Abstract
At a time when the notion of microorganisms did not exist, our ancestors empirically established methods for the production of various fermentation foods: miso (bean curd seasoning) and shoyu (soy sauce), both of which have been widely used and are essential for Japanese cooking, and sake, a magical alcoholic drink consumed at a variety of ritual occasions, are typical examples. A filamentous fungus, Aspergillus oryzae, is the key organism in the production of all these traditional foods, and its solid-state cultivation (SSC) has been confirmed to be the secret for the high productivity of secretory hydrolases vital for the fermentation process. Indeed, our genome comparison and transcriptome analysis uncovered mechanisms for effective degradation of raw materials in SSC: the extracellular hydrolase genes that have been found only in the A. oryzae genome but not in A. fumigatus are highly induced during SSC but not in liquid cultivation. Also, the temperature reduction process empirically adopted in the traditional soy-sauce fermentation processes has been found to be important to keep strong expression of the A. oryzae-specific extracellular hydrolases. One of the prominent potentials of A. oryzae is that it has been successfully applied to effective degradation of biodegradable plastic. Both cutinase, responsible for the degradation of plastic, and hydrophobin, which recruits cutinase on the hydrophobic surface to enhance degradation, have been discovered in A. oryzae. Genomic analysis in concert with traditional knowledge and technology will continue to be powerful tools in the future exploration of A. oryzae.
Key words: Aspergillus oryzae, genome sequence, solid-state culture, secretory hydrolase, non-syntenic block, comparative genomics
1. Historical backgrounds
Aspergillus oryzae is a fungus widely used in traditional Japanese fermentation industries, including soy sauce, sake, bean curd seasoning and vinegar production. Filamentous fungi generally have the ability to produce various and vast amounts of enzymes in a secretory manner. Among filamentous fungi, A. oryzae is known to have prominent potential for the secretory production of various enzymes. In addition, developments in genetic engineering technology have led to the application of A. oryzae in the production of industrial enzymes in modern biotechnology. Aspergillus oryzae was used for the first example of commercial production of heterogonous enzyme, the lipase for laundry detergent in 1988.1
1.1. Solid-state cultivation
One of the distinctive features of the use of A. oryzae in traditional Japanese fermentation is the use of solid-state cultivation (SSC) (rice grain, soybean and wheat bran). This style of fermentation is thought to have originated 3000–2000 years ago in China.2,3 The technology was imported into Japan during the Yayoi period (B.C. 10th–A.D. 3rd).2 Inocula from filamentous fungi for fermentation have been commercially available as koji since the 13–15th century (Heian and Muromachi period).4 This indicates that koji was cultivated without the knowledge that it is composed of a microorganism. Thus, the word, koji, indicates both the material fermented by A. oryzae in the form of SSC and the A. oryzae microorganism itself (koji mold). A key technology enabling the industrial production and distribution of A. oryzae was the production of conidiospores whilst keeping them alive and uncontaminated. Traditionally, this technology involved the use of hardwood leaves burned to white ashes in poor aeration. The conidiospores packed in paper bags were layered with the ashes between them in a box and stored. The technology was indispensable to the avoidance of contamination by other microorganisms in a period when desiccant or air conditioning was unavailable.4 This technology led to the discovery that leaf ashes added to steamed rice also allowed reliable production of conidiospores. Conidiospores prepared industrially for sake brewing are called Moyashi. Signboards with three Japanese characters, ‘moyashi’ (or fermentation starter), indicate the supplier of A. oryzae conidiospores for sake brewers (Fig. 1). It is currently known that alkaline pH, produced by the addition of the ash, prevents contamination by other microorganisms and that minerals contained with the ash enhance the formation of conidiospores.2,4,5 This technology has been applied to the production of Trichoderma spores as a biological pesticide by Akita Konno Co., Ltd, a biotech company whose origins stem from the traditional fermentation industry.
Figure 1.
A historical signboard of a producer of A. oryzae conidiospores. Aspergillus oryzae conidiospores are industrially produced and are distributed to fermentation companies. Two suppliers were established ∼600 years ago (Muromachi period). No other suppliers were established before A.D. 17–18th. The figure shows a photograph of an original signboard, Kuro-ban (black stamp), prepared under the license of Koji-za, the association of A. oryzae conidiospores suppliers during the Muromachi period. Currently there are five major distributors in Japan supplying A. oryzae conidiospores to 4500 sake (Japanese alcoholic beverage, ca. 1900 brewers), miso (soybean paste, ca. 1200 brewers) and shoyu (soy sauce, ca. 1500 brewers) brewers in Japan, excluding several of the biggest soy-sauce companies. The three characters called Hiragana, which were originally developed in Japan; on the signboard have pronunciations, “mo”, “ya” and “shi” from top to bottom. Moyashi means the A. oryzae conidiospores used mainly by sake brewers.
The ability of secretory production of proteins is further enhanced in solid-state culture compared with submerged culture. For example, A. oryzae can produce ∼50 g of α-amylase from 1 kg of wheat bran, which is roughly equivalent to 1 L of liquid culture medium. The first example of the industrial application of SSC to enzyme production by A. oryzae was Taka-diastase, developed as a stomach medicine by Jokichi Takamine in 1894. Recently, solid-state fermentation systems using the industrial fungi A. oryzae and A. sojae have been introduced for the production of various industrial enzymes (cf. amylases, proteases, lipases) and speciality chemicals.6 In spite of the extensive use of SSC, much less is known about SSC than for submerged cultivation, which is widely used in modern biotechnology owing to its easier automatic handling. Gene expression analysis revealed important factors affecting secretion of enzymes in SSC, physical barrier and low water activity.7 Recently, practical approaches to liquid cultivation containing solid materials succeeded in the effective production of enzymes in a secretory manner allowing the degradation of raw materials with a similar efficiency to SSC.8 Analysis of the mechanism for the secretory production of enzymes in SSC may be important to the understanding of highly efficient secretion.
