Abstract
Escherichia coli has dispensable genome regions and eliminating them may improve cell use by reducing unnecessary metabolic pathways and complex regulatory networks. Although several strains with reduced genomes have already been constructed, there have been no reports of strains constructed with deletions assayed for influence on growth. To retain robust growth and fundamental metabolic pathways, the growth of each deletion strain and combination effects of deletions were checked using M9 minimal medium. Then a new strain, MGF-01, with a 1 Mb reduced genome was constructed by integrating deletions that did not affect growth. MGF-01 grew as well as the wild type in the exponential phase and continued growing after the wild type had entered the stationary phase. The final cell density of MGF-01 was 1.5 times higher than that of the wild-type strain. Using MGF-01 as a production host, a 2.4-fold increase in l-threonine production was achieved.
1. Introduction
Escherichia coli is one of the most widely used organisms for the industrial production of recombinant proteins, amino acids and other chemical products.1–5 But it is necessary to remove some regulatory mechanisms such as depression by the product to improve its productivity.6 Furthermore, there are >400 function unknown genes.7 Reducing dispensable genome regions may decrease such complicated regulatory mechanisms and function unknown genes. Functional analysis and modeling, therefore, may be easier. Several groups have already established E. coli cells with fewer genes.8–11 Also, reduced genomes have been designed mainly in silico using gene annotations and information from comparative or functional genomics to identify minimal gene sets.9,10 Hashimoto et al.10 eliminated regions between essential genes as much as possible. Sixteen regions with a total length of 1.38 Mb were deleted from the E. coli K-12 MG1655 chromosome, the resulting strain being designated as Δ16. Although the genome size of Δ16 is the smallest, it exhibits an aberrant cell morphology and an increased doubling time relative to the number of deletions. Alternatively, Pósfai et al.11 selected strain-specific genomic regions as deletion targets by comparing six sequenced E. coli genomes. Forty-three segments were sequentially deleted from the MG1655 chromosome, resulting in 0.71 Mb genome reduction. The resulting strain, MDS43, had lost all mobile elements such as insertion sequences (ISs) and prophages. A related MDS41 strain with 41 deletions showed 20–25% lower mutation than the wild type because of its IS-free genotype. This strain as well as MG1655 exhibited good growth in minimal medium. Another strain, MDS42, with 42 deletions exhibited twice the electroporation efficiency of MG1655. The in silico design for MDS strains worked well to produce reduced genome strains with unique properties; however, to reduce the genome more than that of MDS43, it is necessary to delete common genomic regions of several E. coli strains.12 The opportunity of managing growth is higher in common regions, which include multiple metabolic pathway and regulatory genes. Single-gene knockout mutants have been constructed but little is known about how much each of those affects the growth of E. coli in minimal medium.13 Furthermore, little is known about the cumulative effects of deletions.
Here, we report construction of a reduced genome E. coli cell by assessing the growth on M9 minimal medium of all intermediate deletion mutants. In order to maintain robust cell growth, only deletions that did not affect cell growth were used to construct a reduced genome strain. The total length of the eliminated regions was 1 Mb, and the resulting strain was designated as MGF-01. MGF-01 showed unexpected but beneficial growth on M9 minimal medium. We believe this strain can be used as a tool for functional analysis of E. coli and as a host for industrial production.
