rickettsiales, O. tsutsugamushi is an obligate intracellular parasite. In nature, the bacterium lives in trombiculid mites, which are called ‘tsutsugamushi’ in Japanese. In infected mites, the bacterium is found in the host cell cytosol of various organs including the oocytes, and is efficiently passed on to the offspring via transovarial transmission.2–6 Trombiculid mites suck the tissue fluid of mammals only once in their life cycle at the larval stage, when the bacterium is transferred to the animal to induce scrub typhus. Reverse transfer, from infected animals to mites, occurs inefficiently,7,8 and the bacterium transmitted in such a way is rarely passed on to the offspring.7–9 Accordingly, limited numbers of mite lines retain O. tsutsugamushi. However, O. tsutsugamushi or mites carrying this bacterium are widely distributed in Asia and the Western Pacific islands; one billion people are estimated to be at risk for scrub typhus, and one million cases occur annually.10
In infected animals and humans, O. tsutsugamushi invades the macrophages and vascular endothelial cells, escapes from the phagosomes, and propagates in the cytosol like other Rickettsiaceae family members, inducing an acute febrile state. It often causes a deadly illness if not treated with appropriate antibiotics, but the molecular pathogenicity is still poorly understood and no vaccine is currently available.
Orientia tsutsugamushi strains are serologically classified into several subgroups based on the antigenic variations of a major surface protein called ‘56-kDa protein’.11 The results of our pulsed-field gel electrophoresis (PFGE) analysis indicate that the genome size of O. tsutsugamushi is much larger than those of other members of the order Rickettsiales, and that the genome exhibits a significant strain-to-strain size variation, ranging from 2.0 to 2.7 Mb (Tamura et al., unpublished data). However, no known techniques for genetic manipulation are applicable to this bacterium, and only a few pieces of genetic information have so far been accumulated; only the DNA sequences of the genes for the 56-kDa protein from various serotypes have been intensively analyzed.12,13 In order to better understand the genetic features of this bacterium, we determined the complete genome sequence of O. tsutsugamushi strain Ikeda. Here, we present the genomic features of the microorganism revealed by genome sequence analysis of the strain Ikeda and genomic comparison with other sequenced members of the family Rickettsiacea. Some of our results obtained by genomic comparison of the strain Ikeda and other recently sequenced O. tsutsugamushi strain Boryong14 are also presented.
2. Materials and methods
2.1. Bacterial strain
Orientia tsutsugamushi strain Ikeda was originally isolated from a patient in 1979 in Niigata Prefecture, Japan. The strain belongs to the Japanese Gilliam serotype and is highly virulent in mice.15 The mite vector of the strain was not identified, but Japanese Gilliam serotype strains are usually carried by Leptotrombidium pallidum or Leptotrombidium scutellare.12,15–18 The strain has been kept in the laboratory by repeated propagation on L929 cells and passages in mice since the initial isolation, which may have generated multiple sub-strains containing some genomic alterations. To avoid the usage of heterogeneous genomic DNA derived from such sub-strains for genome sequencing, we performed single plaque purification on L929 cells as previously described.19 The cloned strain was propagated in L929 cells, and bacterial cells were purified on a percoll gradient as described previously.20
2.2. DNA preparation and sequencing
Bacterial DNA was obtained from the purified cells by standard procedures.21 A single batch of DNA preparation was used for all the experiments described hereafter. The genomic DNA was randomly sheared by means of a Hydroshear (GeneMachines) and used for the construction of genomic libraries. We prepared two pUC18-based random genomic libraries with insert sizes of 1–1.5 or 4–5 kb. A BAC library with an insert size of 11–15 kb was also constructed using the pIndigoBAC-5 vector (Epicentre). Sequencing was carried out using the BigDye v3.1 chemistry on ABI370 or ABI3730 sequencers (ABI), or the ET chemistry on MegaBACE4500 sequencers (GE Healthcare).
Because the genome sequence of O. tsutsugamushi was extremely rich in repetitive sequences, we employed a sequencing strategy comprising the following four steps. We first produced 32 000 shotgun reads from the 1–1.5 kb library and 14 000 reads from the 4–5 kb library using the forward sequencing primer. The Phred/Phrap software package22 was used for base-calling, quality assessment, and sequence assembly. The assemblies were visualized in order to count the base variations and detect misassembly using the Consed software.23 Based on the assembled data from 46 000 shotgun reads, we selected 6700 informative clones from both libraries, which were predicted to be located at the contig ends. The reverse end-sequences of these clones were determined using the reverse primer, and were assembled with the 46 000 forward sequences. The redundancy of the random shotgun sequences we obtained up to this stage was ∼13. At the third stage, the end sequences of 2383 BAC clones were determined, and 142 clones which were predicted to bridge the contigs were selected. These clones were individually sequenced and used as scaffolds in the sequence assembly. After re-assembly, 26 additional BAC clones were selected, sequenced individually, and added to the assembly. This assembly yielded 15 major contigs ranging in length from 20.4 to 296.0 kb. Finally, in order to close the 15 gaps, genomic PCR was carried out for five gaps using an LA long PCR kit (Takara), and 14 BAC clones were retrieved to close the remaining 10 gaps. The sequences of the five PCR products (3.7–25.4 kb) and the 14 BAC clones were individually determined. By adding these sequences to the assembly, we obtained the complete genome sequence of O. tsutsugamushi strain Ikeda.
2. 3. Validation of the sequence assembly
To validate the sequence assembly, we digested the chromosomal DNA of strain Ikeda with SmaI or EagI, or double-digested it with these two enzymes, and analyzed the digested DNA by PFGE as described previously.24 The digestion patterns we obtained were in good agreement with those deduced from the assembled sequence (Supplementary Fig. S2). In addition, we designed 18 pairs of PCR primers that amplified the 18-genomic segments that covered a 69-kb region of the chromosome (nucleotide positions: 611 095–670 870 bp) and performed a PCR scanning analysis.25 In this analysis, all primer pairs yielded PCR products of expected sizes (data not shown).
2. 4. Gene identification, sequence annotation, and data analyses
The identification of potential protein coding sequences (CDSs) and their annotation was done by using GenomeGambler, version 1.5.26 We first identified CDSs longer than 50 amino acids. Then, by searching all the intergenic regions for CDSs showing sequence similarities to known proteins with the BLASTP program,27 we identified 69 CDSs shorter than 50 amino acids. Orthologous genes were identified using the MBGD (Microbial Genome Database for Comparative Analysis) database (http://mbgd.genome.ad.jp/).28
Repetitive sequences were identified through sliding window analysis (window size, 100 bp; step size, 3 bp). All 100-bp sequence windows were searched against the entire genome sequence using the BLASTN program.27 The ssearch3 program29 was also used to define the borders of each repetitive sequence. The determination of the consensus sequences of each repetitive sequence and their classification were carried out by manual inspection based on the multiple alignments generated by the ClustalW program.30 Repeated gene families, which were defined in this study as the genes whose gene products exhibited at least 90% amino acid sequence identity over 60% of the alignment length, were identified by all-to-all BLASTP analysis of the O. tsutsugamushi gene products. The KEGG (Kyoto Encyclopedia of Genes and Genomes) database (http://www.genome.jp/kegg/)31 was used to analyze the metabolic pathways of O. tsutsugamushi. The annotated genome sequence of O. tsutsugamushi strain Ikeda has been deposited at the DDBJ/EMBL/GenBank databanks under accession number AP008981.