1.2. Isolation and identification of A. oryzae
Sequencing the genome of A. oryzae RIB40 (ATCC-42149) was completed in 2005.9 The sequenced strain is a wild-type strain, most similar to those used for sake brewing but still has the ability of strong production of proteases, which is one of the most important characteristics for soy-sauce fermentation. Aspergillus oryzae was first isolated from koji by H. Ahlburg in 1876 when he was invited to the Japanese Medical College. Its original name, Eurotium oryzae, was later renamed A. oryzae by F. Cohn because he found that it lacked the ability of sexual reproduction. It is said that A. oryzae is a domesticated species and, therefore, that A. oryzae may be found only in a domesticated form but not in nature. On the other hand, it has been stated, in a historical literature, that koji should be isolated from the mold growing on an ear of rice. Some reports about the isolation of A. oryzae from soil, plants and food10 support the natural isolation of A. oryzae for fermentation. However, these species might now be distributed in nature following domestication many years ago.
There are two scenarios to be considered for the origination of koji fermentation. In the first scenario, A. oryzae may have been isolated from nature independently in Japan. A literature describing naturally fermented alcoholic beverage was found in a historical document, Harima no Kuni Fudoki, edited in AD 715.2 Another historical document described that koji must be isolated from the mold growing on an ear of rice. This means that A. oryzae may have existed in nature before domestication and might be isolated from other dangerous species such as A. flavus through the method indicated in the historical literatures above. In the second scenario, A. oryzae may have been imported from China during the Yayoi period. The material used for the fermentation in East Asia was a block of hardened raw wheat or rice powder. The filamentous fungi that grew on the block is thought to have been Mucor or Rhizopus.2 On the other hand, the material used in the production of Japanese alcoholic beverage was steamed rice grain, thus, proteinous components in rice are considerably denatured. Mucor and Rhizopus, which have a low productivity of proteases allowing the assimilation of denatured proteins as nitrogen sources, grow poorly on steamed rice. On the contrary, A. oryzae, producing a considerable amount of proteases, could predominantly grow in an environment where a mixture of spores of these heterologous species exists. The names of the traditional fermented materials, which indicate yellow color, but not dark gray as Mucor and Rhizopus, also suggests that A. oryzae was first used in Japanese fermentation industries. Therefore, the second scenario may not be well supported.
1.3. Safety
The long history of extensive use of A. oryzae in food fermentation industries prompted industrial applications of A. oryzae to be listed as Generally Recognized as Safe (GRAS) by the Food and Drug Administration (FDA) in the USA.11,12 The safety of this organism is also supported by World Health Organization (WHO).13 Although A. oryzae is genetically very close to A. flavus, which is known to produce the most potent natural carcinogen, aflatoxin, A. oryzae has no record of producing aflatoxin or any other carcinogenic metabolites.14 Fermented foods produced by A. oryzae including A. sojae, the close relative to A. oryzae in section Flavi, have been shown to be aflatoxin free.15,16 The two species, A. oryzae and A. flavus were traditionally distinguished based on morphological, physiological and culture-based characteristics.17 Recent DNA-based techniques have enhanced the potential for distinction.18–20 Zhang et al. reported that homologs of aflatoxin biosynthesis gene cluster of A. oryzae were not expressed even under the conditions that are favorable to aflatoxin expression in A. flavus and A. parasiticus.21
We have received huge benefits from A. oryzae for thousands of years and may continue to receive them in the future. At the same time, we are facing various mysteries regarding A. oryzae: unique cultivation method, species origin, isolation and so forth, as described above. In spite of economical importance and scientific interest, the fact that A. oryzae forms multinucleate conidia and lacks a sexual life cycle have prevented extensive basic studies by conventional approaches. It is very interesting to see whether and to what extent genomics elucidates the subjects concerning ancestor, molecular mechanism for high secretion and safety, and the potential of A. oryzae in new fields where A. oryzae has never yet been applied.
2. Lessons from genomics
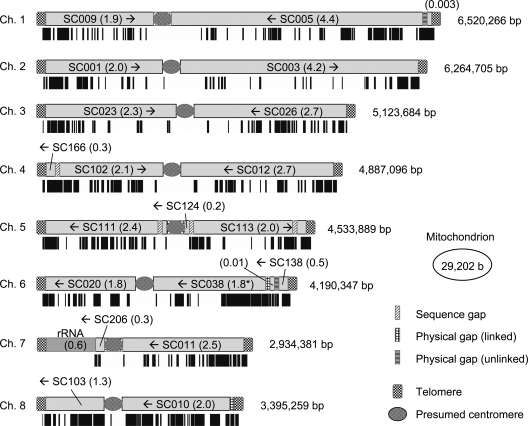
The A. oryzae genome consists of eight chromosomes with an entire genome size of 37.6 Mb (Fig. 2). Comparison of the A. oryzae genome with two other Aspergilli sp., A. nidulans and A. fumigatus, sequenced in the similar timelines revealed that A. oryzae had a 25–30% bigger genome size than the other two species.9 Accordingly, A. Oryzae had 2000–3000 more genes than the other two species, indicating similar gene densities of the three species. The increase of gene number, in accordance with the increase of genome size, mainly derived from the gene expansion of metabolic genes. This includes secretory hydrolases, transporters and primary and secondary metabolism genes, among which the expansion of secondary metabolism genes is most prominent9 (see another review22 for detail). Aspergillus oryzae is known to have a wide variety of hydrolytic enzymes and a strong capacity for degrading various materials. The gene expansion of secretory hydrolases explains these abilities well. The multiple transporters may be used for efficient degradation or further conversion of degraded materials, but their main role may be to excrete toxic compounds for A. oryzae to grow. In fermentation, A. oryzae is exposed to vast amounts and high concentrations of compounds derived from a single material, rice, soybean, wheat, etc., which may include phytoalexin or other compounds that inhibit growth. The unusual environment that was artificially introduced might allow A. oryzae to acquire additional transporters.
Figure 2.
