Abstract
The gram-negative anaerobic bacterium Porphyromonas gingivalis is a major causative agent of chronic periodontitis. Porphyromonas gingivalis strains have been classified into virulent and less-virulent strains by mouse subcutaneous soft tissue abscess model analysis. Here, we present the whole genome sequence of P. gingivalis ATCC 33277, which is classified as a less-virulent strain. We identified 2090 protein-coding sequences (CDSs), 4 RNA operons, and 53 tRNA genes in the ATCC 33277 genome. By genomic comparison with the virulent strain W83, we identified 461 ATCC 33277-specific and 415 W83-specific CDSs. Extensive genomic rearrangements were observed between the two strains: 175 regions in which genomic rearrangements have occurred were identified. Thirty-five of those genomic rearrangements were inversion or translocation and 140 were simple insertion, deletion, or replacement. Both strains contained large numbers of mobile elements, such as insertion sequences, miniature inverted-repeat transposable elements (MITEs), and conjugative transposons, which are frequently associated with genomic rearrangements. These findings indicate that the mobile genetic elements have been deeply involved in the extensive genome rearrangement of P. gingivalis and the occurrence of many of the strain-specific CDSs. We also describe here a very unique feature of MITE400, which we renamed MITEPgRS (MITE of P. gingivalis with Repeating Sequences).
Key words: Porphyromonas gingivalis, whole genome sequence, genome rearrangement, conjugative transposon, MITE
1. Introduction
Periodontal disease, the major cause of tooth loss in the general populations of industrial nations,1,2 is a chronic inflammatory disease of the periodontium that leads to erosion of the attachment apparatus and supporting bone for teeth3 and is one of the most frequently occurring infectious diseases in humans.4 Recently, a number of epidemiological studies have shown significant relationships between periodontal diseases and cardiovascular diseases.5–8 Several periodontal pathogens, including Porphyromonas gingivalis, have been found in atherosclerotic plaques.9,10
Porphyromonas gingivalis is a gram-negative anaerobic bacterium that is classified in the genus Porphyromonas, family Porphyromonadaceae, order Bacteroidales, class Bacteroides, phylum Bacteroidetes.11 The bacterium, which is often found in deep periodontal pockets of humans, is asaccharolytic and highly proteolytic. Porphyromonas gingivalis produces a broad array of potential virulence factors involved in tissue colonization and destruction as well as host defense perturbation. Potential virulence factors of P. gingivalis have been extensively described in several reviews.12–14 Among these, fimbriae (FimA fimbriae and Mfa1 fimbriae), which are responsible for attachment of bacterial cells to host cell surfaces, and proteolytic enzymes such as Arg-gingipain (Rgp) and Lys-gingipain (Kgp), which degrade various host proteins, have been studied in detail.15–17 However, no systematic analysis of P. gingivalis virulence factors has yet been carried out.
Using the mouse subcutaneous soft tissue abscess model, P. gingivalis strains are divided into virulent and less-virulent strains. Virulent strains such as W83 typically induce a necrotic lesion at the site of injection within 24 h. An extending gangrene-like necrosis spreads, secondarily along fascial planes into the abdominal and thoracic areas between 24 and 48 h, producing a foul-smelling and bloody exudate under the animal's skin. Sloughing and scab formation from necrosis of the epidermis, weight and hair loss, and animal death from microbial sepsis also are common clinical features. Less-virulent strains such as ATCC 33277 produce only a localized abscess 3 days after subcutaneous inoculation.18 W83 has already been genome-sequenced.19 Strain ATCC 33277 is the type strain of P. gingivalis and has been widely used for characterization of pathophysiological features of the microorganism. We have used molecular genetic techniques to study P. gingivalis and have constructed a number of mutants from strain ATCC 33277 to investigate the roles of various genes in the pathogenicity of P. gingivalis.20–25 In the present study, we determined the whole genome sequence of strain ATCC 33277 and performed a genomic comparison of ATCC 33277 and W83. Our findings showed that extensive genome rearrangements have taken place between the two strains. Transposable elements appear to have played central roles in the generation of these genome rearrangements, which have in turn created many strain-specific genes, including several potentially virulence-related genes.
2. Materials and methods
2.1. Genome sequencing
Porphyromonas gingivalis ATCC 33277 was obtained from the American Type Culture Collection (ATCC) and has been kept for more than 20 years. Porphyromonas gingivalis W83 was obtained from Dr M. J. Duncan (Department of Molecular Genetics, The Forsyth Institute). Porphyromonas gingivalis GAI7802 was obtained from Dr E. Hoshino (Niigata University School of Dentistry), TDC60, TDC117, and TDC275 were obtained from Dr K. Ishihara (Tokyo Dental College), and SU63 was obtained from Dr M. Yoneda (Fukuoka Dental College). For preparing the genomic DNA, a single colony of each P. gingivalis strain was grown at 37°C anaerobically (10% CO2, 10% H2, 80% N2) in brain heart infusion broth (BD Bioscience, San Jose, CA, USA) supplemented with 5 µg of hemin and 0.5 µg of menadione per ml.