3. Results and discussion
3. 1. General features of the genome
Orientia tsutsugamushi has a circular chromosome consisting of 2 008 987 bp (Table 1 and Fig. 1). No plasmids or prophages were detected. A putative origin of replication (ori) was assigned through cumulative GC skew analysis, because no clear skew in the leading and lagging strands was observed through conventional GC skew analysis (Supplementary Fig. S3).32 The overall features of the O. tsutsugamushi genome are summarized in Table 1, and a comparison with selected members of the order Rickettsiales is given in Supplementary Table S1.
Table 1.
General features of the genome of O. tsutsugamushi strain IKEDA
| Chromosome (bp) | 2 008 987 |
| G + C content (%) | 30.5 |
| tRNA genes | 34 |
| rRNA genesa | 3 |
| Other RNA genes | 1 |
| CDSsb | 1967 (803) |
| Function assigned | 1445 (651) |
| Hypothetical | 522 (152) |
| Repeated genes | 1196 (766) |
| Function assigned | 895 (619) |
| Hypothetical | 301 (147) |
| Singleton genes | 771 (37) |
| Function assigned | 550 (32) |
| Hypothetical | 221 (5) |
| Repetitive elements | 969 |
| 1. OtAGEs | 185 |
| 2. Transposable elements | 619 |
| ISOt1 | 113 |
| mISOt1 | 61 |
| ISOt2 | 17 |
| mISOt2a | 30 |
| mISOt2b | 65 |
| ISOt3 | 33 |
| ISOt4 | 124 |
| mISOt4 | 55 |
| ISOt5 | 70 |
| GIIOt1 | 51 |
| 3. Short repeats | 165 |
| SROt1 | 77 |
| SROt2 | 50 |
| SROt3 | 23 |
| SROt4 | 6 |
| SROt5 | 5 |
| SROt6 | 2 |
| SROt7 | 2 |
aO. tsutsugamushi contains one set of rRNA genes (5S, 16S, and 23S), but the 16S rRNA gene is located separately from the others.
bThe numbers of pseudogenes are indicated in parentheses.
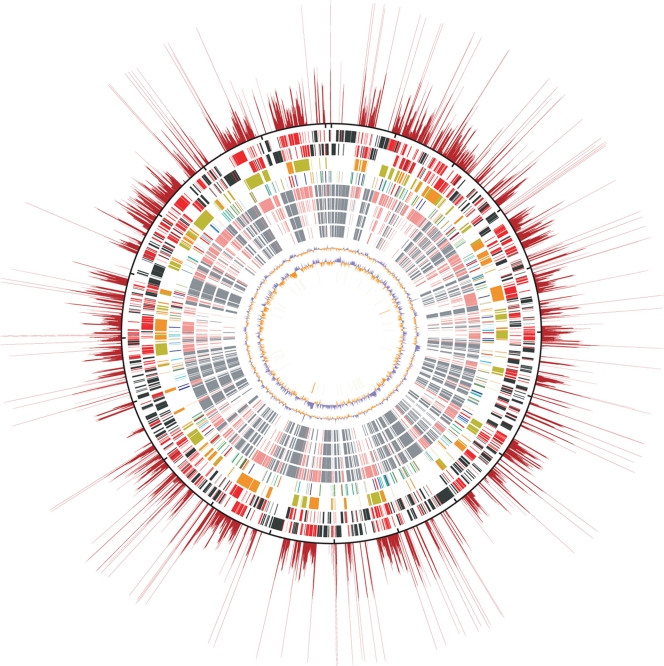
Figure 1.
Structural features of the O. tsutsugamushi genome. The putative origin of replication is located at the top. The outermost red rays show the repetition rates of each chromosomal region. The repetition rates were determined by sliding window analysis (window size, 100 bp; step size, 3 bp). The total number of 100-bp windows retrieved from the entire genome sequence was 669 663. All the 100-bp window sequences were searched for homologous regions using the BLASTN program. The repetition rate was defined as the total number of BLASTN hits (threshold: >120 BLASTN similarity score, excluding hits to their own sequences). The longest ray corresponds to the 108-folds repetition. The outermost black circle is a scale where the short dashes mark 100-kb intervals. The circles inside the scale show, from the outside in: (i) genes on the forward strand (singleton genes in black and repeated genes in red), (ii) genes on the reverse strand (singletons in black and repeated genes in red), (iii) OtAGE sequences on the forward (orange) and reverse (greenish yellow) strands, (iv) transposable elements (ISOt1 and mISOt1, green; ISOt2 and mISOt2, light blue; ISOt3, blue; ISOt4 and mISOt4, orange; ISOt5, dark blue; GIIOt1, red), (v) singleton genes (gray) and repeated genes (light red) whose homologues are present in R. bellii, (vi) R. felis, (vii) R. conorii, (viii) R. prowazekii, (ix) G + C content [%: blue, higher than the average of the whole genome (30.5%); orange, lower than the average], (x) GC skew [(G–C)/(G + C): blue, ≥0; orange, <0], and (xi) tRNA (green) and rRNA (orange) genes.
Although the G + C content and the number of rRNA and tRNA genes do not largely differ from those of other Rickettsiales, the O. tsutsugamushi genome contains a much higher number of protein-coding genes (1967 CDSs) and has the largest genome size in the order Rickettsiales. Of the 1967 CDSs, as many as 1196 are repeated genes constituting 85 families (O. tsutsugamushi Repeated Gene families: OtRG1–OtRG85), where the members in each family exhibit more than 90% amino acid sequence identity (see Section 2 for the definition of repeated genes; all other genes that do not belong to any of the repeated gene families are referred to as ‘singleton genes’ in this study). This unusual gene duplication is associated with the explosive amplification of several repetitive sequences, as described below. Since massive gene decay has also taken place in these repeated genes, as much as 64% of the 1196 repeated CDSs (766) are pseudogenes (truncated, split, or degraded). In contrast, smaller numbers of pseudogenes were identified among the singleton genes (4.8%, or 37 out of the 771 CDSs). The proportion of pseudogenes among the singleton genes is, however, within a range similar to those of other Rickettsia genomes33–37 known to harbor relatively larger numbers of split genes, a signature of progressive genome reduction.