Contigs and NSBs/SBs maps Contigs (SCnnn; n, decadecimal number) and NSBs (black bars) are mapped on the A. oryzae chromosomes.9 Arrows and values in parentheses indicate direction and length of the contigs in mega base, respectively. Adapted from Machida et al.9
2.1. Insights into the molecular mechanism of safety
Interestingly, the expanded genes indicated above were highly enriched on the non-syntenic blocks (NSBs) that were determined by the comparison of gene orders on the genomes between A. oryzae and A. nidulans or A. fumigatus. The NSBs were distributed through the entire A. oryzae genome in a mosaic manner. The expressed sequence tag (EST) analysis23 showed that the transcriptional expression of the NSB genes was considerably weaker than the genes on syntenic blocks (SBs) (P = 4.1 × 10−134).9 The difference of absolute levels of the gene expression between the two blocks was confirmed by DNA microarray.24 Therefore, the secondary metabolism genes, which are highly enriched on NSBs, are not expressed in the culture conditions examined including SSC. Consequently, the EST analysis showed a striking contrast in expression of the aflatoxin biosynthesis gene homologs; whereas ESTs for all the 25 gene homologs25 were found in A. flavus.26 No ESTs except for aflJ and norA were found in A. oryzae.23 These results suggest that the long history of industrial use of A. oryzae has removed genes unfavorable for human consumption. Alternatively, A. oryzae, by some means, might have been selected as a safe mutant from the beginning. Another industrial species, A. sojae, which is genetically very close to A. parasiticus, the aflatoxin producer, lacks the ability to produce aflatoxin, and so is similar to A. oryzae, although it harbors aflatoxin gene cluster homologs.27 One of the possible reasons explaining the inability of the expression of the aflatoxin biosynthesis gene homologs in the two species is mutation of the aflR gene. However, Takahashi et al. showed that complementation of the mutated A. sojae aflR gene (truncation by nonsense mutation) was insufficient for the expression of aflatoxin cluster homologs.27 Further, base substitutions in the two elemental sequences (AreA and FacB binding sites) in the A. oryzae aflR promoter alone do not seem to be key for the absence of aflR mRNA.28 These results suggest a silencing mechanism similar to that observed in the regulation of aflatoxin biosynthesis gene homologs in A. sojae.15,27,28 Recently, a global regulator of secondary metabolism genes, laeA,29 was found. The fact that the predicted laeA gene seems intact in A. oryzae may suggest the existence of a global regulation mechanism of secondary metabolism by other transcriptional regulatory factor(s). Greater than 40% of the A. oryzae strains deposited in the National Research Institute of Brewing lack more than half of the entire cluster of aflatoxin biosynthesis gene homologs (Group 2 and other strains), but the remaining strains have the near intact cluster (Group 1 strains).30 It is thought that the latter strains have at least double or more safety locks preventing induction of aflatoxin biosynthesis gene homologs outside the cluster.
2.2. Secrets of solid-state fermentation
Transcriptome analysis using cDNA microarray revealed that the carbohydrate metabolism showed typical characteristics of gluconeogenesis in SSC in spite of the presence of rich nutrients in koji culture.31 The lower expression levels of catabolic genes of the glycolytic pathway and the tricarboxylic acid cycle in solid-state culture led to a release from catabolite repression, and consequently to high-level expression of hydrolytic enzymes. Low water activity may cause inefficient degradation and uptake of nutrients resulting in starvation.
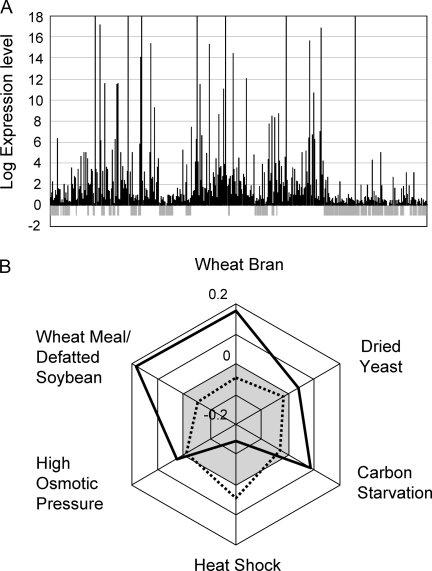
Recently, the expression regulation of the NSB genes were analyzed by DNA microarray comprised more than 11 000 oligonucleotide probes of the A. oryzae genes. The significantly lower expression level of the NSB genes when compared with the SB genes in an average was confirmed by the DNA microarray experiments (Fig. 3A). Besides the difference in absolute expression levels between NSBs and SBs, the expression regulation showed striking difference between the two blocks.24 The genes on NSBs were significantly repressed at heat shock (42°C), whereas those on SBs were induced in an average. Interestingly, the genes on NSBs appeared globally up-regulated in SSC, but those on SBs did not display global characteristics of up- or down-regulation under the same conditions (Fig. 3B).24 High induction ratios of the genes encoding extracellular enzymes, which belong to the metabolism of the COG meta-category, contributed most to the global induction of the NSB genes. In contrast, the global up-regulation of the SB genes at heat shock was mainly due to the induction of the genes belonging to the meta-category of Information Storage and Processing. The two meta-categories played an important role for the characteristic global up-regulation of the genes on each block showed obviously bigger occupation in comparison between the two blocks. Furthermore, the NSB genes that did not have genes with sequence similarity to the genes on SBs showed higher induction ratios on average compared with those from the NSB genes that had homologs on SBs. Therefore, the genes that had similarity between the two blocks after the removal of the genes specific in each block showed global expression profiles which are more similar than using entire genes on each block.24
Figure 3.
Characteristic transcriptional expression of the genes on NSBs and SBs. (A) Approximately 1500 genes on chromosome 6 were mapped with even spacing with their absolute expression levels at 42°C in YPD medium, measured by DNA microarray. The gray boxes under the horizontal zero axis indicate NSBs. (B) Averages of the logarithmic (log2) expression ratios of the genes on the SBs (dotted line) and NSBs (bold line). Wheat Meal/Defatted Soybean, Wheat Meal/Defatted Soybean against CD; Wheat Bran, Wheat Bran against CD; Dried Yeast, CD containing 2% dried yeast against CD; Carbon Starvation, CD without glucose against YPD; Heat Shock, YPD at 42°C against YPD at 30°C; High Osmotic Pressure, CD containing 0.8 M NaCl against CD. See further details in the previous reports. Reproduced from Tamano et al., 2008.24
Hydrolases occupied a major part of the genes highly induced in two SSC (9 out of the top 29 induced NSB genes and 10 out of the top 47 induced SB genes). The genes include those encoding hydrolases [cf. leucine aminopeptidase (Chien et al., 2002)32 and neutral protease II (Tatsumi et al., 1991)]33 on NSBs and alkaline protease (Murakami et al., 1991)34, glucoamylase (Hata et al., 1998)35 on SBs, which play an important role in the degradation of proteins and starch in the raw materials for fermentation. Another notable feature of the highly induced genes in SSC is the high abundance of transporters in the major facilitator superfamily (MFS). These transporters might be necessary for the growth in vast amounts of the limited variety of raw materials, where trace amounts of toxic materials in the raw materials might be condensed. As mentioned above, the transcriptional level of the NSB genes are significantly lower than the SB genes. However, some NSB genes, neutral protease (AO090103000310) and MFS transporter (AO090010000493) genes, for example, showed an expression strength comparable to alkaline protease (AO090003001036) and translation elongation factor (AO090120000080), which showed the highest expression levels among the SB genes (∼4 sigma bigger than the average of entire genes). Thus, the genes above may make a large contribution to the degradation of raw materials. The bigger induction ratios of the genes specific to NSBs suggest that the products of these genes might be important to the effective degradation of the raw materials through breaking minor chemical linkage, which loosen the conformation of polymers for example. During solid-state fermentation, the materials with A. oryzae growing on their surface are mixed upside down for a while. This process, which is called Teire, is necessary to keep the temperature below ∼40°C and to improve aeration during fermentation while A. oryzae is growing vigorously. The results of the expression analysis above suggest that this process is important in preventing the A. oryzae-specific homologs of various hydrolases on NSBs from being repressed by temperature increase.