The genomic DNA was randomly sheared by Hydroshear (GeneMachines) and used for genomic library construction. We prepared two pTS1-based random genomic libraries with insert sizes of 1–2 kb or ∼10 kb. Sequencing was carried out using BigDye v3.1 chemistry on ABI 3700 or ABI 3730 sequencers (Applied Biosystems) or ET chemistry on MegaBACE 4500 sequencers (GE Healthcare). The whole genome sequence was obtained by assembling 36 394 reads (9.5-fold coverage) from both shotgun libraries. The Phred/Phrap software package26 was used for base-calling, quality assessment, and sequence assembly. Assemblies were visualized for counting-based variations and detecting misassembly using Consed software.27 Numbers and lengths of the NotI fragments of the ATCC 33277 chromosome predicted from the determined nucleotide sequence agreed well with those observed in pulsed field gel electrophoresis analysis of NotI-digested ATCC 33277 genomic DNA.28
2.2. Sequence analysis
Protein-coding sequences (CDSs) were identified by using the combination of GENOME GAMBLER v1.51,29 CRITICA,30 GENEHACKER,31 and GLIMMER v2.0 programs.32 The sequences of 3′ terminal regions of all 16S ribosomal RNA genes in W83, and ATCC 33277 were identical to that of Bacteroides fragilis (AGAAAGGAGG, the accession number M61006). Each CDS was thus reviewed manually for the presence of a start codon (ATG, TTG, or GTG) and a potential ribosome-binding sequence that should be related to a part of the AGAAAGGAGG sequence. Functional annotation of the CDSs was made on the basis of results of homology searches against public protein (nr) database from NCBI (http://www.ncbi.nlm.nih.gov/) by the BLASTP program.33 tRNA genes were identified by the tRNAscan-SE program.34 All-to-all BLASTP analysis of CDSs was performed between W83 and ATCC 33277 to identify conserved and strain-specific CDSs. Since each genome contained a number of multi-copied CDSs such as transposase genes, we first grouped these multi-copied CDSs in each genome using BLASTCLUST33 (The threshold used was ≥90% amino-acid sequence identity.). The largest CDS in each multi-copied CDS group was used as the representative of each group for the identification of conserved and strain-specific CDSs. In the present study, we defined conserved CDSs as ones that had 60–140% of the length of a query sequence and showed ≥90% sequence identity in bidirectional best-hit analysis. The MUMmer program35 was used to define conserved genomic regions, inversions, and translocations between the two genomes. The Pip Maker program36 was also used for DNA sequence alignment.
2.3. PCR analysis
The total genomic DNA of ATCC 33277 and two pairs of PCR primers (CTnPg1-left and CTnPg1-right, CTnPg1-up and CTnPg1-down, Supplementary Table S1) were used to detect the excision of a conjugative transposon (CTn), CTnPg1. These primer pairs were designed to amplify the attP and attB regions for CTnPg1, respectively. PCR amplification was performed by using 100 ng of the genomic DNA and LA Taq (Takara Shuzo, Tokyo, Japan) in the following setting: preheating (94°C for 1 min) and 30 cycles of DNA denaturation (94°C for 20 s), primer annealing (55°C for 30 s) and DNA extension (68°C for 2 min). The amplified fragments were subjected to direct sequencing to determine the nucleotide sequences of the attP and attB sites.
2.4. RNA isolation and real-time PCR
Porphyromonas gingivalis cells were grown to the mid-exponential phase. Total RNA was isolated from the harvested cells using an RNeasy Mini Kit (Qiagen Sciences, Valencia, CA, USA) and reverse-transcribed in a reaction mixture containing a random primer (Promega Co., Madison, WI, USA), dNTP mixture, RNase inhibitor (Wako Pure Chemical Industries, Ltd., Osaka, Japan), dithiothreitol, Superscript II (Invitrogen, Carlsbad, CA, USA), and DEPC-treated water. Real-time quantitative PCR was performed using Full Velocity SYBR Green QPCR Master Mix (Stratagene, La Jolla, CA, USA) according to the manufacturer's instructions. Primer sequences for the real-time PCR are listed in Supplementary Table S1. PCR amplification was performed in the following setting: preheating (95°C for 5 min) and 30 cycles of DNA denaturation (95°C for 10 s), and primer annealing/DNA extension (60°C for 30 s). The expression level of each targeted gene was normalized to that of the 16S rRNA gene. The comparative cycle threshold method37 was used for relative quantification.
2.5. Nucleotide sequence accession numbers
The fully annotated genome sequence of P. gingivalis strain ATCC 33277 has been deposited in GenBank/EMBL/DDBJ databases under the accession number AP009380. Nucleotide sequences of the glucose kinase-encoding genes (glk) of five P. gingivalis strains have been deposited under the accession numbers AB293447 (strain TDC60), AB293448 (TDC117), AB293449 (TDC275), AB293450 (SU63), and AB293451 (GAI7802).
3. Results and discussion
3.1. General features of the ATCC 33277 genome
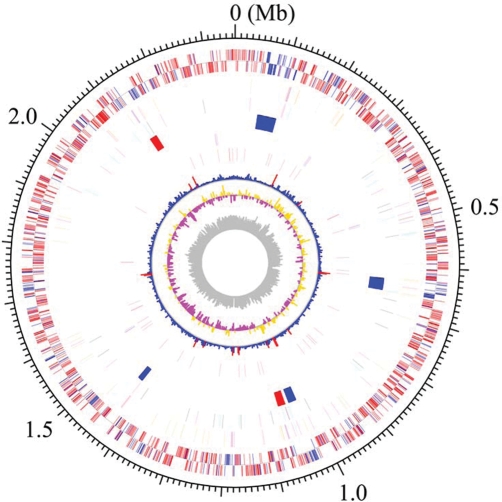
The genome of ATCC 33277 comprised a single circular chromosome of 2 354 886 bp with an average G + C content of 48.4% (Fig. 1). The size was almost the same as that of W83 (2 343 476 bp). The ATCC 33277 genome contained 2090 CDSs (PGN No.) with an average size of 970 bp, covering 86.1% of the whole chromosome sequence. It contained 4 RNA operons (rrn, 5S rRNA-23S rRNA-tRNAAla-tRNAIle-16S rRNA) and 53 tRNA genes that provide specificity for all kinds of amino acids. The numbers of rrn operons and tRNA genes were identical to those of W83. By the χ2 analysis, we identified 13 regions with atypical nucleotide composition on the ATCC 33277 chromosome (Fig. 1, 8th circle). Many genes in the regions exhibited higher similarity to the genes in other bacterial species such as B. fragilis than those in strain W83, suggesting that they have been introduced to ATCC 33277 by horizontal gene transfer.