Another striking feature of the O. tsutsugamushi genome is that it exhibits very poor colinearity to any of the previously sequenced Rickettsia genomes including that of R. bellii, which exhibits exceptionally little colinearity to other Rickettsia genomes33 (Supplementary Fig. S4). This suggests that extensive genome shuffling has taken place in the evolutionary course of O. tsutsugamushi. The massively amplified repetitive sequences may have mediated this genome shuffling.
3.2. Explosive amplification of repetitive sequences
We identified 18 types of repetitive sequences in the O. tsutsugamushi genome. They have explosively been amplified and scattered throughout the genome (Fig. 1). Although massive decay has also taken place in these repetitive sequences, their total length now reaches 933.8 kb (46.5% of the entire genome). Among the bacteria so far sequenced, Wolbachia pipientis wMel and Anaplasma phagocytophilum, both of which are obligate intracellular bacteria belonging to the order Rickettsiales, contain the highest proportion of repetitive sequences (14.2 and 12.7%, respectively),38 but much lower than O. tsutsugamushi (Supplementary Fig. S5). Of interest is that the size of the O. tsutsugamushi genome divested of these repetitive sequences is almost the same as that of the R. prowazekii genome (1070 and 1116 kb, respectively).
The identified repetitive sequences were categorized into three types: (i) a genetic element which we named ‘O. tsutsugamushi amplified genetic element’ (OtAGE), (ii) transposable elements, and (iii) others (Table 1).
3.2.1. OtAGE
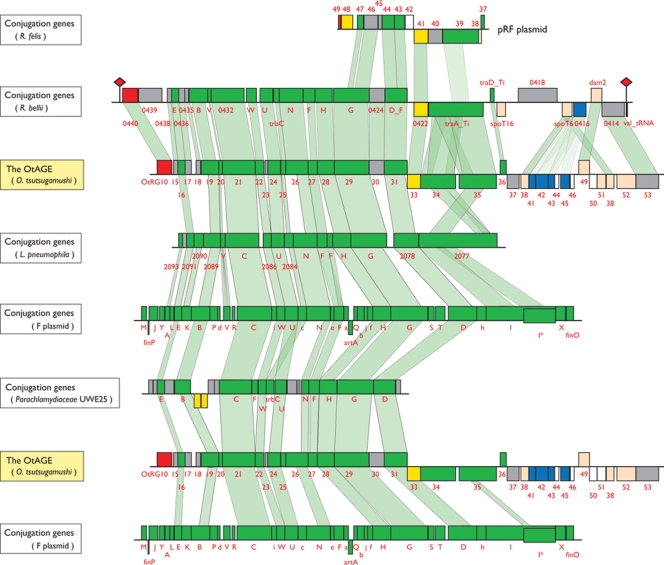
The OtAGE is a large genetic element about 33 kb in size and encodes 33–38 genes. This element has explosively been amplified in the O. tsutsugamushi genome, and each copy has extensively been degenerated by various types of deletion and insertion. However, we could reconstitute a probable intact form of the OtAGE by comparing the structures of relatively large copies (Fig. 2). Based on the reconstituted structure, we identified a total of 185 remnants of the OtAGE in the O. tsutsugamushi genome. The sum of their lengths is ∼694 kb (34.6% of the entire genome). We could not define their integration junctions because of the high sequence variation and accumulation of transposable elements in the possible junction regions.
Figure 2.
Gene organization of OtAGEs (O. tsutsugamushi amplified genetic elements). The gene organizations of the longest 30 OtAGEs identified in the O. tsutsugamushi genome are shown. The structure of the intact OtAGE that we deduced from the structures of these 30 OtAGEs is shown at the top. Arrows depicted by solid lines indicate intact genes and those depicted by dotted lines indicate degraded ones. Genes for conjugal transfer are depicted in green, integrase genes in red, genes for ATPase domain-containing proteins in blue, genes for other functions in light orange, genes for transposases in yellow, conserved hypothetical genes in white, O. tsutsugamushi-specific hypothetical genes whose gene products exhibited no significant sequence similarities to known protein sequences in gray, and genes that were probably trapped or inserted in OtAGEs in black. Four types of integrase genes and three types of gene cassettes encoding ATPase domain-containing proteins are present. Balloon marks indicate the insertion sites of IS elements (the ISOt number is labeled for each IS element). Deleted regions are indicated by broken dotted lines.
The OtAGE encodes an integrase gene (int) at the left end, which is followed by a set of genes for conjugative transfer (tra genes) similar to those in various conjugative plasmids represented by the F plasmid (Fig. 3). The OtAGE can thus be regarded as a member of the integrative and conjugative element (ICE) group, a recently recognized group of mobile genetic elements that disseminate by conjugative transfer like conjugative plasmids and integrate themselves into the genome like temperate bacteriophages.39 The tra gene product sequences are highly conserved between OtAGEs (>90% amino-acid sequence identity), but four types of int genes were clearly distinguished (OtRG10–OtRG14; OtRG12 and OtRG13 represent one split gene, Fig. 2). These integrases have significantly diverged in sequence (36–38% identity to each other), indicating that they have different origins.
Figure 3.
Comparison of the OtAGE with other integrative and conjugative elements (ICEs). The genetic organizations of the OtAGE and five other ICEs are drawn to scale. Only the regions required for conjugal transfer are shown, except for the OtAGE and an OtAGE-like ICE of R. bellii. The R. bellii element is integrated into a valine tRNA gene with 54-bp sequence duplication (indicated by red rhomboids). The R. felis plasmid contains only few genes related to conjugation. Genes for conjugal transfer are depicted by green boxes, those for integrase by red, those for transposase by yellow, those for ATPase domain-containing proteins by blue, conserved hypothetical genes by white, and O. tsutsugamushi-specific hypothetical genes by gray. The homologous regions are indicated by light green shading. Homologous regions were identified by the BLASTP program (e-value threshold: ≤1e−10). To identify the homologous regions in the traA, traE, and traV gene products, which showed higher sequence diversity between elements, we used the ClustalW program.30
In the right one-third of the OtAGE, genes for various functions reside (Fig. 2). These include two genes encoding almost identical SpoT (ppGpp hydrolase)-like proteins, a gene cassette encoding multiple ATPase domain-containing proteins, and genes for DNA methylase and DNA helicase. As for the gene cassette encoding ATPase domain-containing proteins, three types of cassettes, each encoding two to four ATPase domain-containing proteins, were identified. The encoded ATPase domain-containing proteins exhibited significant sequence diversities, not only between the three types of cassettes but also within the same cassette (up to 49% amino-acid sequence identity, but in most cases less than 40%), indicating a complex evolutionary history of the gene cassettes. In addition, several genes, whose gene products are also apparently not related to conjugative transfer, such as peroxiredoxin and ankyrin repeat (AR)-proteins, were found to be inserted into various sites in the OtAGE.