Interestingly, no genes encoding polyketide synthase (PKS) or non-ribosomal peptide synthetase (NRPS) (Cramer et al., 2006)36 on NSBs were induced in SSC, although on average the NSB genes were highly induced. Similarly, of the SB genes, no genes encoding PKS and NRPS were induced except one PKS gene (AO090701000530). In contrast, the genes encoding hydrolases and transporters, which were highly enriched on NSBs as well, were induced as described above.24 This indicates that the expression profiles of the NSB genes are classified into at least two groups and might suggest the mechanism of gene regulation of A. oryzae in acquiring a high potential for degrading raw materials with a high level of safety. Aspergilli possess more sensor histidine kinases (13–15) than yeast (1–3), whereas the numbers of histidine-containing phosphotransfer factors and response regulators are basically the same. Aspergilli have HK8 histidine kinase family, which is absent in other sequenced filamentous fungi belonging to Sordariomycetes and Dothideomycetes.9 Further, A. oryzae generally has greater numbers of HK6, MAPK, MAPKK and MAPKKK than other filamentous fungi.9 The complex signal transduction cascade may be necessary in order to regulate genes expanded in the A. oryzae genome and may acquire the ability to adapt various growth conditions.
The genes specifically expressed in SSC have been cloned and sequenced by preparing the subtracted cDNA library in which the genes specific to SSC were enriched. Approximately half of the genes expressed specifically in SSC encoded secretory or intracellular enzymes and transporter proteins. Most of the remaining genes were functionally unknown.23,37 Recent proteomic analysis showed that A. oryzae produced a much larger amount of total proteins in SSC (wheat bran) than in submerged cultivation (wheat bran suspension supplemented with ammonium sulfate and phosphate) although more variety of secreted proteins over 50 kDa was produced in submerged culture than in solid-state culture.38 Secretion of a large amount of enzymes was thought to be necessary in order to obtain nutrients under the low water activity. More than half of the entire amount of the visible proteins in the 2D gel electrophoresis is α-amylases and their proteolytic products.
Interestingly, A. oryzae mycelia grown on a nylon membrane placed over a Czapek–Dox medium plate showed significant induction of the alternative glucoamylase gene, glaB, which is known to be one of the typical genes specific to SSC. Small pore size (0.2 µm) of the membrane and high concentration of maltose (50%) in the medium were important for strong induction.39 Environmental factors responsible for transcriptional induction specific to SSC are thought to have low water activity, low diffusion of nutrients and gases, continuity of medium distribution and physical barriers for hyphal extension.39–41 This method is very useful because of its SSC-like conditions and mycelia can be much more easily prepared than native SSC.
2.3. Genome evolution of A. oryzae
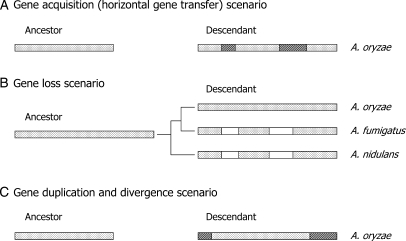
The comparison of three Aspergillus spp. genomes from spp. A. oryzae, A. nidulans and A. fumigatus revealed expansion of genes and genome size. The expansion reached ∼20% of the A. oryzae genome. At least two scenarios must be taken into consideration accounting for the mechanism of the expansion; (i) the ancestor might have a small genome size similar to A. nidulans and A. fumigatus, and might have acquired genetic materials to make the A. oryzae genome bigger, (ii) the ancestor might have a genome size similar to A. oryzae, and the other two species might have lost their genes (Fig. 4A and B). Phylogenomic analysis of the three species revealed that A. nidulans branched off earlier than speciation of A. oryzae and A. fumigatus.42 Therefore, if A. nidulans and A. fumigatus lost their genes as in the second scenario, they have lost the genes independently after speciation. However, considering that the gene organization between the two species seems very similar as shown by the synteny map of the two species and that the number of genes lost from the ancestor reaches nearly 20%,9 the second scenario is unlikely to be accepted. Mosaic distribution of phylogenetically anomalous genes on the chromosomes and apparently similar phylogenetic distances of the expanded genes to their homologs on SBs suggest a horizontal gene transfer (HGT) mechanism for the gene acquisition.9 Recently, Fedorova et al. proposed a gene duplication and divergence (GDD) mechanism43,44 for the acquisition of the A. fumigatus specific genes.45 Nearly half of the A. fumigatus specific genes with paralogs localized in the subtelomeric regions that occupied <20% of the entire genome. The result together with the significant decrease of gene size and intron number of the A. fumigatus specific genes strongly suggests the GDD scenario.45 This suggests that most of the A. oryzae expanded genes might have been acquired by this mechanism (Fig. 4C). Apparent localization of NSBs to the chromosomal ends (see Fig. 2) could support the idea. Nevertheless, there is a clear exception involving two A. oryzae specific genes clustered into the bacterial clades with the highest similarity to Agrobacterium tumefaciens genes (AGR_L_1864, AGR_L_1866).9 Large number of the extra genes in the A. oryzae genome might have been expanded by the combination of the two mechanisms, HGT and GDD.
Figure 4.