2. Materials and methods
2.1. Bacterial strains and growth conditions
Escherichia coli strain KM22 with recBCD genes replaced by λ Red recombination functions (ΔrecBCD::Plac-bet exo kan) was obtained from Dr Kenan C. Murphy.14 A chromosomal region including λ Red genes was introduced into wild-type strain W3110, using P1 transduction, the resulting strain being designated as W3110red (W3110 ΔrecBCD::Plac-bet exo kan). A l-threonine production strain was constructed by replacing the homoserine O-succinyltransferase gene, metA, with the desensitized (canceled feedback regulation of l-threonine) aspartokinase/homoserine dehydrogenase gene, thrA345; homoserine kinase gene, thrB; threonine synthase gene, thrC; and chloramphenicol acetyltransferase gene, cat (ΔmetA::thrA345BC cat).15 Luria–Bertani (LB) medium and M9 minimal medium are described elsewhere.16 For the growth test, 10 mg/L FeSO4 20 g/L CaCO3 and 10 g/L glucose were added to the M9 minimal medium. A seed culture was grown at 37°C for 14 h in a test tube containing 5 mL LB medium. Two hundred microliters of seed culture was inoculated into a 300 mL flask with baffles (IWAKI, Tokyo, Japan) containing 20 mL M9 minimal medium. The cultivation conditions were 37°C and 250 rpm. During the cultivation, 5 g/L glucose was added at 20 h to avoid glucose depletion. Two hundred microliters of culture was diluted with 1.8 mL 0.1 N HCl to evaluate growth. For l-threonine production, strains W3110_thr and MGF-01_thr were each grown in a 300 mL flask with baffles containing 20 mL fermentation medium. The fermentation medium was composed of 70 g of glucose, 20 g of (NH4)2SO4, 1 g of KH2PO4, 0.5 g of MgSO4·7H2O, 5 mg of FeSO4·7H2O, 5 mg of MnSO4·4H2O, 2 g of yeast extract, 120 mg of methionine and 30 g/L CaCO3 per liter of water at pH 7.0.6
2.2. Selection of deletion targets
The genome sequences of E. coli K-12 MG1655 (accession no. U00096)17 and Buchnera sp. APS (accession no. NC002528)18 were used for comparative genomics, and E. coli unique genes were selected for elimination. Essential genes reported in the PEC database (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp) were eliminated as deletion candidates.19 The annotations of the remaining candidates were surveyed in the ERGO databases (http://www.integratedgenomics.com/) to judge their necessity for good growth in M9 minimal medium.20 Regions with more than 10 continuous unnecessary genes were chosen for deletion. These candidate regions for deletion are shown in Supplementary Table S1.
2.3. Construction of reduced genome strains
The details of the used markerless deletion method are given in Supplemental Materials. Single-deletion strains as to each candidate region were prepared using a λ Red recombination system and negative selection marker sacB (markerless deletion method) previously reported (Supplementary Fig. S1, Supplementary Table S1).21 Deletions distributed in 50 kb were integrated to construct deletion-unit strains by continual use of the markerless deletion method (Supplementary Table S2). All single-deletion strains and deletion-unit strains were carefully assessed for growth in M9 minimal medium. Deletions that did not affect growth were integrated to construct multiple-deletion strains (Supplementary Fig. S2). Multiple-deletion strains were also assayed for growth in M9 minimal medium.
2.4. Analytical methods
Comparative genomic hybridization was performed as described previously.12 Cells were stained using 4,6-diamidino-2-phenylinodole (DAPI) after fixation for fluorescent microscopic observation of chromosomes.22 Supernatants of cultures were used for l-threonine, acetate and glucose quantification. A high-pressure liquid chromatography system with post-column amino acid labeling and detection ‘Prominence’ (SHIMADZU, Kyoto, Japan) was used to quantify l-threonine. A DX 500 Ion Chromatograph System (DIONEX, Tokyo, Japan) was used to quantify acetate and a 7070 Automatic analyzer (HITACHI, Tokyo, Japan) was used to quantify glucose. The flanking regions of deletion sites were amplified by PCR, and sequenced with the Big Dye terminator method and an ABI3700 sequencer.
Dual-label microarray experiments with TaKaRa E. coli W3110 custom DNA tip (TAKARA SHUZO, Shiga, Japan) were performed as previously described.23 After 21 h cultivation in M9 minimal medium, RNA was prepared using RNAprotect Bacteria Reagent and RNeasy Mini Kit (QIAGEN). Signal intensity of each spot in the microarray was quantified using GenePix™ Pro 4.0 (Axon Instruments) software. Further data analyses were conducted by using computer software programs, Microsoft® Excel and GeneSpring® 5.0.2 (Silicon Genetics, Redwood, CA, USA). Only genes existed in MGF-01 were used to compare gene expression between MGF-01 and W3110red.