Figure 1.
Circular map of the chromosome of P. gingivalis strain ATCC 33277. From the outside, the first and second circles show CDSs on the plus and minus strands, respectively. CDSs conserved in strains ATCC 33277 and W83 are indicated in red and ATCC 33277-specific CDSs in blue. The 3rd to 5th circles show IS elements (orange, ISPg1; light green, ISPg2; magenta, ISPg3; cyan, ISPg4; brown, ISPg5; blue, ISPg6), MITEs (magenta, MITE239; black, MITEPgRS; cyan, MITE700), CTns, and Tns (blue, CTnPg1-a, CTnPg1-b, CTnPg2, and CTnPg3; red, TnPg17), respectively. The 6th and 7th circles show rrn operons and tRNA genes, respectively. The 8th circle shows the result of χ2 analysis of nucleotide composition. Regions exhibiting values of >600 are indicated in red and those of <600 are indicated in blue. The G + C skew and G + C content are shown in the 9th and 10th circles, respectively.
3.2. Strain-specific CDSs
To more precisely compare the CDS sets encoded on the ATCC 33277 and W83 genomes, we reannotated CDSs on the W83 genome by the same criteria as those used for ATCC 33277. We detected 114 CDSs (PGa No.) in the W83 genome that had not been annotated by Nelson et al.19 They included many fragments of transposases of insertion sequences (ISs) but at least 27 function-assignable genes, such as those for translocase SecE subunit, pseudouridine synthases, and lysine-specific cysteine proteinase (Supplementary Table S2). In total, we identified 2023 CDSs in W83.
ATCC 33277 and W83 genomes had a number of multi-copied CDSs such as those for IS transposases. ATCC 33277 and W83 contained 53 and 32 multi-copied CDS groups, respectively; the gene products of each group exhibited ≥90% amino-acid sequence identity (Supplementary Tables S3 and S4). In bidirectional best-hit analysis to identify conserved CDSs between the two strains, we used the largest CDS in each multi-copied CDS group as the representative of each group. By the analysis, we identified 1490 conserved CDSs, 461 ATCC 33277-specific CDSs, and 415 W83-specific CDSs. The strain-specific CDSs are listed in Supplementary Tables S5 and S6, respectively.
Most of the strain-specific CDSs encoded hypothetical proteins of unknown functions, but function-predictable W83-specific CDSs included several genes that may be related to the higher virulence of the strain, such as those for glycosyltransferase (PG0110), a protein required for capsular polysaccharide biosynthesis (PG0111), sensor histidine kinase (PG0719), surface antigen PgaA (PG0742), and thiol protease (PG1055). The two strains encoded different sets of DNA restriction-modification system proteins.
3.3. CTn and transposon
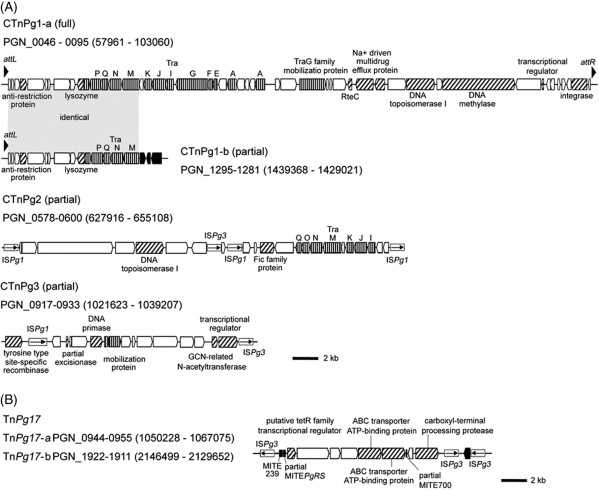
ATCC 33277 contained a variety of mobile genetic elements. We found four copies of CTns that were absent in W83 (Fig. 2). CTnPg1-a is 44.3 kb in size and encodes 50 CDSs (PGN_0046 to PGN_0095), including a set of genes for conjugative transfer and integration as well as those for an Na+-driven multi-drug efflux pump. Several genes showed moderate sequence homologies to the genes of CTns of Bacteroides species.38–40 CTnPg1-b, which is 9.7 kb in size and encodes 15 CDSs (PGN_1281 to PGN_1295), is identical to a part of CTnPg1-a (PGN_0046 to PGN_0060). One end of CTnPg1-b has been disrupted by multiple IS insertions. We identified two additional CTns, CTnPg2 and CTnPg3, but both were also truncated and highly degraded by multiple IS insertions.
Figure 2.