Possible mechanism of gene and genome size expansion of A. oryzae. (A) The common ancestor of the three Aspergillus species (A. oryzae, A. fumigatus and A. nidulans) is assumed to have smaller genome size of A. oryzae and similar size to A. fumigatus and A. nidulans. Aspergillus oryzae might have acquired genetic materials (represented by hatched boxes) during evolution. (B) The common ancestor is assumed to have the genome size similar to A. oryzae. The other two species might have lost genetic materials (represented by open boxes) during evolution. The phylogenetic relationship of the three species42 is indicated. (C) The A. oryzae genome size might have been expanded by the gene duplication followed by divergence of one of the duplicated genes (represented by hatched boxes).
The most recent effort for sequencing the A. flavus genome has made it possible to analyze the history of its genome evolution and to evaluate the potential of cellular function of the two species by comparing the two genomes. Aspergillus flavus had genome size (36.8 Mb) and gene number (12 197) very similar to those of A. oryzae (36.7 Mb, 12 079, respectively). Surprisingly, the number of genes found uniquely in each genome was only ∼350 indicating that the genes of the two species are highly similar in terms of amino acid sequence and organizations.46,47 This clearly indicates that the expanded genes in A. oryzae existed from the beginning, i.e. before domestication. Detailed comparative analysis will shed light on the important and interesting subjects mentioned above. Aspergillus flavus, which utilizes nutrients from plants, may require redundant and various secretory hydrolases. During the long history of domestication, the ancestor of the two species lacked the ability to produce poisonous compounds, aflatoxin for example, and these enzymes might have been applied to effective degradation of raw materials during fermentation.
2.4. Applied genomics of A. oryzae
Genomics has facilitated the application of A. oryzae to recycling of biodegradable plastic. Maeda et al.48 found that A. oryzae could grow with poly-(buthylene succinate) (PBS) as a sole carbon source by degrading it with cutinase, CutL1. They applied DNA microarray31 to investigate the additional factor(s) involved in efficient PBS degradation, and found that the rolA (rodA-like) gene was highly transcribed specifically in the presence of PBS.49 This mechanism appears to involve cutinase recruitment to a solid surface, dependent on the conformation of the RolA bound to the hydrophobic surface. Interestingly, they found that RolA bound to the solid PBS surface and specifically recruits CutL1 to the surface, which stimulated hydrolysis of PBS by CutL1.49 Specific interactions between RolA adsorbed on the hydrophobic surface, and CutL1 was confirmed by quartz crystal microbalance50 and immunostaining methods. The interaction dramatically facilitated the PBS degradation catalyzed by CutL1. Furthermore, they found that high levels of fluorescence recovery was observed after photobleaching51 of fluorescein isothiocyanate-labeled RolA, suggesting lateral mobility of RolA on the PBS surface.49 The mechanism could imply a novel tactic for fungal invasion of plants, insects and other animals. Without the genomic approach, RolA, which lacks enzymatic activity that can be measured by conventional biochemistry, could be hardly determined without consuming much time and labor.
Recently, disruption of genes involved in the non-homologous end joining (NHEJ) pathway, ku70 and/or ku80, was found to enhance the gene targeting efficiency in Kluyveromyces lactis and Neurospora crassa.52,53 The novel finding was immediately applied to industrial filamentous fungi, A. oryzae and A. sojae.54 Namely, disruption of ku70 or ku80 enhanced targeting efficiency for A. oryzae from 10% to greater than 60%. However, the efficiency reproducibly was lower than that of ku-disruptants of K. lactis and N. crassa (nearly 100% for both) and suggests the existence of an unknown NHEJ pathway in A. oryzae and A. sojae.54 Nonetheless, the use of ku-disruptant has dramatically enhanced the performance of analyzing gene function by a systematic disruption approach. Most recently, deletion of DNA ligase IV (LigD), another factor involving the NHEJ pathway, resulted in a targeting efficiency as high as 100% in N. crassa and A. oryzae.55,56 The ligD mutant was successfully applied to a comprehensive gene disruption of MAP kinases in A. oryzae.
Aspergillus oryzae and A. fumigatus were revealed to have over 200 genes implicated in the fungal mating process and an alpha and an HMG mating-type gene, respectively. This suggests that both species may be capable of sexual reproduction.42 The sequenced strain of A. fumigatus was found to have an HMG gene. A PCR-based study revealed the existence of A. fumigatus strains with the alpha mating-type gene.57 However, those genes were perfectly associated with vegetative (heterokaryon) compatibility groups (VCGs) among the strains examined, that is, the strains in the same VCG have a single mating-type gene (Natalie C. Fedrova, personal communication). Comparison of the genomes of A. oryzae and A. flavus, considering their close genetic relationship, should be of great interest in addressing the capability of sexual development of A. oryzae.
The novel technology has been applied to the deletion of a large DNA region up to 200 kb.58 This allows the engineering of these species by completely deleting industrially undesirable genes to further improve productivity and safety. Recent technologies, high-resolution mass spectrometry and the next-generation DNA sequencers will further facilitate our understanding of the mechanism that produces various metabolites and will elucidate mutations indispensable for industrial use. To date sequencings of ∼50 fungal genomes have been completed or are near completion (see Table 1 for resources). Comparative genomics and functional genomics in combination with knowledge gained from traditional fermentation technologies should be extremely valuable as a means for both the basic and industrial research of A. oryzae.
Table 1.
Internet resources for fungal genomics
| Organization | URL |
|---|---|
| Broad Institute | http://www.broad.mit.edu/annotation/fgi/ |
| Genoscope | http://www.genoscope.cns.fr/spip/Fungi-sequenced-at-Genoscope.html |
| Joint Genome Institute (JGI) | http://genome.jgi-psf.org/euk_home.html |
| J. Craig Venter Institute (JCVI) | http://pfgrc.jcvi.org/ |
| National Center for Biotechnology Information (NCBI) | http://www.ncbi.nlm.nih.gov/genomes/FUNGI/funtab.html |
| National Institute of Technology and Evaluation (NITE) | http://www.bio.nite.g.o.jp/dogan/Top |
| Sanger Center | http://www.sanger.ac.uk/Projects/Fungi/ |
| Washington University Genome Sequencing Center | http://genome.wustl.edu/sub_genome_group_index.cgi?GROUP=5 |
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Supplementary Material
Acknowledgements
The authors are grateful to Dr Yuichiro Murai of Kojiya Sanzaemon, Co., Ltd and Harunatsuakifuyu Sousyo for providing the photograph of the historical signboard. We wish to thank Dr Hiroshi Konno of Akita Konno Co., Ltd for the introduction on traditional technologies for the industrial production of Moyashi. We also thank Dr Katsumi Isono for critical reading and extensive review.