Among the tra gene clusters so far identified in various conjugative plasmids and ICEs, the gene cluster in the OtAGE is most similar in sequence and gene organization to that identified in the R. bellii genome33 (Fig. 3). The R. bellii tra gene cluster is also preceded by an int gene. Furthermore, the R. bellii int/tra gene cluster is located in a 38.7-kb element that is integrated into a valine tRNA gene with a terminal sequence duplication of 54 bp.40 The R. bellii element also contains a set of genes similar but not identical to those on the right part of the OtAGE, indicating that it is an ICE closely related to the OtAGE. Among the Rickettsia species so far sequenced, R. massiliae, which has very recently been sequenced,40 also contains an OtAGE-like element (data not shown) and R. felis contains a few tra genes or their remnants in a plasmid.34 We could not estimate the time point when the OtAGEs invaded O. tsutsugamushi. However, the presence of four distinct int genes and three different types of gene cassettes encoding ATPase domain-containing proteins among the OtAGEs may suggest that invasion of O. tsutsugamushi by similar but different types of OtAGEs took place at least four times.
A sex pilus-like structure was observed on the cell surface of R. bellii, suggesting that the R. bellii ICE may still be active in conjugation.33 In contrast, all the OtAGEs contained various types of deletions, and many of the remaining tra genes have become pseudogenes by base substitution and deletion, or insertion of transposable elements (Fig. 2). Thus, all the OtAGEs currently present in O. tsutsugamushi strain Ikeda appear to be no longer transmissible by themselves.
Although an increasing number of ICE have been identified in a wide range of prokaryotes, the amplification of ICEs in a single genome has never been reported so far. Thus, the amplification of the OtAGE is a quite unique phenomenon. The mechanism of this unusual amplification is not known. However, once a mutation affecting the inhibition mechanism of invasion by the same type of ICE, similar to TraT-mediated surface exclusion and TraS-mediated entry exclusion in the F plasmid,41,42 was introduced, reciprocal transfer of the OtAGE could have taken place between the O. tsutsugamushi cells densely inhabiting the host cell cytosol. Such reciprocal transfers may have induced the unusual amplification of the OtAGE in O. tsutsugamushi.
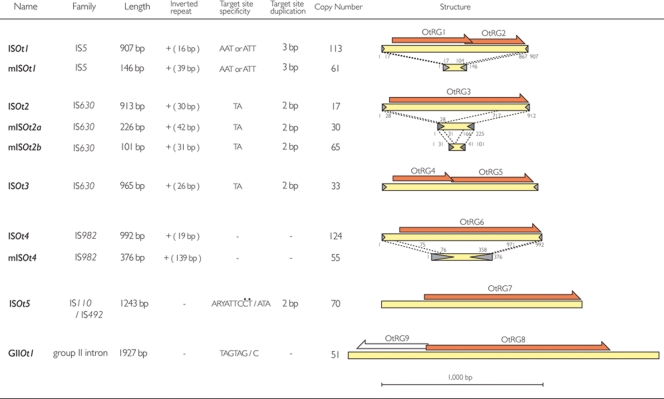
3.2.2. Transposable elements
We identified five types of insertion sequence (IS) elements (named ISOt1–ISOt5), four types of miniature inverted-repeat transposable elements (MITEs), and a Group II (GII) intron (Fig. 4). None of these transposable elements is conserved in other sequenced Rickettsia genomes. Each type of transposable element has also been explosively amplified, and a total of 621 copies including their fragments were identified, altogether representing 13.0% of the genome (261 kb in total). The amplification of transposable elements has been described also in other obligate intracellular bacteria with reduced genomes, such as Wolbachia pipientis wMel [seven types of IS elements (51 copies in total) and four types of GII introns (17 copies)],38 Parachlamydia sp. UWE25 [82 IS transposases (TPases)],43 R. felis (82 TPases),34 and R. bellii (39 TPases).33 However, the number of copies in O. tsutsugamushi is more than 10 times higher than those in these bacteria, and comparable to that in Shigella dysenteriae, which contains the highest number of IS elements among the prokaryotes sequenced to date (701 copies in a 4469-kb chromosome and a 183-kb plasmid).44
Figure 4.
Transposable elements of O. tsutsugamushi strain Ikeda. The genetic organizations of the ransposable elements identified in the genome of O. tsutsugamushi strain Ikeda are drawn to scale. Family names, lengths and other information for each type of transposable element are also shown. ISOt1, ISOt2, and ISOt4 have cognate miniature inverted-repeat transposable elements (MITEs) namely mISOt1, mISOt2, and mISOt4, respectively. There are two variants of mISOt2 (mISOt2a and ISOt2b). Regions with almost identical sequences between IS elements and MITEs are indicated by dotted lines. Transposases and reverse transcriptases are depicted by orange arrows, and terminal inverted repeats by grey arrow heads. ISOt1 is most closely related to IS1381 among the members of the IS5 family, and ISOt5 to IS621 among the members of the IS630 family.
Among the five IS elements, two (ISOt2 and ISOt3) belong to the IS630 and Tc-1/mariner family. Tc-1/mariner elements are often found in the genomes of nematodes and insects, but rarely in bacteria.45 This may imply that ISOt2 and ISOt3 were transferred from chiggers. Among the four types of MITEs, one contains the same terminal inverted repeat sequences (TIRs) as ISOt1, and is therefore named ‘mISOt1’ (miniature ISOt1). Two (namely ‘mISOt2a’ and ‘mISOt2b’) share the same or very similar TIRs with ISOt2, and the remaining one (mISOt4) with ISOt4. The massive amplification of these MITEs (201 copies in total) suggests that they are (or at least were) very actively mobilized by transposases encoded in trans in the cognate IS elements. Although numerous MITEs have been found in many eukaryotic genomes,46 no such amplification of MITEs has been observed in prokaryotes. Similarly, to the best of our knowledge, O. tsutsugamushi contains the highest number of GII introns (51 copies) among the prokaryotes thus far sequenced, with Thermosynechococcus elongates BP-1 being the next with 28 copies (http://www.fp.ucalgary.ca/group2introns/).47 Many of the transposable elements identified in the O. tsutsugamushi genome have been truncated by deletion or fragmented by insertion of other transposable elements (Supplementary Fig. S6).
3.2.3. Other repetitive sequences
We additionally identified seven types of repetitive sequences in the O. tsutsugamushi genome (165 copies in total) as summarized in Supplementary Table S2. These repetitive sequences, namely ‘Short Repeats of O. tsutsugamushi’ (SROt1–SROt7), are not related to the Rickettsia palindromic elements (RPEs) widely distributed in Rickettsia genomes.35 Among the seven SRs, only SROt2 (110 bp in length) was found to contain TIR-like sequences of 21 bp, suggesting that it may also be a type of MITE, although no cognate IS element was identified. The functions and origins of other SRs are unknown.