Novel CTns and Tn identified in the ATCC 33277 genome. (A) Structures of CTnPg1-a, CTnPg1-b, CTnPg2, and CTnPg3. (B) Structure of TnPg17. CDSs are depicted by arrows and IS elements by open boxes (vertically striped arrow, tra or mob genes; thin arrow in box, IS transposase; black arrow, partial transposase; hatched arrow, other functionally annotated CDS; white arrow, hypothetical protein). Black triangles in CTnPg1-a and CTnPg1-b indicate direct repeat sequences, and black boxes in TnPg17 MITEs. The regions of CTnPg1-a and CTnPg1-b indicated by gray shading have an identical sequence.
We identified two identical copies of a novel composite transposon (Tn) named TnPg17-a and TnPg17-b in the ATCC 33277 genome. TnPg17 is 16.8 kb in size and has ISPg3 at both ends. Target site duplications of 4 and 7 bp were found for TnPg17-a and TnPg17-b, respectively. TnPg17 carries genes for a tetR family transcriptional regulator, ABC transporter ATP-binding proteins, and a carboxyl-terminal processing protease.
3.4. IS and miniature inverted-repeat transposable element
A total of 93 IS elements (including 38 partial copies) and 48 miniature inverted-repeat transposable elements (MITEs) (including 18 partial copies) were found in ATCC 33277 (Table 1). The IS elements identified were classified into six types, ISPg1–ISPg6, all of which are also present in W83.19 MITEs comprise a group of small mobile genetic elements and are massively amplified often in plants.41 They do not encode transposases by themselves but have terminal inverted repeats (TIRs) that are the same as or very similar to those of some IS elements, and they are thus transposable by the action of transposase provided in trans by the cognate IS element. In ATCC 33277, we identified three types of MITEs, MITE239, MITE700, and MITE464, all of which have also been identified on the W83 genome.19 The structure of MITE239 was well conserved between the copies, 239 bp in length and with the same TIR as that of ISPg3. However, MITE700 and MITE464 exhibited highly variable structures, and their structural features have not been described in detail in the previous report.19 Therefore, we first determined the structural features of these two MITEs by comparing the sequences of all of the MITE700 and MITE464 copies identified in ATCC 33277.
Table 1.
IS elements and MITEs on the ATCC 33277 and W83 genomes
| Mobile genetic element | Strain |
|
|---|---|---|
| ATCC 33277 Number of intact copies (number of fragments) |
W83a Number of intact copies (number of fragments) |
|
| ISPg1 | 31 (18) | 11 (40) |
| ISPg2 | 2 (4) | 5 (3) |
| ISPg3 | 22 (7) | 4 (5) |
| ISPg4 | 0 (2) | 10 (0) |
| ISPg5 | 0 (4) | 10 (1) |
| ISPg6 | 0 (3) | 1 (2) |
| ISPg7 | 0 (0) | 1 (0) |
| MITE239 | 12 (2) | 5 (0) |
| MITEPgRS | 11 (9) | 14 (7) |
| MITE700 | 7 (7) | 7 (2) |
aDetermined based on the reannotated W83 data.
MITE700 also contains the same TIRs as those of ISPg3, but its internal sequence is not related to that of MITE239. We identified 14 copies of MITE700 (including seven partial copies) in ATCC 33277. Multiple sequence alignment analysis revealed that MITE700 is ∼720 bp in length but exhibits a high sequence variation due to internal deletions/insertions. The sequence of a region located just downstream of the left TIR is highly variable between the copies. We identified three subtypes of MITE700 based on the sequence variation in this region (Supplementary Fig. S1).
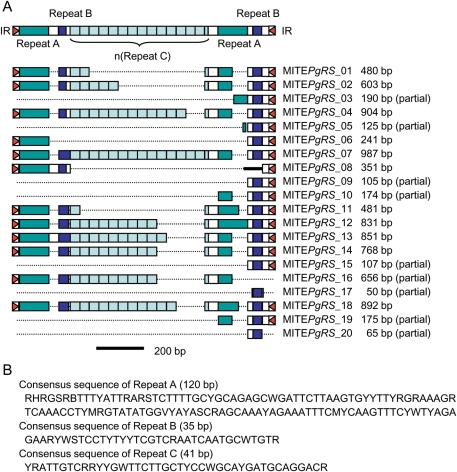
MITE464 has the same TIRs as those of ISPg1. We found that its internal sequence consists of three types of repeat sequences (referred to as Repeats A, B, and C) (Fig. 3 and Supplementary Fig. S2). Repeats A and B exist just inside both TIRs, constituting a direct-repeat-like structure. Between the two Repeat A/B regions, multiple Repeat C regions exist in tandem, but the number of Repeat C regions varies between MITE464 copies, up to 14 repeats. Since no such repeating structure has been found in any MITEs described so far, we propose a new name for this MITE, MITEPgRS (MITE of P. gingivalis with Repeating Sequences).
Figure 3.
MITE in P. gingivalis with Repeating Structure (MITEPgRS). (A) Schematic presentation of the consensus structure of MITEPgRS and the structures of 20 copies of MITEPgRS identified in the ATCC 33277 genome are shown. Three kind of repeat sequences, Repeats A, B, and C, are depicted by colored boxes. Red triangles indicate IR sequences and a black thick line in MITEPgRS_08 a unique nucleotide sequence. (B) Consensus sequences of Repeats A, B, and C are shown.