3. Results
3.1. Candidate regions for deletion
Dispensable regions of E. coli were selected using comparative genomics between E. coli and Buchnera sp. The species Buchnera sp. was thought to share a common ancestor with E. coli, and this symbiont attained a 0.64 Mb small genome through reduction in its size during evolution.18 Therefore, it was suitable for E. coli to compare genome construction with that of Buchnera sp. to extract candidate regions for deletions. Regions that did not exist in Buchnera sp. were selected. Meanwhile, this symbiont was missing the biosynthesis genes for some amino acids, lipopolysaccharides and phospholipids which were required for growth on M9 minimal medium. Consequently, after eliminating E. coli essential genes from the list of candidates, the list was again checked in order to exclude genes necessary for normal growth on M9 minimal medium. Regions of more than 10 continuous unnecessary genes were chosen as candidate regions for deletion. Using these criteria, 83 regions were selected (Δ001–Δ089 including missing numbers). Furthermore, transporter genes, ISs and toxin–antitoxin pairs were selected, and 20 more regions were designed for deletion (Δ090–Δ109). One hundred and three regions were selected and the total length of the candidates was 1830 kb (Supplementary Table S1).
3.2. Construction of an E. coli cell with a reduced genome
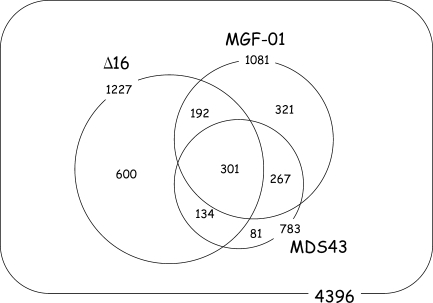
We tried to delete all the target regions by the markerless deletion method. The growth of each deletion mutant was examined by measuring the time-dependent change in cell density and 84 of the 103 deletion mutants showed comparable growth to the wild type. Deletions of normal-growth strains were used for constructing a reduced genome strain. Deletions were accumulated in one strain as described in Fig. 1. Finally, 53 deletions were combined in one strain via 28 cycles of deletion-transfer (Supplementary Fig. S2). This strain was designated as MGF-01. All 53 deletions of MGF-01 and a substitution at the recBCD site, replaced by λ Red recombinase genes, were confirmed by comparative genomic hybridization (Fig. 2). To confirm the construction of each junction, both 1 kb flanking regions of all 53 deletions were sequenced. Comparison of the deletion maps of Δ16, MDS43 and MGF-01 is shown in Fig. 3, and a Venn diagram of Δ16, MDS43 and MGF-01 is shown in Fig. 4. One thousand and eighty-one out of 4396 CDSs were deleted from W3110 to construct MGF-01, and function unknown genes were reduced from 471 to 346 (Supplementary Table S3). Although there are 301 common CDSs deleted in all three strains, unique CDSs were deleted in each strain because deletion targets were selected with different criteria. The total length of deletions in MGF-01 was 1.03 Mb and the GC content of the reduced-size genome was increased by 0.27% and reached to 51.8%.
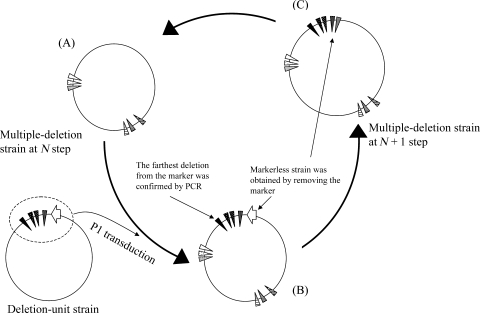
Figure 1.
Method used to integrate deletion-units. (A) Multiple deletion strain at step N was used as the recipient for P1 transduction. A deletion-unit strain was used as the donor of deletions. (B) Integrated new deletion units were selected as to the chloramphenicol resistance of the marker cassette. Transduction of the most distal deletion from the marker cassette was confirmed by PCR. (C) The marker cassette including B. subtilis sacB was replaced by a DNA fragment without the marker cassette using a λ red recombination system or P1 transduction using a P1 phage prepared from a markerless-unit strain.19 A strain with additional deletions (strain at N + 1 step) was selected as to its sucrose resistance. Each circle represents an E. coli genome and wedges denote deletions. The white arrow represents the sacB_cat marker cassette.