References
- 1.Christensen T., Woeldike H., Boel E., Mortensen S. B., Hjortshoej K., Thim L., Hansen M. T. High level expression of recombinant genes in Aspergillus oryzae. Bio/Technology. 1988;6:1419–1422. [Google Scholar]
- 2.Murakami H. Koji-gaku (in Japanese) Brewing Society of Japan.; 1980. [Google Scholar]
- 3.Gomi K., Abe K. The Aspergilli: Genomics, Medical Applications, Biotechnology, and Research Methods (Mycology) Boca Raton, FL: CRC Press; 2007. Food products fermented by Aspergillus oryzae; pp. 427–437. [Google Scholar]
- 4.Murai T. Tane-Koji Konjaku Monogatari (in Japanese) Shushi-Kenkyu. 1989;7:39–44. [Google Scholar]
- 5.Hata Y., Ishida H. Glucoamylase-encoding genes of Aspergillus oryzae—Monograph (in Japanese) Seibutsu-kogaku. 2000;78:120–127. [Google Scholar]
- 6.Yu J., Proctor R., Brown D., Abe K., Gomi K., Machida M., Hsegawa F., Bhatnagar D., Cleveland T. E. Genomics of economically significant Aspergillus and Fusarium species. In: Arora D. K., Khachatourians G. G., editors. Applied Mycology and Biotechnology: Fungal Genomics. v. Elsevier; 2003. pp. 249–283. [Google Scholar]
- 7.Hata Y., Ishida H., Kojima Y., Ichikawa E., Kawato A., Suginami K., Imayasu S. Comparison of two glucoamylases produced by Aspergillus oryzae in solid-state culture (koji) and in submerged culture. J. Ferment. Technol. 1997;84:532–537. [Google Scholar]
- 8.Shoji H., Sugimoto T., Hosoi K., Shibata K., Tanabe M., Kawatsura K. Simultaneous production of glucoamylase and acid-stable alpha-amylase using novel submerged culture of Aspergillus kawachii NBRC4308. J. Biosci. Bioeng. 2007;103:203–205. doi: 10.1263/jbb.103.203. [DOI] [PubMed] [Google Scholar]
- 9.Machida M., Asai K., Sano M., Tanaka T., Kumagai T., Terai G., Kusumoto K., Arima T., Akita O., Kashiwagi Y., Abe K., Gomi K., Horiuchi H., Kitamoto K., Kobayashi T., Takeuchi M., Denning D. W., Galagan J. E., Nierman W. C., Yu J., Archer D. B., Bennett J. W., Bhatnagar D., Cleveland T. E., Fedorova N. D., Gotoh O., Horikawa H., Hosoyama A., Ichinomiya M., Igarashi R., Iwashita K., Juvvadi P. R., Kato M., Kato Y., Kin T., Kokubun A., Maeda H., Maeyama N., Maruyama J., Nagasaki H., Nakajima T., Oda K., Okada K., Paulsen I., Sakamoto K., Sawano T., Takahashi M., Takase K., Terabayashi Y., Wortman J. R., Yamada O., Yamagata Y., Anazawa H., Hata Y., Koide Y., Komori T., Koyama Y., Minetoki T., Suharnan S., Tanaka A., Isono K., Kuhara S., Ogasawara N., Kikuchi H. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 10.Klich M. A. Biogeography of Aspergillus species in soil and litter. Mycologia. 2002;94:21–27. [PubMed] [Google Scholar]
- 11.Tailor M. J., Richardson T. Applications of microbial enzymes in food systems and in biotechnology. Adv. Appl. Microbiol. 1979;25:7–35. doi: 10.1016/s0065-2164(08)70144-8. [DOI] [PubMed] [Google Scholar]
- 12.Abe K., Gomi K., Hasegawa F., Machida M. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia. 2006;162:143–153. doi: 10.1007/s11046-006-0049-2. [DOI] [PubMed] [Google Scholar]
- 13.FAO_WHO. Committee on Food Additives 31. Geneva: World Health Organization Technical Report Series; 1987. [PubMed] [Google Scholar]
- 14.Barbesgaard P., Heldt-Hansen H. P., Diderichsen B. On the safety of Aspergillus oryzae: a review. Appl. Microbiol. Biotechnol. 1992;36:569–572. doi: 10.1007/BF00183230. [DOI] [PubMed] [Google Scholar]
- 15.Matsushima K., Chang P. K., Yu J., Abe K., Bhatnagar D., Cleveland T. E. Pre-termination in aflR of Aspergillus sojae inhibits aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2001;55:585–589. doi: 10.1007/s002530100607. [DOI] [PubMed] [Google Scholar]
- 16.Matsushima K., Yashiro K., Hanya Y., Abe K., Yabe K., Hamasaki T. Absence of aflatoxin biosynthesis in koji mold (Aspergillus sojae) Appl. Microbiol. Biotechnol. 2001;55:771–776. doi: 10.1007/s002530000524. [DOI] [PubMed] [Google Scholar]
- 17.Murakami H. Classification of the koji mold. J. Gen. Appl. Microbiol. 1971;17:281–309. [Google Scholar]
- 18.Gomi K., Tanaka A., Iimura Y., Takahashi K. Rapid differentiation of four related species of koji molds by agarose gel electrophoresis of genomic DNA digested with SmaI restriction enzyme. J. Gen. Appl. Microbiol. 1989;35:225–232. [Google Scholar]
- 19.Lee C. Z., Liou G. Y., Yuan G. F. Comparison of the aflR gene sequences of strains in Aspergillus section Flavi. Microbiology. 2006;152:161–170. doi: 10.1099/mic.0.27618-0. [DOI] [PubMed] [Google Scholar]
- 20.Chang P. K., Bhatnagar D., Cleveland T. E., Bennett J. W. Sequence variability in homologs of the aflatoxin pathway gene aflR distinguishes species in Aspergillus section Flavi. Appl. Environ. Microbiol. 1995;61:40–43. doi: 10.1128/aem.61.1.40-43.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y. -Q., Wilkinson H., Keller N. P., Tsitsigiannis D. Secondary metabolite gene clusters. In: An Z., editor. Handbook of Industrial Microbiology. New York: Marcel Dekker; 2005. pp. 355–386. [Google Scholar]
- 22.Kobayashi T., Abe K., Asai K., Gomi K., Juvvadi P. R., Kato M., Kitamoto K., Takeuchi M., Machida M. Genomics of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2007;71:646–670. doi: 10.1271/bbb.60550. [DOI] [PubMed] [Google Scholar]