On the basis of the structural features of each MITE, we rescreened IS elements and MITEs on the W83 genome, and we identified 93 IS elements (including 51 partial ones) and 35 MITEs (including nine partial ones) (Table 1 and Supplementary Table S7). The total numbers of IS elements and MITEs of the two strains were very similar, but the compositions showed a significant difference. In ATCC 33277, ISPg1, ISPg3, and MITE239 have been significantly expanded, whereas no intact copies of ISPg4 and ISPg5 are present. A previous study also showed that the numbers of copies and insertion sites of ISPg1 (formerly IS1126) markedly varied among P. gingivalis strains.42 The composition of IS elements and MITEs may be an important feature to distinguish strains of P. gingivalis.
3.5. Genome rearrangement
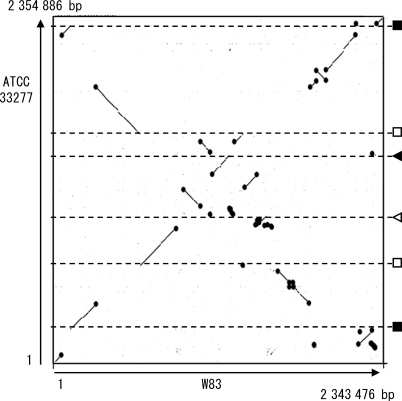
Whole genome sequence alignment analysis by using the MUMmer program revealed that extensive genomic rearrangements have taken place between ATCC 33277 and W83 (Fig. 4). A number of X-shaped structures were observed, indicating that symmetrical inversions repeatedly occurred around the replication axis. We identified a total of 175 genomic regions in which genomic rearrangements took place in either strain. Inversions or translocations were observed in 35 regions, and simple insertions, deletions, or replacements were observed in 140 regions. Remarkably, about two-thirds of these genomic rearrangements were associated with the presence of mobile genetic elements (IS, MITE, Tn, CTn, and a not well-defined large mobile element of W83). Large inversions or translocations have occurred between two rrn operons, duplicated DNA regions coding for a histone-like family DNA-binding protein (PGN_0614 and PGN_1407), a hypothetical protein (PGN_0615 and PGN_1406), elongation factor P (PGN_0616 and PGN_1405), two identical 11 bp sequences (TAATCATAATA), and two similar 12/13 bp sequences (TTTTC(GCC/AATG)AAAA). DNA sequences similar to the 12/13 bp sequences were also present in the att sites for CTnPg1 described in the following section. These rearrangements appear to be deeply involved in the generation of strain-specific CDSs: 60% of ATCC 33277-specific CDSs and 68% of W83-specific CDSs were created by these genomic rearrangements.
Figure 4.
The DNA sequence identity plot of P. gingivalis ATCC 33277 and W83 chromosomes. The dnaA gene is located at the left and bottom corner. Black circles indicate mobile genetic elements (CTn Tn, IS, MITE, or a not-well-defined large mobile element of W83). The chromosomal locations of other genetic elements that mediated inversions or translocations are shown in the right: rrn operons (black squares), duplicated regions coding for a histone-like DNA binding protein, a hypothetical protein and elongation factor P (open squares), 12/13 bp repeat sequences (black triangle), and 11 bp repeat sequences (open triangle).
ATCC 33277 and W83 both contain four rrn operons of identical nucleotide sequences, but chromosomal locations of the four rrn operons differ markedly between the strains. By comparing the rrn operon-flanking regions in the two strains, we found that an inversion had taken place between rrn1 and rrn4 (see Supplementary Fig. S3). Additional genomic rearrangements that have occurred in the genomic loci other than rrn operons further altered the relative locations of the four rrn operons on the two genomes. We analyzed the structures of rrn operon-flanking regions in five other strains of P. gingivalis (TDC60, TDC117, TDC275, SU63, and GAI7802) using a set of orientation-specific primer pairs, and we found that all of the rrn operon-flanking regions of these five strains have the same structures as those of ATCC 33277 (Supplementary Fig. S3). This result suggests that inversions between rrn1 and rrn4 have taken place specifically in the W83 strain lineage among the strains tested.
The genomic regions for the biosynthesis of cell surface molecules have also significantly diverged between the two strains. They included the regions for FimA fimbrilin, Mfa1 fimbrilin, capsular polysaccharides, RagA and RagB antigens, and glycosyl transferase.42–45 Among these, the difference in the locus for capsular polysaccharide biosynthesis (GP1 locus) is particularly important because capsular polysaccharide is known to be one of the major virulence factors of P. gingivalis. The GP1 locus of ATCC 33277 (PGN_0223-PGN_0236) is identical to that reported for strain 38143 except that one nonsense mutation was found in the PGN_0223-homolog of strain 381 (Supplementary Fig. S4).
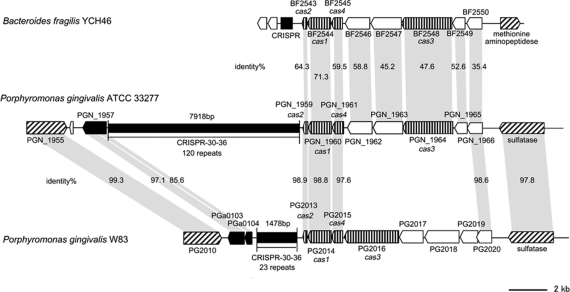
The clustered regularly interspaced short palindromic repeats (CRISPR) locus also exhibits notable structural difference between the two strains. The numbers of repeats in the repeat/spacer region of the CRISPR-30-36 locus are 120 in ATCC 33277 and 23 in W83 (Fig. 5). The nucleotide sequences of the repeats are identical, but there is no homology in the spacer regions. A part of the CRISPR-associated gene (cas) encoding region has been replaced by very different sequences. Of interest is that the gene organization of the cas-encoding region of ATCC 33277 is nearly identical to that of B. fragilis YCH46 but very different from that of W83. The gene products also exhibit a high level of similarity of amino-acid sequences to those of B. fragilis YCH46. These results suggest that these genes may have been horizontally transferred between P. gingivalis and B. fragilis.