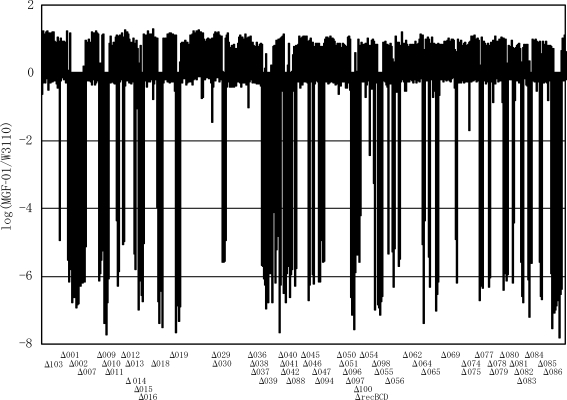
Figure 2.
Map of deletions detected in MGF-01 on microarray analysis. Each bar represents the log ratio of normalized signal intensities (MGF-01 ORF signal/W3110 ORF signal). Deleted regions were determined when the signal ratios were below –1.8. The signal ratios were plotted against ORF locations of JW0001 to JW4366. The corresponding deletion identification numbers are indicated at the bottom of the figure.
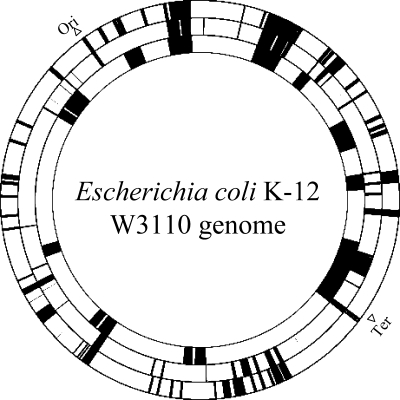
Figure 3.
A schematic illustration of the reduced genomes of E. coli. Rings represent the E. coli W3110 genome. From the center, the deleted regions of Δ0169, MDS4310 and MGF-01 are indicated by black boxes. Strains Δ16 and MDS43 were derived from MG1655. The parental strain of MGF-01 was W3110 possessing a large inversion from rrnD to rrnE. In the figure, the order of the genes of Δ16 and MDS43 between rrnD and rrnE is inverted according to that of W3110 in order to compare deletions in this region with those in MGF-01. Ori and Ter indicate the origin and terminus domains for replication, respectively.
Figure 4.
A Venn diagram of data sets of deleted CDSs from strains Δ16, MDS43 and MGF-01. Strains Δ16 and MDS43 were derived from MG1655, and MGF-01 is derived from W3110. The number of deleted CDSs in each strain was determined.
3.3. Phenotypes of MGF-01
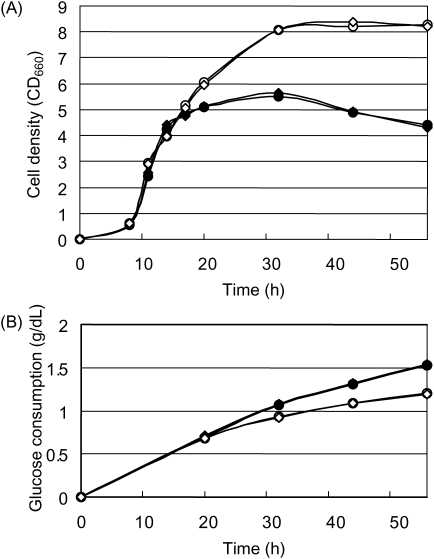
Time courses of growth of MGF-01 and W3110red in M9 minimal medium are shown in Fig. 5A. MGF-01 grew as the wild-type strain in the exponential phase and continued growing after the wild-type strain had entered the stationary phase. The final cell density of MGF-01 was 1.5 times higher than that of the wild-type strain. Colony forming units (CFU/mL) of MGF-01 and W3110red at 30 h incubation were 9.6 × 109 and 6.3 × 109, respectively. The glucose consumption speed after 20 h was lower for MGF-01 compared with the wild-type strain despite its higher cell density (Fig. 5B). Under aerobic batch culture conditions, the glucose catabolism of E. coli often overflows, leading to the accumulation of acetate that they inhibit E. coli growth.24 The levels of acetate accumulation in MGF-01 and the wild type were 0.50 and 1.37 g/L, respectively. The correlation among the high cell density, low glucose consumption and low acetate accumulation indicates that MGF-01 may utilize glucose more efficiently than the wild type.
Figure 5.