3.3. Repeated genes
The massive amplification of repetitive sequences in the O. tsutsugamushi genome is directly or indirectly linked to the generation of a large number of repeated genes, which were classified into 85 OtRG families (Supplementary Table S3). Among these, nine are TPases and a reverse transcriptase/maturase in the IS elements and the GII intron, and 45 are the core components of the OtAGE mentioned above. The remaining 31 OtRG families are located at various sites in the chromosome, but are frequently associated with the ISs/MITEs and/or OtAGEs (Supplementary Fig. S7). This suggests that these mobile elements have been involved in the proliferation of many of these OtRG families. In fact, eight of the 31 OtRG families have a single orthologue in other rickettsial species. These genes may have been trapped in or between these mobile elements, and duplicated along with the amplification of the mobile elements. The eight OtRGs include a unique two-component system sensor histidine kinase/response regulator hybrid protein fused with a sodium/proton transporter domain (OtRG55) and a peroxiredoxin (OtRG59). These proteins may be involved in sensing and/or coping with osmotic or oxidative stresses which the bacterium encounters in the mite cells or during the infection of mammals. Most of the other OtRG families are hypothetical protein families with unknown functions, but their amplification may also have conferred some advantages to O. tsutsugamushi, by adapting them to the host environments. In this regard, of particular interest are several intramolecular repeat-containing proteins that have also been amplified (see Section 3.5).
3.4. Singleton genes and the metabolic capacity of O. tsutsugamushi
The number of singleton genes (771 genes) identified in the O. tsutsugamushi genome is lower than the total number of CDSs (834) in R. prowazekii, which has the smallest genome among the Rickettsia species so far sequenced.36 As shown in Table 2, the genome comparison of O. tsutsugamushi and R. prowazekii revealed 542 genes that are shared by the two species. Most of them (519 genes, 95.8%) are conserved in all other sequenced Rickettsia species (Table 2), and are therefore likely to represent the core gene set of the family Rickettsiaceae. This set includes various housekeeping genes and also a set of genes for the type IV secretion system (T4SS) similar to the virB system of Agrobacterium tumefaciens. Despite the extensive genome shuffling, the organization of T4SS genes is highly conserved between O. tsutsugamushi and Rickettsia. A similar set of T4SS genes is also conserved in the family Anaplasmataceae (Supplementary Fig. S8). This strongly suggests that T4SS plays an essential role in maintaining the intracellular life style of the members of the order Rickettsiales.
Table 2.
Conservation of the O. tsutsugamushi singleton genes in the sequenced five Rickettsia speciesa
| Conservation patternsb | Number of genesc |
|---|---|
| Ot (+)/Rp (−) | 195 (149) |
| Absent in all the five Rickettsia | 161 (131) |
| Variably present in the five Rickettsia | 34 (18) |
| Ot (+)/Rp (+) | 542 (84) |
| Present in all the five Rickettsia | 519 (80) |
| Variably present in the five Rickettsia | 23 (4) |
| Ot (−)/Rp (+) | 257 (125) |
| Present in all the five Rickettsia | 178 (75) |
| Variably present in the five Rickettsia | 79 (50) |
aThe five Rickettsia species examined here include R. prowazekii, R. conorii, R. felis, R. typhi, and R. bellii.
bOt(+)/Rp(−), O. tsutsugamushi singleton genes that are not present in R. prowazekii; Ot(+)/Rp(+), O. tsutsugamushi singleton genes that are present in R. prowazekii; Ot(−)/Rp(+), R. prowazekii genes that are not present in O. tsutsugamushi.
cThe numbers of genes encoding hypothetical proteins of unknown functions are indicated in parentheses.
It has been previously reported that O. tsutsugamushi cells do not contain lipopolysaccharide (LPS) and peptidoglycan, while the members of genus Rickettsia contain these cellular components.48 Consistent with this, the electron-microscopic appearance of the outer membrane of O. tsutsugamushi is very different from those of the members of the genus Rickettsia, and these differences were the major reasons to separate O. tsutsugamushi from the genus Rickettsia.49 The higher sensitivity to various physical stresses observed in O. tsutsugamushi may also be attributable to these differences. Unexpectedly, a set of genes required for the biosynthesis and degradation of peptidoglycan was identified in O. tsutsugamushi, although those for LPS biosynthesis were completely missing. In our preliminary proteome analysis, we confirmed that at least 10 of these peptidoglycan biosynthesis-related genes were expressed in the O. tsutsugamushi cells grown in L929 cells (data not shown).
Through the genomic comparison of O. tsutsugamushi and R. prowazekii, we also identified 195 O. tsutsugamushi singleton genes that are absent or degraded in R. prowazekii (referred to as ‘O. tsutsugamushi-specific’ genes), and 257 R. prowazekii genes that are absent or degraded in O. tsutsugamushi (referred to as ‘R. prowazekii-specific’ genes) (Table 2, Supplementary Table S4). Of the 195 O. tsutsugamushi-specific genes, 161 are absent in all other sequenced Rickettsia species, and 94 genes exhibit no significant homology to known proteins. This finding suggests that a significant portion of the O. tsutsugamushi-specific genes have foreign origins. It is noteworthy that these genes are often clustered in the O. tsutsugamushi genome (data not shown), suggesting that they have been brought into O. tsutsugamushi as blocks. A prominent example is a genomic region about 19 kb in size located around the 550-kb position in the O. tsutsugamushi chromosome (Fig. 1). This region exhibits a higher G + C composition than the average of the O. tsutsugamushi genome, and 23 Orientia-specific genes are accumulated. Two major surface antigens of O. tsutsugamushi, i.e. the 56- and 22-kDa proteins (OTT_0945 and OTT_1548, respectively), and a 56-kDa protein-like protein (OTT_0946) are also O. tsutsugamushi specific.
Among the 257 R. prowazekii-specific genes, 178 are conserved in all the Rickettsia species so far sequenced. Of the 178 genes, 139 have their orthologues in a wide range of alpha-Proteobacteria. Thus, at least these 139 genes can be regarded as genes that have been deleted or degraded in the lineage leading to O. tsutsugamushi after separation from the Rickettsia family. It is noteworthy that they include a significant number of genes for various housekeeping functions, such as the formation of a cell envelope, energy metabolism, fatty-acid metabolism, nucleotide metabolism, and DNA recombination and repair (indicated by red arrows in Fig. 5; see also Supplementary Table S3). Several genes for RNA synthesis and modification, such as the RNA polymerase omega subunit and RNA helicase RhlE, those for stress response, such as the HslUV protease, ClpB chaperone, BipA GTPase and the OsmY protein, and those for drug sensitivity, such as the EmrA and EmrB proteins, are also specifically missing in O. tsutsugamushi. In addition, the genes for hemolysin A and patatin-like phospholipase, both of which are thought to be involved in the pathogenesis of Rickettsia species, are missing in O. tsutsugamushi.