Figure 5.
Comparison of CRISPR-30-36 regions of P. gingivalis and B. fragilis. Locations and directions of CDSs (arrows) and repeat regions (black rectangles) are drawn to scale. Homologous CDSs are indicated by gray shading, and their amino-acid sequence identities are also shown. CDSs for IS transposases are indicated by black arrows, cas genes by vertically striped arrows, other functionally annotated CDSs by hatched arrows, and CDSs for hypothetical proteins by white arrows. The identity between PGN_1964 and PG2016 is 15.7%.
Among the bacterial species so far sequenced, Bacteroides species are phylogenically most closely related to P. gingivalis. However, two sequenced B. fragilis strains show no such extensive genomic rearrangement as seen in P. gingivalis.40,46 Shigella flexneri, a pathogen for dysentery, contains a large number of IS elements and the bacterium has induced extensive genomic rearrangements among strains, which may create differences in virulence and epidemicity.47,48 In P. gingivalis, the genomic rearrangements induced by IS and other mobile genetic elements may also have been involved in the generation of strain-to-strain difference in virulence. In this context, a recent finding that treatment of P. gingivalis cells with H2O2 induces expression of the ISPg1 transposase gene is noteworthy.49 The fact that the number of copies of ISPg1 varies among P. gingivalis strains and the fact that ISPg1 is frequently associated with genome rearrangements suggest that oxidative stress-induced expression of the ISPg1 transposase gene results in transposition of ISPg1 that may mediate genomic rearrangements in P. gingivalis, and such rearrangements may contribute to the adaptation of P. gingivalis strains to an oxygen concentration-changeable environment in the gingival crevice.
3.6. Excision of CTnPg1
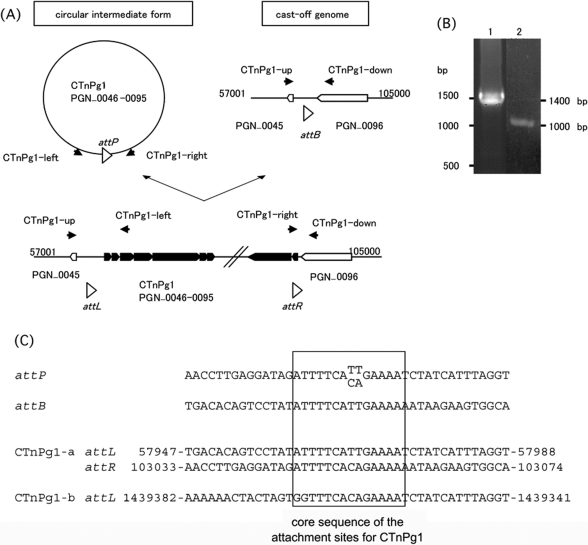
We could not exactly determine the attachment (att) site of CTnPg1 by comparing the genome sequences of ATCC 33277 and W83 because integration of CTnPg1-a has induced a genomic rearrangement and half of CTnPg1-b has been deleted. We therefore examined whether CTnPg1 can be excised from the chromosome and form a circular intermediate. By PCR analysis of the chromosomal DNA of ATCC 33277 using two primers targeting the left and right ends of CTnPg1 (Fig. 6A), we obtained a PCR product of 1500 bp in size (Fig. 6B). We further investigated whether a cast-off genome can be generated by the excision of CTnPg1 using two PCR primers targeting the CTnPg1-a-flanking regions, and we obtained a PCR product of 1000 bp in size (Fig. 6A and B). By comparing the sequences of the two PCR products with the genome sequence of ATCC 33277, we identified the core sequence of the att site for CTnPg1, ATTTTCA(CA/TT)GAAAA (Fig. 6C). The same sequence was also found at one end of CTnPg1-b.
Figure 6.
Excision of CTnPg1-a. (A) Schematic presentation of the structure of CTnPg1-a and the strategy to detect the excised circular intermediate and cast-off chromosome. Locations of PCR primers are indicated by black arrow heads. CDSs on CTnPg1 are depicted by black arrows and other CDSs by open arrows. Open triangles indicate att regions of CTnPg1. (B) Agarose gel electrophoresis of PCR products obtained by the primer pairs CTnPg1-right/CTnPg1-left (lane 1) and CTnPg1-up/CTnPg1-down (lane 2). (C) Sequence alignment of attP, attB, attL, and attR regions of CTnPg1. The 14 bp core sequence is indicated by a box.