Cell density and glucose consumption of MGF-01 in M9 minimal medium. (A) Growth in M9 minimal medium. (B) Glucose consumption in M9 minimal medium. The results of duplicate experiments are shown in the figure. W3110red, filled circle and filled diamond; MGF-01, open circle and open diamond.
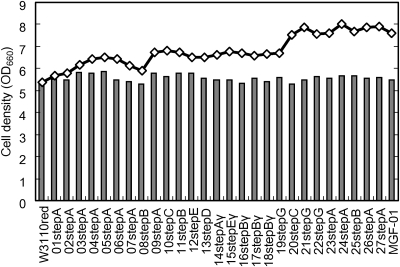
The final cell densities of the multiple-deletion strains and corresponding deletion-unit strains or single-deletion strains at each deletion step are shown in Fig. 6. Although no individual single-deletion strain or deletion-unit strain showed a significant increase in final cell density, multiple-deletion strains with stepwise genome reduction showed increased final cell densities. Consequently, the phenotype of increasing cell density may be brought about by combination(s) of deletions.
Figure 6.
Final cell densities of multiple deletion strains in M9 minimal medium. Direct lineage multiple deletion strains of MGF-01, deletion unit strains and single deletion strains used to construct MGF-01, were horizontally ordered according to the integration lineage of deletions. The final cell densities in M9 minimal medium are indicated as follows. Bars: final cell densities of deletion-unit strains or single-deletion strains. Diamonds: final cell densities of direct-lineage strains of MGF-01.
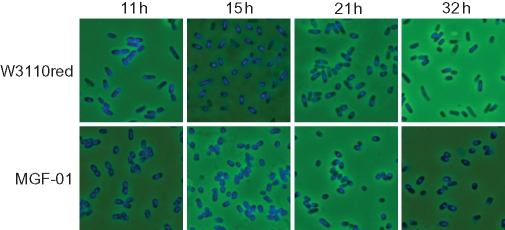
Chromosomal DNA was stained with DAPI to visualize the intracellular distribution of the chromosome (Fig. 7). The shape of MGF-01 was slightly rounder but there were little prolonged or anucleate cells. The normal cell shape of MGF-01 and the CFU result indicate that the optical density represented the cell density of MGF-01, which was 1.5-fold higher than that of the wild type. The intracellular distribution of the chromosome was apparently the same as in the wild type. Therefore, the growth of MGF-01 was not affected by the multiple deletions from the viewpoints of division and intracellular distribution of the chromosome.
Figure 7.
Microscopic images of DAPI-stained cells. Cells cultivated in M9 minimal medium were harvested at 11, 15 and 32 h, and stained using DAPI.
3.4. Threonine production of MGF-01
MGF-01 showed good growth in M9 minimal medium. To evaluate the production capability of MGF-01, an l-threonine production unit was introduced. l-Threonine is one of the major amino acids produced in fermentation processes. In the l-threonine biosynthesis pathway, the key enzyme, aspartokinase/homoserine dehydrogenase, encoded by thrA is feedback-regulated and depressed by the product, l-threonine, and a mutation to remove this feedback inhibition has been reported (thrA345).15 The methionine biosynthesis pathway is a branch pathway of l-threonine biosynthesis. To block this route, we substituted metA, which encodes the first enzyme of this branch pathway, by an l-threonine production unit containing the desensitized thrA gene (ΔmetA::thrA345BC_cat). This l-threonine production unit was transferred to W3110red and MGF-01 by P1 transduction, and the resulting strains were designated as W3110red_thr and MGF-01_thr, respectively. The production medium was different from M9 minimal medium, and contained yeast extract (Materials and methods). In this medium, both strains showed higher cell densities than in M9 minimal medium; however, the cell density of MGF-01 was only 1.2 times higher than that of wild type (1.5 times higher in M9 minimal medium). The l-threonine production after 48 h cultivation is summarized in Table 1. Interestingly, the l-threonine production and the yield of MGF-01_thr were 2.44 and 1.69 times higher than those of W3110_thr. A high glucose consumption rate (1.44 times) and a low byproduct (acetate) synthesis rate (0.09 time) contributed to this high productivity and high yield. MGF-01 also showed beneficial properties for l-threonine production.
Table 1.