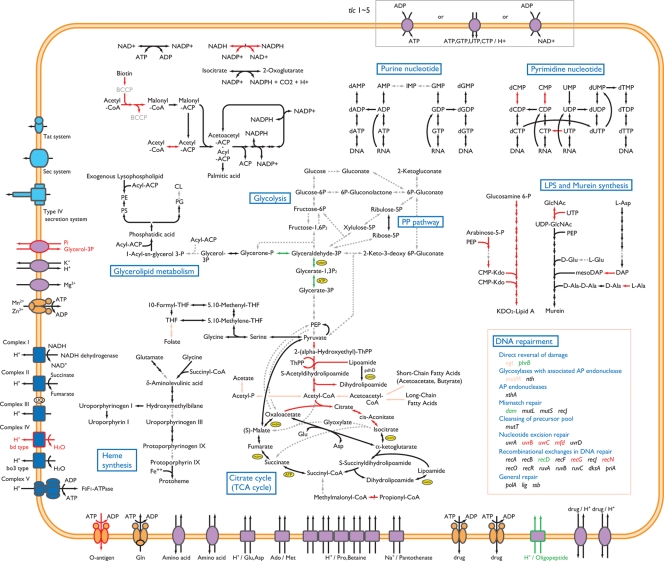
Figure 5.
Metabolic pathways of O. tsutsugamushi and R. prowazekii. The metabolic pathways of O. tsutsugamushi and R. prowazekii are compared. Pathways identified in both bacteria are indicated by black arrows, those identified in neither bacterium by gray broken-lined arrows, those identified only in O. tsutsugamushi by green arrows, and those identified only in R. prowazekii by red arrows. Pink arrows indicate the pathways that exhibit variable distribution in the genus Rickettsia but are absent in O. tsutsugamushi. Genes for DNA repair functions that were identified in O. tsutsugamushi and R. prowazekii are listed in the box.
Many genes for nucleotide metabolism, including those for de novo synthesis pathways, are absent in O. tsutsugamushi, as in Rickettsia species (Fig. 5). The genes for CTP synthase and cytidylate kinase are additionally missing in O. tsutsugamushi. This may have rendered the bacterium more dependent on the host cell functions for the pyrimidine nucleotides. All these defects are probably compensated by the presence of five types of nucleotide transporters, as shown in Chlamydia trachomatis.50 This may also be the case for the nicotinamido adenine dinucleotide (NAD) metabolism.51
DNA recombination and repair systems are relatively well conserved in Rickettsia species (Fig. 5). But a significant number of genes for these functions have specifically been deleted in O. tsutsugamushi, such as those for the UvrB, UvrC, RecN and RecG proteins and the transcription-repair coupling factor Mfd. This may be related to the acquisition and amplification of mobile genetic elements, as well as to the maintenance of an extremely repeat-rich genome.
Of particular interest is the lack of pyruvate dehydrogenase due to the deletion of the pdhABC genes and a second copy of pdhD. Other pathways generating acetyl-CoA are also missing (Fig. 5). Thus, the bacterium appears to be unable to synthesize acetyl-CoA. This defect is probably linked to the incomplete tricarboxylic acid (TCA) cycle of the bacterium, where the citrate synthase and aconitate hydratase genes are inactivated by an authentic frame shift mutation and a premature stop codon, respectively (Fig. 5). However, these defects may be bypassed by asparate aminotransferase, which catalyzes the oxaloacetate/alpha-ketoglutarate interconversion. Pyruvate may also be introduced into the TCA cycle by malate oxidoreductase. In addition, the glycoxylate cycle is missing in O. tsutsugamushi, as in Rickettsia species.
The inability to synthesize acetyl-CoA probably is further linked to the deletion of the genes for acetyl-CoA carboxylase, which catalyzes the transfer of carboxyl groups to acetyl-CoA to generate malonyl-CoA, the first step in fatty-acid biosynthesis (Fig. 5). The genes for biotin-apoprotein ligase (birA) and biotin transporter (bioY), which are required for the formation of biotin carboxyl carrier protein, are also specifically missing in O. tsutsugamushi. In addition, the fabH gene encoding KAS III, which catalyzes the condensation of the acetyl-CoA starter unit with malonyl-ACP to yield acetoacetyl-CoA, the first step in the fatty-acid elongation pathway, is missing. Although the gene for KAS I is also absent as in Rickettsia species, the genes for malonyl-CoA:ACP transacylase and KAS II (fabD and fabF) are conserved. It is thus most likely that fatty-acid biosynthesis in O. tsutsugamushi is initiated by these two enzymes using host-derived malonyl-CoA.
Another distinguishing feature of O. tsutsugamushi is the lack of genes for lipoprotein biosynthesis. Although a gene for lipoprotein-specific leader peptidase (lsp) was identified, other genes for the modification and localization of lipoproteins (lgt, lnt, lolA/B/C/E/D) are missing (Supplementary Table S3). This may raise the possibility that O. tsutsugamushi lacks lipoproteins or has a very unique lipoprotein biosynthesis system. Consistent with this, of the five O. tsutsugamushi proteins whose homologues have been predicted to be lipoproteins in Rickettsia species, none contains the typical lipobox required for processing and lipid-modification of lipoproteins. This finding, together with the absence of LPS, suggests that a simple cell envelope may be advantageous to the intracellular life of O. tsutsugamushi.
All these genomic features of O. tsutsugamushi indicate that high levels of horizontal gene transfer (HGT) and gene amplification have occurred in O. tsutsugamushi after it diverged from the genus Rickettsia. On the other hand, O. tsutsugamushi has also undergone further reductive genome evolution and established an intracellular lifestyle that is more dependent on its host cell functions than Rickettsia species.
3.5. Repeat-containing proteins
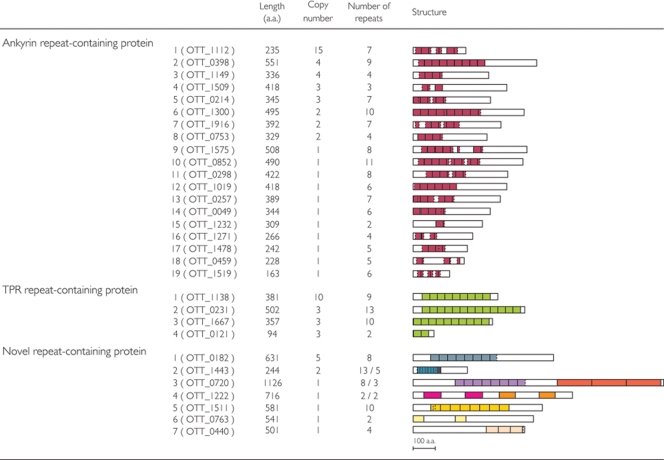
As briefly mentioned in Section 3.3, the OtRG families include eight types of ankyrin repeat-containing proteins (AR proteins), four tetratricopeptide repeat-containing proteins (TPR proteins), and two novel repeat-containing proteins (Fig. 6). We additionally identified 11 singleton genes encoding different types of AR proteins and five singleton genes encoding novel repeat-containing proteins. Ankyrin repeats have been found in many eukaryotic proteins and are known to mediate various protein–protein interactions.52 However, they are rarely found in prokaryotes. Several bacteria, most of which are intracellular bacteria, such as R. felis, R. bellii, W. pipientis, Legionella pneumonia, and Coxiella burnetii, are exceptionally enriched in AR proteins (18–25 proteins), and these proteins are suspected to be involved in host–bacterial interactions. Orientia tsutsugamushi contains 20 types of genes for AR proteins, and the total number of copies (46) greatly exceeds those of other intracellular bacteria. As all these AR proteins have no signal sequence for the general secretion pathway, they could be candidates for the effecter proteins secreted by T4SS. And they may be involved in the modulation of host-cell functions by O. tsutsugamushi, which is required for the bacterium to develop and maintain a symbiotic relationship in mite cells and/or survive and propagate in mammals. Other repeat-containing proteins also do not possess apparent signal sequences, suggesting that they would also be T4SS effector candidates.