- 23.Akao T., Sano M., Yamada O., Akeno T., Fujii K., Goto K., Ohashi-Kunihiro S., Takase K., Yasukawa-Watanabe M., Yamaguchi K., Kurihara Y., Maruyama J., Juvvadi P. R., Tanaka A., Hata Y., Koyama Y., Yamaguchi S., Kitamoto N., Gomi K., Abe K., Takeuchi M., Kobayashi T., Horiuchi H., Kitamoto K., Kashiwagi Y., Machida M., Akita O. Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Res. 2007;14:47–57. doi: 10.1093/dnares/dsm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamano K., Sano M., Yamane N., Terabayashi Y., Toda T., Sunagawa M., Koike H., Hatamoto O., Umitsuki G., Takahashi T., Koyama Y., Asai R., Abe K., Machida M. Transcriptional regulation of genes on the non-syntenic blocks of Aspergillus oryzae and its functional relationship to solid-state cultivation. Fungal. Genet. Biol. 2008;45:139–151. doi: 10.1016/j.fgb.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Yu J., Bhatnagar D., Cleveland T. E. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 2004;564:126–130. doi: 10.1016/S0014-5793(04)00327-8. [DOI] [PubMed] [Google Scholar]
- 26.Yu J., Whitelaw C. A., Nierman W. C., Bhatnagar D., Cleveland T. E. Aspergillus flavus expressed sequence tags for identification of genes with putative roles in aflatoxin contamination of crops. FEMS Microbiol. Lett. 2004;237:333–340. doi: 10.1016/j.femsle.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi T., Chang P. K., Matsushima K., Yu J., Abe K., Bhatnagar D., Cleveland T. E., Koyama Y. Nonfunctionality of Aspergillus sojae aflR in a strain of Aspergillus parasiticus with a disrupted aflR gene. Appl. Environ. Microbiol. 2002;68:3737–3743. doi: 10.1128/AEM.68.8.3737-3743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tominaga M., Lee Y. H., Hayashi R., Suzuki Y., Yamada O., Sakamoto K., Gotoh K., Akita O. Molecular analysis of an inactive aflatoxin biosynthesis gene cluster in Aspergillus oryzae RIB strains. Appl. Environ. Microbiol. 2006;72:484–490. doi: 10.1128/AEM.72.1.484-490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bok J. W., Keller N. P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y. H., Tominaga M., Hayashi R., Sakamoto K., Yamada O., Akita O. Aspergillus oryzae strains with a large deletion of the aflatoxin biosynthetic homologous gene cluster differentiated by chromosomal breakage. Appl. Microbiol. Biotechnol. 2006;72:339–345. doi: 10.1007/s00253-005-0282-5. [DOI] [PubMed] [Google Scholar]
- 31.Maeda H., Sano M., Maruyama Y., Tanno T., Akao T., Totsuka Y., Endo M., Sakurada R., Yamagata Y., Machida M., Akita O., Hasegawa F., Abe K., Gomi K., Nakajima T., Iguchi Y. Transcriptional analysis of genes for energy catabolism and hydrolytic enzymes in the filamentous fungus Aspergillus oryzae using cDNA microarrays and expressed sequence tags. Appl. Microbiol. Biotechnol. 2004;65:74–83. doi: 10.1007/s00253-004-1608-4. [DOI] [PubMed] [Google Scholar]
- 32.Chien H. C., Lin L. L., Chao S. H., Chen C. C., Wang W.C., Shaw C. Y., Tsai Y. C., Hu H. Y., Hsu W. H. Purification, characterization, and genetic analysis of a leucine aminopeptidase from Aspergillus sojae. Biochim. Biophys. Acta. 2002;1576:119–126. doi: 10.1016/s0167-4781(02)00307-x. [DOI] [PubMed] [Google Scholar]
- 33.Tatsumi H., Murakami S., Tsuji R. F., Ishida Y., Murakami K., Masaki A., Kawabe H., Arimura H., Nakano E., Motai H. Cloning and expression in yeast of a cDNA clone encoding Aspergillus oryzae neutral protease II, a unique metalloprotease. Mol. Gen. Genet. 1991;228:97–103. doi: 10.1007/BF00282453. [DOI] [PubMed] [Google Scholar]
- 34.Murakami K., Ishida Y., Masaki A., Tatsumi H., Murakami S., Nakano E., Motai H., Kawabi H., Arimura H. Isolation and characterization of the alkaline protease gene of Aspergillus oryzae. Agric. Biol. Chem. 1991;55:2807–2811. [PubMed] [Google Scholar]
- 35.Hata Y., Ishida H., Ichikawa E., Kawato A., Suginami K., Imayasu S. Nucleotide sequence of an alternative glucoamylase-encoding gene (glaB) expressed in solid-state culture of Aspergillus oryzae. Gene. 1998;207:127–134. doi: 10.1016/s0378-1119(97)00612-4. [DOI] [PubMed] [Google Scholar]
- 36.Cramer R. A., Jr, Stajich J. E., Yamanaka Y., Dietrich F. S., Steinbach W. J., Perfect J. R. Phylogenomic analysis of non-ribosomal peptide synthetases in the genus Aspergillus. Gene. 2006;383:24–32. doi: 10.1016/j.gene.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Akao T., Gomi K., Goto K., Okazaki N., Akita O. Subtractive cloning of cDNA from Aspergillus oryzae differentially regulated between solid-state culture and liquid (submerged) culture. Curr. Genet. 2002;41:275–281. doi: 10.1007/s00294-002-0314-y. [DOI] [PubMed] [Google Scholar]
- 38.Oda K., Kakizono D., Yamada O., Iefuji H., Akita O., Iwashita K. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 2006;72:3448–3457. doi: 10.1128/AEM.72.5.3448-3457.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishida H., Hata Y., Ichikawa E., Kawato A., Suginami K., Imayasu S. Regulation of the glucoamlase-encoding gene (glaB), expressed in solid-state culture (koji) of Aspergillus oryzae. J. Ferment. Bioeng. 1998;86:301–307. doi: 10.1016/s0378-1119(97)00612-4. [DOI] [PubMed] [Google Scholar]
- 40.Gervais P., Bensoussan M. Solid-state fermentations of the genus Aspergillus. In: Smith J. E., editor. Aspergillus. Plenum Press; 1994. pp. 101–140. [Google Scholar]
- 41.Ishida H., Hata Y., Kawato A., Abe Y., Suginami K., Imayasu S. Identification of functional elements that regulate the glucoamylase-encoding gene (glaB) expressed in solid-state culture of Aspergillus oryzae. Curr. Genet. 2000;37:373–379. doi: 10.1007/s002940000118. [DOI] [PubMed] [Google Scholar]