3.7. Glucose kinase-encoding gene
Disability of saccharolysis is one of the major characteristics of P. gingivalis. Consistent with this, the glucose kinase-encoding gene (glk) (PG1737) has a nonsense mutation in strain W83 (Supplementary Fig. S5). Nelson et al.19 suggested that the defect of glk accounts for asaccharolysis of P. gingivalis. In ATCC 33277, the glk gene (PGN_0380) has also been disrupted by an insertion of MITE239 but contains no nonsense mutation. Therefore, we analyzed the glk genes of other P. gingivalis strains to know which type of genetic defect is generally observed in P. gingivalis. Unexpectedly, however, the glk genes of five strains examined (strains TDC60, TDC117, TDC275, SU63, and GAI7802) were all intact. We detected no nonsense mutation or MITE239 insertion although a few amino-acid substitutions were observed in these glk genes (Supplementary Fig. S5B). To determine whether the glk gene is expressed in the five strains, we quantified the mRNA of the glk gene using the Kgp-encoding gene (kgp) as a control (Supplementary Table S8). In this analysis, a large amount of the glk gene transcript was detected in all of the five strains. On the other hand, all the strains used in this study (ATCC 33277, W83, and five other strains) were confirmed to be asaccharolytic (data not shown). Thus, these data suggest that defects of glk, which were detected in W83 and ATCC 33277, cannot always account for asaccharolysis of P. gingivalis. Further work is needed to clarify physiological roles of glucose kinase in P. gingivalis and what is responsible for asaccharolysis of P. gingivalis.
3.8. Conclusion
In this study, we determined the whole genome sequence of ATCC 33277, a less-virulent P. gingivalis strain, and carried out a genomic comparison with a virulent strain, W83. Although the genome size and GC content are almost the same, we detected extensive rearrangements between the two strains, many of which have been induced by various mobile genetic elements (IS, MITE, Tn, and CTn). Such structural alterations of the P. gingivalis genomes generated many strain-specific CDSs and may be closely associated with difference in virulence of the two strains.
Funding
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas “Comprehensive Genomics” (No. 17020007) to M.H.; “Applied Genomics” (No. 18018032) to K.N. from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Supplementary Material
Acknowledgements
We thank K. Oshima, K. Furuya, C. Yoshino, H. Inaba, K. Motomura, and Y. Hattori (University of Tokyo), A. Tamura and N. Itoh (Kitasato University), and Y. Kikuchi (Matsumoto Dental College) for their technical support.
References
- 1.Papapanou P. N. Epidemiology of periodontal diseases: an update. J. Int. Acad. Periodontol. 1999;1:110–116. [PubMed] [Google Scholar]
- 2.Irfan U. M., Dawson D. V., Bissada N. F. Epidemiology of periodontal disease: a review and clinical perspectives. J. Int. Acad. Periodontol. 2001;3:14–21. [PubMed] [Google Scholar]
- 3.Armitage G. C. Periodontal diseases: diagnosis. Ann. Periodontol. 1996;1:37–215. doi: 10.1902/annals.1996.1.1.37. [DOI] [PubMed] [Google Scholar]
- 4.Oliver R. C., Brown L. J., Loe H. Periodontal diseases in the United States population. J. Periodontol. 1998;69:269–278. doi: 10.1902/jop.1998.69.2.269. [DOI] [PubMed] [Google Scholar]
- 5.Mattila K. J., Valtonen V. V., Nieminen M., Huttunen J. K. Dental infection and the risk of new coronary events: prospective study of patients with documented coronary artery disease. Clin. Infect. Dis. 1995;20:588–592. doi: 10.1093/clinids/20.3.588. [DOI] [PubMed] [Google Scholar]
- 6.Beck J., Garcia R., Heiss G., Vokonas P. S., Offenbacher S. Periodontal disease and cardiovascular disease. J. Periodontol. 1996;67:1123–1137. doi: 10.1902/jop.1996.67.10s.1123. [DOI] [PubMed] [Google Scholar]
- 7.Joshipura K. J., Rimm E. B., Douglass C. W., Trichopoulos D., Ascherio A., Willett W. C. Poor oral health and coronary heart disease. J. Dent. Res. 1996;75:1631–1636. doi: 10.1177/00220345960750090301. [DOI] [PubMed] [Google Scholar]
- 8.Morrison H. I., Ellison L. F., Taylor G. W. Periodontal disease and risk of fatal coronary heart and cerebrovascular diseases. J. Cardiovasc. Risk. 1999;6:7–11. doi: 10.1177/204748739900600102. [DOI] [PubMed] [Google Scholar]
- 9.Chiu B. Multiple infections in carotid atherosclerotic plaques. Am. Heart J. 1999;138:S534–S536. doi: 10.1016/s0002-8703(99)70294-2. [DOI] [PubMed] [Google Scholar]
- 10.Kozarov E. V., Dorn B. R., Shelburne C. E., Dunn W. A., Jr, Progulske-Fox A. Human atherosclerotic plaque contains viable Actinbacillus actinomycetemcomitans and Porphyromonas gingivalis. Arterioscler. Thromb. Vasc. Biol. 2005;25:e17–e18. doi: 10.1161/01.ATV.0000155018.67835.1a. [DOI] [PubMed] [Google Scholar]
- 11.Boone D. R., Castenholtz R. W. Bergey's Manual of Systematic Bacteriology. 2nd Ed. Vol. 1. Springer-Verlag: New York; 2002. [Google Scholar]
- 12.Mayrand D., Holt S. C. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol. Rev. 1988;52:134–152. doi: 10.1128/mr.52.1.134-152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamont R. J., Jenkinson H. F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holt S. C., Kesavalu L., Walker S., Genco C. A. Virulence factors of Porphyromonas gingivalis. Periodontology. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 15.Amano A. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front. Biosci. 2007;12:3965–3974. doi: 10.2741/2363. [DOI] [PubMed] [Google Scholar]
- 16.Umemoto T., Hamada N. Characterization of biologically active cell surface components of a periodontal pathogen: the roles of major and minor fimbriae of Porphyromonas gingivalis. J. Periodontol. 2003;74:119–122. doi: 10.1902/jop.2003.74.1.119. [DOI] [PubMed] [Google Scholar]