Figure 6.
Repeat-containing proteins of O. tsutsugamushi. The repeat-containing proteins identified in the O. tsutsugamushi genome and their characteristics are shown. The structure of each repeat-containing protein is drawn to scale. Repeating units identified in each protein or family are depicted by colored boxes. The same repeating units are indicated by the same color.
3.6. Genome comparison with O. tsutsugamushi strain Boryong
While we were preparing this report, the genome sequence of O. tsutsugamushi strain Boryong isolated in Korea was reported.14 The genome size of strain Boryong (2 127 051 bp) is 118 kb longer than that of IKEDA, but the G + C content and the numbers of rRNA and tRNA genes are the same as those of IKEDA. Homology search of the 542 genes that are shared by strain IKEDA and R. prowazekii using BLASTN revealed that 540 genes are also conserved in strain Boryong. This indicates that the genome backbone is highly conserved between the two strains. The homologues exhibited high level of sequence conservation (97.5% identity on average) while notable sequence variations were observed in several genes for surface proteins.
Search of the mobile elements identified in IKEDA using BLASTN revealed that all the elements, including OtAGE, are also present and have explosively been amplified in strain Boryong. Although exact numbers of each element in Boryong are yet to be determined, this result suggests that these elements invaded into O. tsutsugamushi before the separation of the two strains. More importantly, the result of our preliminary dot-plot analysis of the two genomes suggested that intensive genome shuffling has also taken place between the two O. tsutsugamushi strains. Although more careful and intensive comparison of the two genomes is required, the amplified mobile elements seem to have been deeply involved in the genome shuffling.
3.7. Conclusion
In O. tsutsugamushi, high level of gene loss has taken place like in other obligate intracellular bacteria, but massive amplification of various mobile elements has also taken place, which has induced intensive genome shuffling and generated a large number of repeated genes. Although the timing of the acquisition and/or amplification of these elements are yet to be elucidated, the extremely narrow population bottleneck created by the unique life cycle of the bacterium appears to have allowed such a very unique genome evolution. Thus, the genome sequence of O. tsutsugamushi strain Ikeda will be not only the genetic basis for a better understanding of the biology of the bacterium and pathogenesis of the ‘Tsutsugamushi’ disease, but also a very attractive material for studying the processes of genome evolution and the high plasticity of bacterial genomes.
Funding
This work was supported by the Research for the Future Program of the Japan Society for the Promotion of Science (JSPS-RFTF00L01411) and Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Supplementary Material
Acknowledgements
We thank Eiichi Ohtsubo and Kenji Ichiyanagi for their help in our analysis of the IS elements and GII introns. We also thank Seigo Yamamoto, Yoshiro Terawaki and Hiroshi Yoshikawa for their advice and encouragement, Hidehiro Toh, Akemi Yoshida, Yumiko Takeshita, Noriko Kanemaru and Nobuko Fujii for their technical assistance, and Yumiko Hayashi for her linguistic assistance.
References
- 1.Tamura A., Ohashi N., Urakami H., et al. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int. J. Syst. Bacteriol. 1995;45:589–591. doi: 10.1099/00207713-45-3-589. [DOI] [PubMed] [Google Scholar]
- 2.Rai J., Bandopadhyay D. Vertical transmission in chigger borne rickettsiosis. Indian J. Med. Res. 1978;68:31–38. [PubMed] [Google Scholar]
- 3.Rapmund G., Dohany A. L., Manikumaran C., et al. Transovarial transmission of Rickettsia tsutsugamushi in Leptotrombidium (Leptotrombidium) arenicola Traub (Acarina: Trombiculidae) J. Med. Entomol. 1972;9:71–72. doi: 10.1093/jmedent/9.1.71. [DOI] [PubMed] [Google Scholar]
- 4.Rapmund G., Upham R. W., Jr., Kundin W. D., et al. Transovarial development of scrub typhus rickettsiae in a colony of vector mites. Trans. R. Soc. Trop. Med. Hyg. 1969;63:251–258. doi: 10.1016/0035-9203(69)90155-2. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi M., Murata M., Nogami S., et al. Transovarial transmission of Rickettsia tsutsugamushi in Leptotrombidium pallidum successively reared in the laboratory. Jpn. J. Exp. Med. 1988;58:213–218. [PubMed] [Google Scholar]
- 6.Urakami H., Takahasi M., Hori E., et al. An ultrastructural study of vertical transmission of Rickettsia tsutsugamushi during oogenesis and spermatogenesis in Leptortombidium pallidum. Am. J. Trop. Med. Hyg. 1994;50:219–228. doi: 10.4269/ajtmh.1994.50.219. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi M., Murata M., Misumi H., et al. Failed vertical transmission of Rickettsia tsutsugamushi (Rickettsiales: Rickettsiaceae) acquired from rickettsemic mice by Leptotrombidium pallidum (Acari: trombiculidae) J. Med. Entomol. 1994;31:212–216. doi: 10.1093/jmedent/31.2.212. [DOI] [PubMed] [Google Scholar]
- 8.Walker J. S., Chan C. T., Manikumaran C., et al. Attempts to infect and demonstrate transovarial transmission of R. tsutsugamushi in three species of Leptotrombidium mites. Ann. N Y. Acad. Sci. 1975;266:80–90. doi: 10.1111/j.1749-6632.1975.tb35090.x. [DOI] [PubMed] [Google Scholar]
- 9.Traub R., Wisseman C. L., Jones M. R., et al. The acquisition of Rickettsia tsutsugamushi by chiggers (trombiculid mites) during the feeding process. Ann. N Y. Acad. Sci. 1975;266:91–114. doi: 10.1111/j.1749-6632.1975.tb35091.x. [DOI] [PubMed] [Google Scholar]
- 10.Watt G., Parola P. Scrub typhus and tropical rickettsioses. Curr. Opin. Infect. Dis. 2003;16:429–436. doi: 10.1097/00001432-200310000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Enatsu T., Urakami H., Tamura A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol. Lett. 1999;15:163–169. doi: 10.1111/j.1574-6968.1999.tb08791.x. [DOI] [PubMed] [Google Scholar]
- 12.Qiang Y., Tamura A., Urakami H., et al. Phylogenetic characterization of Orientia tsutsugamushi isolated in Taiwan according to the sequence homologies of 56-kDa type-specific antigen genes. Microbiol. Immunol. 2003;47:577–583. doi: 10.1111/j.1348-0421.2003.tb03420.x. [DOI] [PubMed] [Google Scholar]
- 13.Tamura A., Yamamoto N., Koyama S., et al. Epidemiological survey of Orientia tsutsugamushi distribution in field rodents in Saitama Prefecture, Japan, and discovery of a new type. Microbiol. Immunol. 2001;45:439–446. doi: 10.1111/j.1348-0421.2001.tb02643.x. [DOI] [PubMed] [Google Scholar]