- 42.Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R., Batzoglou S., Lee S. I., Basturkmen M., Spevak C. C., Clutterbuck J., Kapitonov V., Jurka J., Scazzocchio C., Farman M., Butler J., Purcell S., Harris S., Braus G. H., Draht O., Busch S., D'Enfert C., Bouchier C., Goldman G. H., Bell-Pedersen D., Griffiths-Jones S., Doonan J. H., Yu J., Vienken K., Pain A., Freitag M., Selker E. U., Archer D. B., Penalva M. A., Oakley B. R., Momany M., Tanaka T., Kumagai T., Asai K., Machida M., Nierman W. C., Denning D. W., Caddick M., Hynes M., Paoletti M., Fischer R., Miller B., Dyer P., Sachs M. S., Osmani S. A., Birren B. W. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 43.Kimura M., Ota T. On some principles governing molecular evolution. Proc. Natl. Acad. Sci. USA. 1974;71:2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurland C. G., Canback B., Berg O. G. Horizontal gene transfer: a critical view. Proc. Natl. Acad. Sci. USA. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fedorova N. D., Khaldi N., Joardar V., Maiti R., Amedeo P., Anderson M. J., Crabtree J., Silva J. C., Badger J. H., Albarraq A., Angiuoli S., Bussey H., Bowyer P., Cotty P. J., Dyer P. S., Egan A., Galens K., Fraser C., Haas B., Inman J., Kent R., Lemieux S., Malavazi I., Orvis J., Roemer T., Ronning C. M., Sundaram J. P., Sutton G., Turner G., Venter J. C., White O. R., Whitty B. R., Yougman P., Wolfe K. H., Goldman G. H., Wortman J. R., Jiang B., Denning D. W., Nierman W. C. Genomic Islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genomics. 2008 doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Payne G. A., Nierman W. C., Wortman J. R., Pritchard B. L., Brown D., Dean R. A., Bhatnagar D., Cleveland T. E., Machida M., Yu J. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 2006;44:9–11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 47.Payne G. A., Yu J., Nierman W. C., Machida M., Barhtnagar D., Cleveland T. E., Dean R. A. A first glance into the genome sequence of Aspergillus flavus. In: Osmani S. A., Goldman G. H., editors. The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods. CRC Press; 2007. pp. 12–24. [Google Scholar]
- 48.Maeda H., Yamagata Y., Abe K., Hasegawa F., Machida M., Ishioka R., Gomi K., Nakajima T. Purification and characterization of a biodegradable plastic-degrading enzyme from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2005;67:778–788. doi: 10.1007/s00253-004-1853-6. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi T., Maeda H., Yoneda S., Ohtaki S., Yamagata Y., Hasegawa F., Gomi K., Nakajima T., Abe K. The fungal hydrophobin RolA recruits polyesterase and laterally moves on hydrophobic surfaces. Mol. Microbiol. 2005;57:1780–1796. doi: 10.1111/j.1365-2958.2005.04803.x. [DOI] [PubMed] [Google Scholar]
- 50.Sato Y., Sagami I., Shimizu T. Identification of caveolin-1-interacting sites in neuronal nitric-oxide synthase. Molecular mechanism for inhibition of NO formation. J. Biol. Chem. 2004;279:8827–8836. doi: 10.1074/jbc.M310327200. [DOI] [PubMed] [Google Scholar]
- 51.Giese B., Au-Yeung C. K., Herrmann A., Diefenbach S., Haan C., Kuster A., Wortmann S. B., Roderburg C., Heinrich P. C., Behrmann I., Muller-Newen G. Long term association of the cytokine receptor gp130 and the Janus kinase Jak1 revealed by FRAP analysis. J. Biol. Chem. 2003;278:39205–39213. doi: 10.1074/jbc.M303347200. [DOI] [PubMed] [Google Scholar]
- 52.Ninomiya Y., Suzuki K., Ishii C., Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kooistra R., Hooykaas P. J., Steensma H. Y. Efficient gene targeting in Kluyveromyces lactis. Yeast. 2004;21:781–792. doi: 10.1002/yea.1131. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi T., Masuda T., Koyama Y. Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol. Genet. Genomics. 2006;275:460–470. doi: 10.1007/s00438-006-0104-1. [DOI] [PubMed] [Google Scholar]
- 55.Ishibashi K., Suzuki K., Ando Y., Takakura C., Inoue H. Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc. Natl. Acad. Sci. USA. 2006;103:14871–14876. doi: 10.1073/pnas.0604477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fedorova N. D., Khaldi N., Joardar V., Maiti R., Amedeo P., Anderson M. J., Crabtree J., Silva J. C., Badger J. H., Albarraq A., Angiuoli S., Bussey H., Bowyer P., Cotty P. J., Dyer P. S., Egan A., Galens K., Fraser C., Haas B., Inman J., Kent R., Lemieux S., Malavazi I., Orvis J., Roemer T., Ronning C. M., Sundaram J. P., Sutton G., Turner G., Venter J. C., White O. R., Whitty B. R., Yougman P., Wolfe K. H., Goldman G. H., Wortman J. R., Jiang B., Denning D. W., Nierman W. C. Genomic Islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genomic. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paoletti M., Rydholm C., Schwier E. U., Anderson M. J., Szakacs G., Lutzoni F., Debeaupuis J. P., Latge J. P., Denning D. W., Dyer P. S. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 2005;15:1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi T., Jin F. -J., Senou Y., Koyama Y. Artificial generation of large chromosomal deletions in Aspergillus oryzae and Aspergillus sojae by using ku70. 9th European Conference of Fungal Genetics.2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.