- 17.Kadowaki T., Nakayama K., Okamoto K., et al. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J. Biochem. 2000;128:153–159. doi: 10.1093/oxfordjournals.jbchem.a022735. [DOI] [PubMed] [Google Scholar]
- 18.Slots J., Rams E. T. Pathogenicity. In: Shah H. N., Mayrand D., Genco R. J., editors. Biology of the Species Porphyromonas gingivalis. Boca Raton, FL, pp: CRC Press Inc; 1993. pp. 127–131. [Google Scholar]
- 19.Nelson K. E., Fleischmann R. D., DeBoy R. T., et al. Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama K. Rapid viability loss on exposure to air in a superoxide dismutase-deficient mutant of Porphyromonas gingivalis. J. Bacteriol. 1994;176:1939–1943. doi: 10.1128/jb.176.7.1939-1943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama K., Kadowaki T., Okamoto K., Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis: evidence for significant contribution of Arg-gingipain to virulence. J. Biol. Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto K., Nakayama K., Kadowaki T., Abe N., Ratnayake D. B., Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 23.Shi Y., Ratnayake D. B., Okamoto K., Abe N., Yamamoto K., Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis: construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 24.Naito M., Sakai E., Shi Y., et al. Porphyromonas gingivalis-induced platelet aggregation in plasma depends on Hgp44 adhesin but not Rgp proteinase. Mol. Microbiol. 2006;59:152–167. doi: 10.1111/j.1365-2958.2005.04942.x. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe-Kato T, Hayashi JI, Terazawa Y, et al. Isolation and characterization of transposon-induced mutants of Porphyromonas gingivalis deficient in fimbriation. Microb. Pathog. 1998;24:25–35. doi: 10.1006/mpat.1997.0170. [DOI] [PubMed] [Google Scholar]
- 26.Ewing B., Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 27.Gordon D., Desmarais C., Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama K. Determination of the genome size of the oral anaerobic bacterium Porphyromonas gingivalis by pulsed field gel electrophoresis. Dent. Jpn. 1995;32:25–28. [Google Scholar]
- 29.Sakiyama T., Takami H., Ogasawara N., et al. An automated system for genome analysis to support microbial whole-genome shotgun sequencing. Biosci. Biotechnol. Biochem. 2000;64:670–673. doi: 10.1271/bbb.64.670. [DOI] [PubMed] [Google Scholar]
- 30.Badger J. H., Olsen G. J. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 31.Yada T., Nakao M., Totoki Y., Nakai K. Modeling and predicting transcriptional units of Escherichia coli genes using hidden Markov models. Bioinformatics. 1999;15:987–993. doi: 10.1093/bioinformatics/15.12.987. [DOI] [PubMed] [Google Scholar]
- 32.Frishman D., Mironov A., Mewes H. W., Gelfand M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S. F., Madden T. L., Schaffer A. A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe T. M., Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delcher A. L., Phillippy A., Carlton J., Salzberg S. L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz S., Zhang Z., Frazer K. A., et al. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaffl M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacic M., Parker A. C., Stagg J., et al. Genetic and structural analysis of the Bacteroides conjugative transposon CTn341. J. Bacteriol. 2005;187:2858–2869. doi: 10.1128/JB.187.8.2858-2869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonheyo G., Graham D., Shoemaker N. B., Salyers A. A. Transfer region of a bacteroides conjugative transposon, CTnDOT. Plasmid. 2001;45:41–51. doi: 10.1006/plas.2000.1495. [DOI] [PubMed] [Google Scholar]
- 40.Kuwahara T., Yamashita A., Hirakawa H., et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA. 2004;101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feschotte C., Jiang N., Wessler S. R. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 42.Dong H., Chen T., Dewhirst F. E., Fleischmann R. D., Fraser C. M., Duncan M. J. Genomic loci of the Porphyromonas gingivalis insertion element IS1126. Infect. Immun. 1999;67:3416–3423. doi: 10.1128/iai.67.7.3416-3423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aduse-Opoku J., Slaney J. M., Hashim A., et al. Identification and characterization of the capsular (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 2006;74:449–460. doi: 10.1128/IAI.74.1.449-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujiwara T., Morishima S., Takahashi I., Hamada S. Molecular cloning and sequencing of the fimbrilin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem. Biophys. Res. Commun. 1993;197:241–247. doi: 10.1006/bbrc.1993.2467. [DOI] [PubMed] [Google Scholar]
- 45.Hall L. M., Fawell S. C., Shi X., et al. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect. Immun. 2005;73:4253–4262. doi: 10.1128/IAI.73.7.4253-4262.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerdeno-Tarraga A. M., Patrick S., Crossman L. C., et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 47.Nie H., Yang F., Zhang X., et al. Complete genome sequence of Shigella flexneri 5b and comparison with Shigella flexneri 2a. BMC Genomics. 2006;7:173. doi: 10.1186/1471-2164-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei J., Goldberg M. B., Burland V., et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 2003;71:2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diaz P. I., Slakeski N., Reynolds E. C., Morona R., Rogers A. H., Kolenbrander P. E. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J. Bacteriol. 2006;188:2454–2462. doi: 10.1128/JB.188.7.2454-2462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.