Results of l-threonine production
| W3110_thr | MGF-01_thr | MGF-01/W3110 | |
|---|---|---|---|
| Threonine (g/L) | 4.4 | 10.6 | 2.44 |
| Cell density (OD660) | 19.3 | 23.3 | 1.21 |
| Glucose consumption (g/dL) | 4.8 | 6.9 | 1.44 |
| Yield (% of glucose) | 9.1 | 15.4 | 1.69 |
| Acetate (g/L) | 9.4 | 0.85 | 0.09 |
4. Discussion
In order to use reduced genome strains for industrial processes, it is necessary to maintain robust growth and the fundamental metabolic mechanism of E. coli. Therefore, we assessed the growth of deletion mutants in M9 minimal medium. Some reduced genome E. coli strains were previously reported; however, the smallest genome E. coli, Δ16, cannot growth in M9 minimal medium.10 MDS41 with a 0.66 Mb reduced genome can grow in M9 minimal medium as well as the wild type.11 MGF-01 with a 1.03 Mb reduce genome in this study can grow in M9 minimal medium. Moreover, the final cell density in M9 minimal medium of MGF-01 was 1.5 times higher than that of W3110red (Fig. 5). This unexpected but beneficial phenotype was not observed for other reduced genome strains.
Another distinct phenotype in M9 minimal medium was that the glucose consumption speed of MGF-01 was significantly decreased compared with that of its parental strain, W3110red, after 20 h cultivation instead of its higher cell density (Fig. 5B). MGF-01 accumulated less than half the acetate that W3110red did and this lower acetate accumulation of MGF-01 may contribute to the more efficient glucose assimilation. The low acetate accumulation by MGF-01 may be brought about by the high glyoxylate shunt activity which accelerates acetyl-CoA consumption leading not to acetate but to the TCA cycle.25 The gene expression of glyoxylate shunt-related genes, isocitrate lyase (aceA) and malate synthase (aceB), was 6.9 and 4.7 times higher in MGF-01 than those of W3110red in the DNA microarray analysis. Since repressors of the glyoxylate shunt, aerobic respiration control protein (arcA) and acetate operon repressor (iclR), were not eliminated, other regulatory systems may be deleted in MGF-01.26
In l-threonine production medium, MGF-01_thr also showed reduced acetate accumulation from 9.4 g/L (W3110_thr) to 0.85 g/L, and produced twice as much l-threonine as W3110_thr (Table 1). It is known that in an l-threonine production strain, the glyoxylate shunt is activated to provide high amounts of carbon to the l-threonine pathway. Low acetate accumulation and high l-threonine production indicate that the glyoxylate shunt in MGF-01_thr may be activated as in the case of M9 minimal medium.15
In M9 minimal medium, glucose consumption was lower than that of the wild type; however, it was higher than that of wild type in l-threonine production medium. This higher glucose consumption rate may be the result of higher l-threonine production by MGF-01_thr. MGF-01 may have higher ability for taking up glucose when there is a sufficient demand such as for l-threonine production. We believe that cellular metabolism may be activated in MGF-01 and glucose is used efficiently for l-threonine production.
Our next aims are to determine what combinations of deletions affect growth and l-threonine production of MGF-01, and to construct a strain with a further reduced genome.
Supplementary Data
Supplementary material is available at www.dnaresearch.oxfordjournals.org.
Funding
The New Energy and Industrial Technology Development Organization (NEDO).
Supplementary Material
Acknowledgements
We would like to thank Dr T. Fujio and Dr M. Hashimoto for the helpful discussions, and Dr K. C. Murphy for providing E. coli strain KM22. This work was carried out as part of The Project for Development of a Technological Infrastructure for Industrial Bioprocesses on R&D of New Industrial Science and Technology Frontiers of the Ministry of Economy, Trade & Industry (METI).