- 14.Cho N. H., Kim H. R., Lee J. H., et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host–cell interaction genes. Proc. Natl Acad. Sci. USA. 2007;8:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohashi N., Koyama Y., Urakami H., et al. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol. Immunol. 1996;40:627–638. doi: 10.1111/j.1348-0421.1996.tb01120.x. [DOI] [PubMed] [Google Scholar]
- 16.Kawamori F., Akiyama M., Sugieda M., et al. Epidemiology of Tsutsugamushi disease in relation to the serotypes of Rickettsia tsutsugamushi isolated from patients, field mice, and unfed chiggers on the eastern slope of Mount Fuji, Shizuoka Prefecture, Japan. J. Clin. Microbiol. 1992;30:2842–2846. doi: 10.1128/jcm.30.11.2842-2846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitaoka M., Okubo K., Asanuma K. Epidemiological survey by means of complement fixation test on scrub typhus in Japan. Acta. Med. Biol (Niigata). 1967;15:69–85. [PubMed] [Google Scholar]
- 18.Shishido A. Strain variation of rickettsia orientalis in the complement fixation test. Jpn. J. Med. Sci. Biol. 1964;17:59–72. doi: 10.7883/yoken1952.17.59. [DOI] [PubMed] [Google Scholar]
- 19.Urakami H., Tsuruhara T., Tamura A. Electron microscopic studies on intracellular multiplication of Rickettsia tsutsugamushi in L cells. Microbiol. Immunol. 1984;28:1191–1201. doi: 10.1111/j.1348-0421.1984.tb00777.x. [DOI] [PubMed] [Google Scholar]
- 20.Tamura A., Uramaki H., Tsuruhara T. Purification of Rickettsia tsutsugamushi by Percoll density gradient centrifugation. Microbiol. Immunol. 1982;26:321–328. doi: 10.1111/j.1348-0421.1982.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 21.Kumura K., Minamishima Y., Yamamoto S., Ohashi N., Tamura A. DNA base composition of Rickettsia tsutsugamushi determined by reversed-phase high-performance liquid chromatography. Int. J. Syst. Bacteriol. 1991;41:247–248. doi: 10.1099/00207713-41-2-247. [DOI] [PubMed] [Google Scholar]
- 22.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 23.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 24.Ohnishi M., Tanaka C., Kuhara S., et al. Chromosome of the enterohemorrhagic Escherichia coli O157:H7; comparative analysis with K-12 MG1655 revealed the acquisition of a large amount of foreign DNAs. DNA Res. 1999;31:361–368. doi: 10.1093/dnares/6.6.361. [DOI] [PubMed] [Google Scholar]
- 25.Ohnishi M., Terajima J., Kurokawa K., et al. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl Acad. Sci. USA. 2002;99:17043–17048. doi: 10.1073/pnas.262441699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakiyama T., Takami H., Ogasawara N., et al. An automated system for genome analysis to support microbial whole-genome shotgun sequencing. Biosci. Biotechnol. Biochem. 2000;64:670–673. doi: 10.1271/bbb.64.670. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S. F., Madden T. L., Schaffer A. A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007;35(Database issue):D343–D346. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson W. R. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 30.Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 32.Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;15:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogata H., La Scola B., Audic S., et al. Genome sequence of Rickettsia bellii illuminates the role of Amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2:733–744. doi: 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogata H., Renesto P., Audic S., et al. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3:1–12. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogata H., Audic S., Renesto-Audiffren P., et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- 36.Andersson S. G., Zomorodipour A., Andersson J. O., et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 37.McLeod M. P., Qin X., Karpathy S. E., et al. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 2004;186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M., Sun L. V., Vamathevan J., et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:327–341. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrus V., Waldor M. K. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Blanc G., Ogata H., Robert C., et al. Lateral gene transfer between obligate intracellular bacteria: evidence from the Rickettsia massiliae genome. Genome Res. 2007;17:1657–1664. doi: 10.1101/gr.6742107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Audette G. F., Manchak J., Beatty P., et al. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology. 2007;153:442–451. doi: 10.1099/mic.0.2006/001917-0. [DOI] [PubMed] [Google Scholar]
- 42.Sukupolvi S., O'Connor C. D. TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiol. Rev. 1990;54:331–341. doi: 10.1128/mr.54.4.331-341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horn M., Collingro A., Schmitz-Esser S., et al. Illuminating the evolutionary history of chlamydiae. Science. 2004;304:728–730. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- 44.Fan Y., Jian Y., Xiaobing Z., et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsson P., Oyston P. C., Chain P., et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 46.Feschotte C., Jiang N., Wessler S. R. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura Y., Kaneko T., Sato S., et al. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 2002;31:123–130. doi: 10.1093/dnares/9.4.123. [DOI] [PubMed] [Google Scholar]
- 48.Amano K., Tamura A., Ohashi N., et al. Deficiency of peptidoglycan and lipopolysaccharide components in Rickettsia tsutsugamushi. Infect. Immun. 1987;55:2290–2292. doi: 10.1128/iai.55.9.2290-2292.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silverman D. J., Wisseman C. L. Comparative ultrastructural study on the cell envelopes of Rickettsia prowazekii, Rickettsia rickettsii, and Rickettsia tsutsugamushi. Infect. Immun. 1978;21:1020–1023. doi: 10.1128/iai.21.3.1020-1023.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tjaden J., Winkler H. H., Schwöppe C., et al. Two nucleotide transport proteins in Chlamydia trachomatis, One for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 1999;181:1196–1202. doi: 10.1128/jb.181.4.1196-1202.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haferkamp I., Schmitz-Esser S., Linka N., et al. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature. 2004;432:622–625. doi: 10.1038/nature03131. [DOI] [PubMed] [Google Scholar]
- 52.Li J., Mahajan A., Tsai M. D. Ankyrin repeat: a unique motif mediating protein–protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.