References
- 1.Goeddel D. V., Heyneker H. L., Hozumi T., et al. Direct expression in Escherichia coli of a DNA sequence coding for human growth hormone. Nature. 1979;281:544–548. doi: 10.1038/281544a0. [DOI] [PubMed] [Google Scholar]
- 2.Udaka S. Isolation of the arginine repressor in Escherichia coli. Nature. 1970;228:336–338. doi: 10.1038/228336a0. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda M. Amino acid production processes. Adv. Biochem. Eng. Biotechnol. 2003;79:1–35. doi: 10.1007/3-540-45989-8_1. [DOI] [PubMed] [Google Scholar]
- 4.Shibasaki T., Mori H., Ozaki A. Enzymatic production of trans-4-hydroxy-l-proline by region- and sterospecific hydroxylation of L-proline. Biosci. Biotechnol. Biochem. 2000;64:746–750. doi: 10.1271/bbb.64.746. [DOI] [PubMed] [Google Scholar]
- 5.Zhu M. M., Lawman P. D., Cameron D. C. Improving 1,3-propanediol production from glycerol in a metabolically engineered Escherichia coli by reducing accumulation of sn-glycerol-3-phosphate. Biotechnol. Prog. 2002;18:694–699. doi: 10.1021/bp020281+. [DOI] [PubMed] [Google Scholar]
- 6.Lee J. H., Oh J. W., Lee H. H., Hyun H. H. Production of l-threonine by auxotrophs and analogue-resistant mutants of Escherichia coli. Kor. J. Appl. Microbiol. Biotechnol. 1991;19:583–587. [Google Scholar]
- 7.Riley M., Abe T., Arnaud M. B., et al. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu B. J., Sung B. H., Koob M. D., et al. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 2002;20:1018–1023. doi: 10.1038/nbt740. [DOI] [PubMed] [Google Scholar]
- 9.Goryshin I. Y., Naumann T. A., Apodaca J., Reznikoff W. S. Chromosomal deletion formation system based on Tn5 double transposition: use for making minimal genomes and essential gene analysis. Genome Res. 2003;13:644–653. doi: 10.1101/gr.611403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto M., Ichimura T., Mizoguchi H., et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005;55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 11.Pósfai G., Plunkett G., III, Fehér T., et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 12.Fukiya S., Mizoguchi H., Tobe T., Mori H. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 2004;186:3911–3921. doi: 10.1128/JB.186.12.3911-3921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy K. C. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J. H., Lee D. E., Lee B. U., Kim H. S. Global analyses of transcriptomes and proteomes of a parent strain and an l-threonine-overproducing mutant strain. J. Bacteriol. 2003;185:5442–5451. doi: 10.1128/JB.185.18.5442-5451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J., Russell D. W. Molecular Cloning: a Laboratory Manual. 3rd Ed. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Blattner F. R., Plunkett G., III, Bloch C. A., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 18.Shigenobu S., Watanabe H., Hattori M., et al. Genome sequence of the endocellular bacterial symbiont of aphid Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 19.Kato J., Hashimoto M. Construction of consecutive deletions of the Escherichia coli chromosome. Mol. Syst. Biol. 2007;3:132. doi: 10.1038/msb4100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overbeek R., Larsen N., Walunas T., et al. The ERGO genome analysis and discovery system. Nucleic Acids Res. 2003;31:164–171. doi: 10.1093/nar/gkg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizoguchi H., Tanaka-Masuda K., Mori H. A simple method for multiple modification of the Escherichia coli K-12 chromosome. Biosci. Biotechnol. Biochem. 2007;71:2905–2911. doi: 10.1271/bbb.70274. [DOI] [PubMed] [Google Scholar]
- 22.Hiraga S., Niki H., Ogura T., et al. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshima T., Wada C., Kawagoe Y., Ara T., Maeda M., Masuda Y., Hiraga S., Mori H. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 2002;45:673–695. doi: 10.1046/j.1365-2958.2002.03037.x. [DOI] [PubMed] [Google Scholar]
- 24.Luli G. W., Strohl W. R. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 1990;56:1004–1011. doi: 10.1128/aem.56.4.1004-1011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noronha S. B., Yeh H. J., Spande T. F., Shiloach J. Investigation of the TCA cycle and the glyoxylate shunt in Escherichia coli BL21 and JM109 using (13)C-NMR/MS. Biotechnol. Bioeng. 2000;68:316–327. [PubMed] [Google Scholar]
- 26.Cronan J. E., Jr, LaPorte D. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, 2nd Ed. Washington D. C.: American Society for Microbiology; 1996. Tricarboxylic acid cycle and glyoxylate bypass; pp. 206–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.